Back to Journals » Drug Design, Development and Therapy » Volume 18
Validation of Core Ingredients and Molecular Mechanism of Cinobufotalin Injection Against Liver Cancer
Authors Chen S, Li M, Xue C, Zhou X, Wei J, Zheng L, Duan Y, Deng H, Tang F , Xiong W, Xiang B, Zhou M
Received 27 October 2023
Accepted for publication 10 April 2024
Published 23 April 2024 Volume 2024:18 Pages 1321—1338
DOI https://doi.org/10.2147/DDDT.S443305
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Georgios Panos
Shipeng Chen,1– 3 Mengna Li,1– 3 Changning Xue,1– 3 Xiangting Zhou,1– 3 Jianxia Wei,1– 3 Lemei Zheng,1– 3 Yumei Duan,1– 3 Hongyu Deng,1 Faqing Tang,1 Wei Xiong,1– 3 Bo Xiang,1– 3 Ming Zhou1– 3
1NHC Key Laboratory of Carcinogenesis, Hunan Key Laboratory of Oncotarget Gene, Hunan Cancer Hospital and the Affiliated Cancer Hospital of Xiangya School of Medicine, Central South University, Changsha, 410013, People’s Republic of China; 2Cancer Research Institute, Central South University, Changsha, 410078, People’s Republic of China; 3The Key Laboratory of Carcinogenesis and Cancer Invasion of the Chinese Ministry of Education, Central South University, Changsha, 410078, People’s Republic of China
Correspondence: Ming Zhou; Bo Xiang, Email [email protected]; [email protected]
Purpose: Cinobufotalin injection has obvious curative effects on liver cancer patients with less toxicity and fewer side effects than other therapeutic approaches. However, the core ingredients and mechanism underlying these anti-liver cancer effects have not been fully clarified due to its complex composition.
Methods: Multidimensional network analysis was used to screen the core ingredients, key targets and pathways underlying the therapeutic effects of cinobufotalin injection on liver cancer, and in vitro and in vivo experiments were performed to confirm the findings.
Results: By construction of ingredient networks and integrated analysis, eight core ingredients and ten key targets were finally identified in cinobufotalin injection, and all of the core ingredients are tightly linked with the key targets, and these key targets are highly associated with the cell cycle-related pathways, supporting that both cinobufotalin injection and its core ingredients exert anti-liver cancer roles by blocking cell cycle-related pathways. Moreover, in vitro and in vivo experiments confirmed that either cinobufotalin injection or one of its core ingredients, cinobufagin, significantly inhibited cell proliferation, colony formation, cell cycle progression and xenograft tumor growth, and the key target molecules involved in the cell cycle pathway such as CDK1, CDK4, CCNB1, CHEK1 and CCNE1, exhibit consistent changes in expression after treatment with cinobufotalin injection or cinobufagin. Interestingly, some key targets CDK1, CDK4, PLK1, CHEK1, TTK were predicted to bind with multiple of core ingredients of cinobufotalin injection, and the affinity between one of the critical ingredients cinobufagin and key target CDK1 was further confirmed by SPR assay.
Conclusion: Cinobufotalin injection was confirmed to includes eight core ingredients, and they play therapeutic effects in liver cancer by blocking cell cycle-related pathways, which provides important insights for the mechanism of cinobufotalin injection antagonizing liver cancer and the development of novel small molecule anti-cancer drugs.
Keywords: cinobufotalin injection, liver cancer, cinobufagin, network pharmacology, cell cycle, core ingredients
Graphical Abstract:
Introduction
Liver cancer is the fourth leading cause of cancer-related death worldwide,1 and its morbidity and mortality are increasing, which seriously threatens human life and health.2 The onset of liver cancer is complex and hidden, and most patients already have advanced liver cancer when they are diagnosed.3 Chemotherapy is one of the best options for the treatment of liver cancer,4 but transarterial chemoembolization and chemotherapeutic drugs such as sorafenib cause potent toxic effects, side effects and drug resistance;5,6 thus, it is necessary to explore complementary and alternative medicines for liver cancer treatment.
In recent years, with the increasing modernization of traditional Chinese medicine (TCM), the treatment of cancer with integrated traditional Chinese and western medicines has become a common clinical strategy and research hotspot.7 Some TCMs used alone or combined with radiotherapy and chemotherapy can improve the sensitivity of chemotherapy, improve the immune system function and reduce the side effects and complications caused by chemotherapy for improving the quality of life for patients.8 Cinobufotalin injection (CI) is an anti-cancer traditional Chinese medicinal preparation independently researched and developed in China, which contains water-soluble ingredients extracted from the skin of Bufo gargarizans Cantor or Duttaphrynus melanostictus Schneider.9 A large number of clinical trials showed that cinobufotalin injection alone or combined with transcatheter arterial chemoembolization (TACE) had obvious curative effects on liver cancer and less toxicity and side effects than other therapeutic approaches.6,10 However, the mechanism underlying the therapeutic effects of cinobufotalin injection on liver cancer has not been fully clarified due to its complex chemical composition. With the rapid recent proposal and development of virtual screening technology, the integrated analysis of network pharmacology and molecular docking has become a more efficient method to study the mechanism of action of TCMs, as it allows comprehensive analysis of relationships among drugs, diseases, genes and targets, the identification of the interaction mechanisms between drugs and the mechanisms by which active ingredients treat diseases, and the discovery of the key pharmacologically active ingredients.11
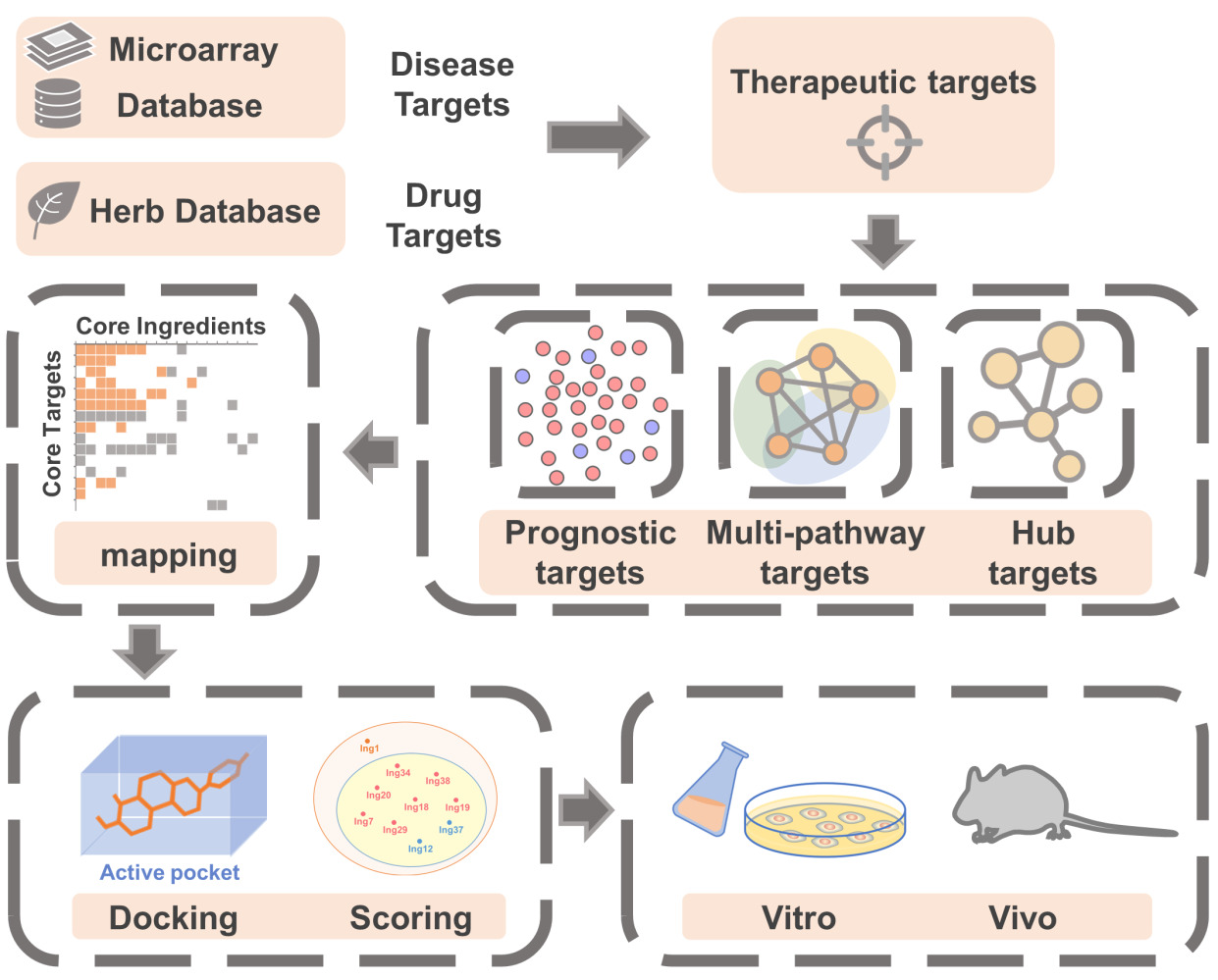
On the basis of traditional network pharmacology, this work explored the possible mechanisms of cinobufotalin injection in treating liver cancer through multidimensional network construction, molecular docking analysis and in vitro and in vivo experimental verification. We finally screened eight core ingredients and ten core target proteins. The results showed that cinobufotalin injection inhibited the expression of cell cycle-related target proteins and blocked cell cycle progression in the treatment of liver cancer, thus blocking the proliferation and division of tumor cells to achieve the purpose of treating liver cancer. Among them, cinobufagin, one of the core ingredients of cinobufotalin injection, played a key role in the treatment against liver cancer. Our work provides potential strategies and a further scientific basis for the treatment of liver cancer.
Materials and Methods
Cell Lines and Drugs
The LM3 and HepG2 cell lines were purchased from Xiangya Central Experiment Laboratory of Central South University (Changsha, China) and maintained in our laboratory. Cells were cultured in DMEM (HyClone, Logan, UT, USA) supplemented with 10% heat-inactivated FBS (Gibco, Invitrogen, Paisley, UK) and antibiotics, including 1% penicillin and 1% streptomycin at 37 °C in an incubator containing 5% CO2. cinobufotalin injection was provided by Anhui Jinchan Pharmaceutical Co., Ltd. (Huaibei, China) and was freeze-dried to obtain its concentrated powder before use. Cinobufagin was purchased from Topscience Co., Ltd. (Shanghai, China).
Compilation of Disease-Related Genes
First, three liver cancer-related microarray datasets (GSE76427 dataset, GSE46408 dataset, GSE54236 dataset) were selected from the GEO database (https://www.ncbi.nlm.nih.gov/geo/), and differential expression analysis was performed (P ≤ 0.05, logFC ≥ 0.5). To ensure the reliability of the data, we screened liver cancer-related genes in three other relevant databases: GeneCards12 (https://www.genecards.org/), Liverome13 (http://liverome.kobic.re.kr/) and OncoDB.HCC14 (http://oncodb.org/). Finally, we selected the overlapping genes among the databases and microarray datasets as liver cancer-related genes.
Screening of Ingredients and Prediction of Drug Targets of Cinobufotalin Injection
We used Herb15 (http://herb.ac.cn/) (HERB000650), and consulted relevant literature, and preliminarily screened 86 ingredients (Supplementary Table 1). Then, we used SwissTargetPrediction16 (http://swisstargetprediction.ch/) to predict the drug targets, and the targets with “probability of ≥ 0.1” were identified and integrated with those in Herb. In addition, due tothe preparation of cinobufotalin injection as an aqueous extract, SwissADME17 (http://www.swissadme.ch/) was used to predict the water solubility and drug-likeness properties of the ingredients. After the compounds with a water solubility level of “Poorly soluble” or “Insoluble” were eliminated, the effective ingredients and putative targets of cinobufotalin injection were obtained by integrating the screening results.
Network Construction and Related Analysis
Network construction was performed using Cytoscape18 3.7.2 (http://www.cytoscape.org). Protein-protein interaction network was constructed with the database STRING19 (https://string-db.org/), in which hub genes were screened with cytoHubba20 (MCC algorithm). Enrichment analysis was carried out with the DAVID21 tool (https://david.ncifcrf.gov/home.jsp) and a bioinformatics platform (http://www.bioinformatics.com.cn/). Based on the microarray data, the 150 target genes were divided into the upregulated group and the downregulated group. The prognostic value of the target genes was evaluated by using GEPIA222 (http://gepia2.cancer-pku.cn/). Here, when Hazard Ratio (HR) ≤ 0.7 or (HR) ≥ 1.3, P ≤ 0.01, we regarded a gene as having prognostic value (Supplementary Table 2). Stage-related expression was analyzed by using UALCAN23 (http://ualcan.path.uab.edu/index.html).
Molecular Docking Analysis
Small molecule compounds were obtained from PubChem (https://pubchem.ncbi.nlm.nih.gov/) and ZINC (https://zinc.docking.org/), and OpenBabel (http://openbabel.org/wiki/Main_Page) was used for file conversion. The structures of the target proteins in complex with the small molecule inhibitors were searched in the PDB database (https://www.rcsb.org/) (CDK1: 4Y72, CDK4: 7SJ3, PLK1: 3THB, CHEK1: 2HOG, TTK: 5N84) and modified with PyMOL (https://pymol.org/2/). To improve the docking accuracy, the scope of the docking box was controlled within the binding pocket of the original protein ligand (Supplementary Table 3). Docking simulations were performed with AutoDock Vina.24
CCK8 Cytotoxicity Assay
Cells were cultured in 96-well plates at a density of 5000 cells/well, and 100 μL of medium with different concentrations of cinobufotalin injection or cinobufagin was added to each well after the cells adhered to the wall. Each concentration was tested with five replicates. After 24 or 48 hours of culture, the viability of cells was determined by measuring the optical density at 450 nm on a Beckman microplate reader (Beckman, Brea, CA, USA).
Colony Formation Assay
Cells were cultured in 6-well plates at a density of 10,000 cells per well. Then, 1000 μL of medium containing drugs with corresponding IC50~24 h concentrations was added to each well after the cells adhered to the wall, respectively. The medium was replaced with fresh medium every 2 days. After 10 days, the culture medium was removed, and the colonies were fixed with 4% paraformaldehyde and stained with crystal violet staining solution (Beyotime, Beijing, China) for 5 minutes. All experiments were performed in triplicate.
Cell Cycle Analysis
Drug-treated cells (The drug concentration in the treatment group was based on IC50~24 h, CI-LM3: 509.9 μg/mL; CI-HepG2: 297.4 μg/mL; Cino-LM3: 1.3 μM; Cino-HepG2: 0.71 μM) were harvested and then resuspended in normal saline, fixed with 70% ethanol at −20 °C for 24 hours, treated with RNase A and stained with propidium iodide (PI).25 Samples were analyzed with a Cytek flow cytometer (Athena), and data were analyzed with ModFit software. All experiments were performed in triplicate.
Surface Plasmon Resonance (SPR) Analysis
Recombinant and purified human CDK1 full-length proteins (1–297aa) with a 6xHis tag at N terminus expressed in E. coli was purchased from CUSABIO (Wuhan, China). Install the NTA chip according to the standard operating procedure of OpenSPR TM instrument (Nicoya, Canada). Prepare dissolved ligand protein, the binding time was 4 minutes. After calculating the binding amount of ligand, remove the analyte. The ligand and protein were naturally dissociated for 360 seconds. Using the software TraceDrawer (Ridgeview Instruments AB, Sweden), the binding affinity (KD) between cinobufagin and CDK1 full-length proteins was detected by One To One analysis model.
Western Blot Analysis
Two cell lines (LM3 and HepG2) in the treatment groups were treated with the IC50 concentrations at 24 hours of two drugs (CI, Cino) for 24 hours and 48 hours, respectively, then the cell lines in the control groups and the treatment groups were digested and lysed respectively. Cell lysates were separated by electrophoresis on 10% SDS-polyacrylamide gels (NCM Biotech) and then transferred to polyvinylidene fluoride (PVDF) membranes (Millipore, Billerica, USA). The membranes were then blocked with 5% skim milk and incubated overnight at 4 °C with the following primary detection antibodies: anti-CCNB1 (dilution 1:1000; Abways, Shanghai, China), anti-CHEK1 (dilution 1:1000; Abways), anti-CCNE1 (dilution 1:400; Abways), anti-CDK1 (dilution 1:1000; Abways), anti-CDK4 (dilution 1:1000; ABclonal, Wuhan, China) and anti-GAPDH (dilution 1:5000; ProteinTech). Next, the membranes were further incubated with species-matched secondary antibodies for one and a half hours at 37 °C and then stained with western blotting substrate (Thermo Scientific, USA). Finally, band signals were detected with an enhanced chemiluminescence detection system according to the manufacturer’s protocol (MiniChemi™ I, Sage Creation, Beijing, China).
Tumor Xenograft Model
All animal studies were approved by the Institutional Animal Care and Use Committee of Central South University (Nq., Changsha, China). The twenty female BALB/c nude mice (ages 4–5 weeks, 18–20 g) used in this experiment were purchased from Hunan Slake Jingda Experimental Animal Co., Ltd. and were randomly divided into four groups (CI-Ctrl: 0.9% NaCl aqueous solution; CI: 2 mg/kg drug dissolved in 0.9% NaCl aqueous solution; Cino-Ctrl: 20% PEG; Cino: 2 mg/kg drug dissolved in 20% PEG). A total volume of 150 μL of LM3 cells (4 × 106 cells) was subcutaneously injected into the right flank of each female BALB/c nude mouse. On the ninth day, when the tumors had a volume of approximately 150 mm3, 200 μL of the above solutions were intraperitoneally injected into the nude mice every two days. Then, the tumor volume (volume = length× width2/2) was recorded every day. After 27 days, the nude mice were sacrificed by cervical dislocation, and the final tumor weights and expression levels of cell cycle-related molecules were measured.
Immunohistochemical Staining
Paraffin-embedded sections of mouse xenograft tissues were subjected to immunohistochemical staining. Briefly, the paraffin-embedded sections were first dewaxed and rehydrated and then subjected to antigen retrieval by high-temperature incubation. Then, the samples were incubated at 4 °C overnight with primary antibodies, including anti-CCNB1 (diluted 1:300; Abways, Shanghai, China), anti-CHEK1 (diluted 1:300; Abways), anti-CCNE1 (dilution 1:300; Abways), anti-CDK1 (dilution 1:300; Abways), and anti-CDK4 (diluted 1:300; ABclonal, Wuhan, China). Immune complexes were visualized using a MaxVision HRP‐polymer IHC Kit Detection System (MaxVision, Fuzhou, China) according to the manufacturer’s protocol. Nuclei were counterstained with hematoxylin (Beyotime Biotechnology). With an optical microscope at 200× magnification, staining was scored as reported in the literature.26 Image-Pro Plus 6.0 software (Media Cybernetics, Inc., Rockville, MD, USA) was used to evaluate the staining area and density and the integrated optical density (IOD) values. The mean densitometric value in the image indicates the relative expression level of the protein.
Statistical Analysis
R 4.3.1 software environment was used to perform statistical analysis. Nonparametric bootstrapping test was performed for comparison between two groups.27 Statistical data are presented as the means ± SEMs. For all analyses, a P value ≤ 0.05 was considered statistically significant. Each experiment was repeated independently at least three times.
Results
Construction and Analysis of the Ingredient-Target Network
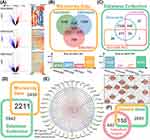
First, we collected 4670 differential expressed genes (336 shared differential expressed genes) related to liver cancer from three microarray datasets (Figure 1A and B). Subsequently, we expanded the analysis with 8153 related genes (53 shared differential expressed genes) from three other databases (Figure 1C). Finally, to ensure the reliability of the data, the 2211 overlapping genes were identified as the liver cancer-related genes (Figure 1D).
Next, we collected 86 ingredients contained in the raw material of cinobufotalin injection, and finally obtained 38 active ingredients contained in cinobufotalin injection based on the water solubility and drug-likeness properties of the ingredients (Supplementary Table 1), and 592 targets were accordingly predicted with Herb and SwissTargetPrediction websites. Subsequently, we constructed the “drug-target” topology network, in which the ingredients with more targets (eg, Ing18, Ing19, and Ing20) may have a broad influence on the regulation of the whole network (Figure 1E). We found that the network contained some key targets, including EGFR, CDK family members, mTOR, etc., all of which play critical roles in the progression of cancer. In-depth analysis also revealed that most drug targets in the network were cancer-related and were involved in numerous pathways, such as cAMP signaling pathway, central carbon metabolism in cancer, and proteoglycans in cancer. We further mapped the target-related genes to the liver cancer-related genes, and a total of 150 genes were primarily identified as the key target-related genes of cinobufotalin injection antagonizing liver cancer (Figure 1F).
Construction and Analysis of the Pathway Network
The related targets in liver cancer accounted for approximately 25% of the targets in the whole therapeutic network (Figure 2A). Enrichment analysis showed that these liver cancer-related target genes were mainly concentrated in pathways in cancer, progesterone-mediated oocyte maturation, central carbon metabolism in cancer, cell cycle, hepatitis B, etc. (Figure 2B and C).
To explore the multiple interactions between ingredients and pathways in the therapeutic network, we selected the first five pathways with the highest correlation with liver cancer and constructed the ingredient-pathway network (Figure 2D), in which eleven ingredients mediated these five pathways simultaneously. Subsequently, we extracted ingredients involved in each pathway separately and constructed single pathway-ingredient networks, where the more central ingredients had more targets to act, suggesting that they may play a more dominant role in the regulation of the pathway (Figure 2E). We screened out the three ingredients with the largest number of acting targets in each pathway, and found a total of seven ingredients, some of which had certain pathway overlapping properties (Figure 2F), such as Ing18 and Ing19, were located in the center of liver cancer-related ingredient-pathway networks. Furthermore, as shown in Figure 2D, 11 ingredients with a degree value of 5 also contain these 7 central ingredients, indicating that these ingredients not only participate in related regulation of multiple pathways at the same time but also mediate multiple related targets in each pathway.
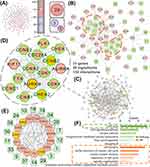
Construction and Analysis of the Hub Target-Ingredient Network
To further determine the prognostic value of these targets in liver cancer, we conducted overall survival (OS)-related prognostic analysis of target-related genes, which showed that in the upregulated group, there were 29 genes whose high expression was associated with poor prognosis (Figure 3A, Supplementary Figure 1A). The downregulated group contained only three genes whose high expression was associated with good prognosis and two other genes whose high expression was associated with poor prognosis (Figure 3A, Supplementary Figure 1B). In summary, there were 31 genes whose high expression was associated with poor prognosis and 3 genes whose high expression was associated with good prognosis (Supplementary Table 2). To determine the importance of interaction between different ingredients and targets at the prognostic level, we constructed a separate prognosis-related ingredient-target network based on the survival analysis results. The results showed that in the whole therapeutic network, the poor prognosis-related targets have greater involvement and that from the perspective of druggability, those ingredients that were able to intervene against the poor prognosis-related targets also have more therapeutic significance (Figure 3B, Supplementary Figure 1C). To ensure the certain prognostic value of the screened ingredients, we selected those ingredients that acted on more prognostic targets for subsequent analysis. At the prognostic level, the core ingredients might play a more important role in the treatment of liver cancer.
To deeply understand the key nodes in the ingredient-target network, in which we further analyzed the genes related to hub targets (Figure 3C and D). Interestingly, we found that all 15 hub genes were from the upregulated group with 76 genes and that 12 of hub genes were poor prognosis-related genes. This result indicated that the targets related to poor prognosis in the upregulated group may play a more critical role in the whole therapeutic network. To determine the functions of the hub targets, enrichment analysis was performed on these hub target-related genes (Figure 3F), and the most significant pathway was found to be “Cell cycle”, and most of the biological process (BP) terms were also related to the cell cycle (Figure 3F). This was also consistent with the top enrichment terms for the whole therapeutic network (Figure 2B), suggesting that in the therapeutic network of cinobufotalin injection, some ingredients might affect the progression of liver cancer by mediating cell cycle progression to exert therapeutic effects. To further understand the relationships between the hub genes and ingredients, we also constructed a hub target-ingredient network (Figure 3E).
Screening and Analysis of Core Ingredients in the Therapeutic Network
To further focus on the core ingredients, we integrated the ingredients previously screened at three different dimensions (Figure 4A), and found that a total of ten ingredients were simultaneously the key ingredients in the three different dimensional networks. We regarded these ingredients as the core ingredients that could regulate the treatment network (Figure 4A). In addition, the previous enrichment analysis showed that the mechanism of cinobufotalin injection in the treatment of liver cancer was mainly focused on the cell cycle pathway (P4) (Figures 2B and 3F), and it was found that all core ingredients acted on P4 pathway-related targets and were relatively concentrated in the network center (Figure 4B and C), suggesting that these ingredients may interfere with the development of liver cancer by mediating a variety of cell cycle-related targets.
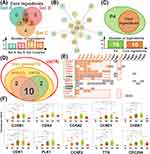 |
Figure 4 Acquisition of core ingredients and core targets. (A) Venn diagram of three sets of ingredients. Set A contains 11 ingredients in Figure 2D that simultaneously act on the top five pathways. Set B contains 13 ingredients in Figure 3B highly correlated with prognosis (degree value ≥4). Set C contains 18 ingredients in Figure 3E acting on hub targets. (B) Cell cycle pathway-ingredient network (orange circle: core ingredients). (C) Venn diagram of the 16 ingredients related to the cell cycle pathway and the 10 core ingredients. (D) Venn diagram of upregulated genes, hub genes, poor prognosis-related genes (PPG) and genes related to the cell cycle pathway (P4). (E) Matrix of hub genes and the corresponding acting ingredients. The ordinate shows the 15 hub target-related genes (red: poor prognosis-related genes, green: genes related to the cell cycle), which are arranged in sequence according to the network weights calculated by the MCC algorithm. The abscissa shows the ingredients acting on these hub targets (orange: core ingredients), which are arranged in sequence according to the number of action targets. The orange rectangle in the matrix represents interactions between the core ingredients and the core targets. (F) Stage-related expression of the core genes. * P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001. |
Subsequently, in the analysis of target-related genes, we found 12 genes related to the P4 (cell cycle) pathway among 15 hub genes, and ten genes associated with poor prognosis among these 12 P4 genes. We regarded these ten genes as core target genes in the ingredient-target network (Figure 4D). We also constructed a hub gene-ingredient matrix to further visualize the interactions between core targets and core ingredients (Figure 4E). In addition, stage-related expression analysis of the core target-related genes (Figure 4F) showed that compared with that in normal tissues, the expression levels of target genes gradually increased from stage I to stage III in liver cancer; however, interestingly, at stage IV, the expression levels of all target genes decreased. This may be because cell division and proliferation are the main events occurring during the early and middle stages of tumor development, while in the middle and late stages, tumor development begins to wane, and invasion and metastasis are enhanced.28 This indicated that cinobufotalin injection might exert greater therapeutic effects in the early and middle stages of liver cancer treatment, based on the core targets.
Docking and Scoring of Active Ingredients and Target Proteins
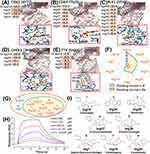
To further verify the reliability of the results, we used molecular docking simulation to predict the affinity between core targets and core ingredients. We selected five core target proteins with reported small molecular inhibitors from PDB database, and obtained their cocrystallization structures (CDK1, CDK4, PLK1, CHEK1 and TTK) for docking. The docking results showed that the predicted binding energies of most of the ingredients were relatively low and were close to those of the corresponding inhibitors; the affinities of some core ingredients (Ing18, Ing34, Ing29, Ing37, etc.) were even higher than those of the corresponding inhibitors (Figure 5A-E). Next, we counted the effective acting targets among the docked ingredients (Figure 5F). Eight core ingredients (docking score ≤ −8) were finally screened (Figure 5G),29 which at least partly accounts for the mechanism of cinobufotalin injection against liver cancer through these eight core ingredients acting on the key targets. Subsequently, in order to further confirm the molecular docking results, we selected the highest-ranked core ingredient cinobufagin (Ing18) (Figure 5F), and detected its affinity with target protein CDK1. Surface plasmon resonance (SPR) assay showed that CDK1 could indeed bind with cinobufagin, with a dissociation equilibrium constant (KD) of 62.5 µM (with an association rate constant (Ka) value of 124 M–1s−1) (Figure 5H). After further verification, eight core ingredients were finally identified that might play a relatively critical role in the therapeutic network of cinobufotalin injection in liver cancer (Figure 5I).
Cinobufotalin Injection and Cinobufagin Were Confirmed to Play Critical Anti-Liver Cancer Roles by Inhibiting the Cell Cycle Pathway
Proliferation and changes in the cell cycle are typical biological characteristics of tumor cells.28 To determine the inhibitory efficiency of cinobufotalin injection in liver cancer cells, we evaluated cell viability and colony-forming ability in both the LM3 and HepG2 cell lines after drug treatment, and the results showed that the viability (Supplementary Figure 2A-D) and the colony-forming ability of liver cancer cells (Figure 6A) decreased significantly with increasing concentration along the tested gradient. To further confirm the core ingredients identified by screening, we selected Ing18 (cinobufagin), which ranked first, to evaluate whether it plays a key role in the anti-liver cancer effect of cinobufotalin injection. The viability and colony-forming ability of liver cancer cells decreased significantly with increasing concentration of cinobufagin along the gradient (Figure 6B, Supplementary Figure 2E-H). Furthermore, the IC50 values (Supplementary Figure 2A-H) of the drugs indicated that cinobufagin has a more efficient anti-tumor effect on liver cancer cells than cinobufotalin injection.
To confirm whether the anti-liver cancer effect of cinobufotalin injection is controlled by inhibition of cell cycle progression, a flow cytometry assay was performed after 24 hours of treatment with the drugs. As expected, the percentages of liver cancer cells in different cell cycle phases changed significantly, and the percentage of G0/G1-phase cells decreased in both LM3 and HepG2 cells, while the percentage of G2/M-phase cells correspondingly increased (Figure 6C), suggesting that cinobufotalin injection blocks G2/M progression in liver cancer cells. We further found that cinobufagin had effects similar to those of cinobufotalin injection on cell cycle progression in liver cancer cells (Figure 6D). In addition, we had previously predicted that there is certain affinities between the core ingredients and targets. We wondered whether these core ingredients could also affect the expression of the protein level of the target itself. Next, we selected five relatively high-ranking targets among the core targets (Figure 4E), ie CCNB1, CHEK1, CCNE1, CDK1 and CDK4 for verification, and the western blot results showed that, interestingly, both cinobufagin and cinobufotalin injection decreased the protein expression levels of the core targets in a generally time-dependent manner (Figure 6E and F). These results indicated that while binding to the target protein, these ingredients may further reduce the stability of the target protein, thus down-regulating the expression of its translation level. In addition, molecular crosstalk caused by ingredient diversity and non-specific binding also provide the possibility for the changes of translation level. As all these targets are involved in cell cycle-related pathways and play key roles in the progression of liver cancer, these results demonstrate that cinobufotalin injection blocks G2/M progression in liver cancer cells by decreasing the expression of cell cycle-related targets, thus exerting anti-tumor effects, and that cinobufagin might be a core ingredient of cinobufotalin injection that exerts these effects on liver cancer cells.
Both Cinobufotalin Injection and Cinobufagin Significantly Inhibit Tumor Growth in vivo
To further confirm the above results in vivo, we established xenograft models by subcutaneous injection of LM3 cells into nude mice (Supplementary Figure 3). As expected, the mice in both the cinobufotalin injection and cinobufagin-treated groups exhibited significant lower growth rates as compared to the corresponding control groups, and xenograft tumor weights under treatment of cinobufagin with the same drug dose showed more significant anti-tumor effect (Figure 7A-E). To further confirm the effects of the drugs on tumor growth and cell cycle progression in vivo, we analyzed the expression levels of the five relatively high-ranking core target molecules CCNB1, CHEK1, CCNE1, CDK1 and CDK4 by immunohistochemistry (Figure 4E and 7F-H). Consistent with the results in vitro, the drug treatment groups showed obviously low expression of cell cycle-related molecules compared with the control groups. These results indicate that cinobufotalin injection exerts its anti-tumor effects by blocking cell cycle progression, and cinobufagin was confirmed to be one of the core ingredients mediating the therapeutic effects of cinobufotalin injection on liver cancer.
Discussion
Liver cancer has a high incidence in China, and clinical results have shown that the Chinese medicinal preparation cinobufotalin injection has a good therapeutic effect on liver cancer.6,30 However, due to its complex composition, the mechanism of cinobufotalin injection in the treatment of liver cancer is unclear, which hinders the clinical application of cinobufotalin injection. In this study, ten core targets and eight core ingredients mediating the anti-liver cancer effects of cinobufotalin injection were identified, and it was found that cinobufotalin injection exerts anti-tumor therapeutic effects on liver cancer by blocking cell cycle progression in a manner dependent on the core targets, which provides scientific evidence for the clinical application of cinobufotalin injection and the development of novel anti-liver cancer drugs.
Almost all cancers are characterized by cell cycle dysregulation.28,31 The cell cycle is controlled mainly through CDKs, cyclins and CDK inhibitors. Studies have shown that abnormal expression of cyclins is closely related to tumorigenesis and tumor development.32,33 A recent study showed that the up-regulation of CDK1 expression was significantly related to the poor prognosis of liver cancer patients.34 Besides, we found that the protein products of many genes in the therapeutic network are FDA-approved drug targets. Through a query in DrugBank, it was found that among the 15 hub genes, there were some targets with corresponding approved inhibitors, such as hesperidin (AURKB),35 ribociclib (CDK4/6),36 fostamatinib (CHEK1),37 proving that these targets are not undruggable targets but instead have certain druggable properties. Therefore, it was speculated that cinobufotalin injection might mediate cell cycle-related pathways through multitargeting characteristics, thus interfering with the development of tumor cells to exert its curative effect.
We searched for the key ingredients on three different levels including multipathway characteristics, prognostic level, and network hub degree, and finally obtained eight core ingredients that are most likely to play a therapeutic role in liver cancer. Among these ingredients, Jin et al found that cinobufagin (Ing18) inhibited the translation and protein production of oncogenes by reducing the expression of genes such as AURKA, which leads to DNA damage and dysregulation of chromosome segregation.38 Cinobufotalin (Ing20) has also shown anti-tumor effects in various types of cancer.39 Meng et al found that cinobufotalin induced G2/M arrest and effectively inhibit the proliferation of liver cancer cells.40 Another study showed that bufarenogin (Ing12) induced apoptosis of colorectal cancer cells to inhibit proliferation and metastasis by regulating Bax and adenine-nucleotide translocator.41 The above reports supported that the eight ingredients might be the core ingredients of cinobufotalin injection antagonizing liver cancer, and that cinobufotalin injection may have therapeutic effects on liver cancer by acting on CDKs and cyclins to induce G2/M arrest.
To further screen core ingredients, we evaluated the affinity between ingredients and targets by molecular docking. Considering the differences of spatial conformations between proteins, not all protein molecules are suitable as drug targets.42,43 Therefore, we only representatively selected the protein molecules (CDK1, CDK4, PLK1, CHEK1, and TTK) that have been reported to have inhibitors for further molecule docking, and all these proteins belong to kinases, which have ATP active sites and definite binding pockets, while most kinase inhibitors belong to ATP competitive inhibitors.44,45 In addition, though those proteins (CCNB1, CCNA2, CCNE1, CCNE2) have no definite drug binding sites, they, as specific molecular chaperones, can form complexes with those kinase proteins with binding sites, further leading to the exposure of catalytic domains.46 Therefore, to some extent, it can still be considered that these protein without definite binding sites can be indirectly regulated by kinase inhibitors.47 Based on the above, we finally narrowed to eight core ingredients. Subsequently, we preferred the first ranked ingredient (lng18 cinobufagin) for further confirmation of the anti-tumor activity and molecular mechanism through in vitro and in vivo experiments. As expected, cinobufagin presented more effective anti-tumor activity compared to the cinobufotalin injection, suggesting that the cinobufagin is one of the core ingredients of cinobufotalin injection antagonizing liver cancer. In the in vivo experiments, we selected 5 mice in each group based on the 3R principle of animal experiments and previous reports.48,49 Although the sample size of mice used in the animal experiments does not comply with the resource equation method (minimum n = 10/k+1, maximum n = 20/k+1, k (the number of groups) = 2),50 the use of bootstrapping for significance test can compensate, at least to some extent, for the small sample size, as it is a powerful method even for very small samples.27
Although cinobufagin was proven to have anti-tumor activity in the treatment of some cancers in previous reports,51 there has not been a complete report on cinobufagin as a core ingredient of cinobufotalin injection that exerts its anti-liver cancer effect by hindering cell cycle progression. This work provides a new idea for further elucidating the therapeutic mechanism of cinobufotalin injection in liver cancer. In addition, the discovery of its key anti-cancer active ingredients provides a critical reference for the development and application of novel clinical anti-cancer drugs.
Abbreviations
BP, biological process; CI, cinobufotalin injection; Cino, cinobufagin; GO, Gene ontology; Ing, ingredients; IC50, half maximal inhibitory concentration; KD, the equilibrium dissociation constant; Ka, association rate constant; KEGG, Kyoto Encyclopedia of Genes and Genomes; OS, overall survival; P4, cell cycle pathway; SPR, surface plasmon resonance; TCM, traditional Chinese medicine; TACE, transcatheter arterial chemoembolization.
Data Sharing Statement
The data used to support the findings of this study are available from the corresponding author (Ming Zhou) on request.
Ethics Statement
The requirement for ethical review for the use of public database data was specifically exempted by the authorization of the Ethics Committee of Central South University. The animal study was approved by the Institutional Animal Care and Use Committee (IACUC) of central south university (No. KTZXM-202204200002, Changsha, China). The study was conducted in accordance with the local legislation and institutional requirements.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This work was supported by the grants from the National Natural Science Foundation of China (82172592), the Hunan Provincial Key Research and Development Program (2023SK2008), the Programme of Introducing Talents of Discipline to Universities (111-2-12) and the Fundamental Research Funds for the Central Universities of Central South University (2022ZZTS0935).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Brown ZJ, Tsilimigras DI, Ruff SM, et al. Management of Hepatocellular Carcinoma: a Review. JAMA Surgery. 2023;158(4):410–420. doi:10.1001/jamasurg.2022.7989
2. Devarbhavi H, Asrani SK, Arab JP, Nartey YA, Pose E, Kamath PS. Global burden of liver disease: 2023 update. J Hepatol. 2023;79(2):516–537. doi:10.1016/j.jhep.2023.03.017
3. Liu Z, Suo C, Mao X, et al. Global incidence trends in primary liver cancer by age at diagnosis, sex, region, and etiology, 1990-2017. Cancer. 2020;126(10):2267–2278. doi:10.1002/cncr.32789
4. Ganesan P, Kulik LM. Hepatocellular Carcinoma: new Developments. Clin Liver Dis. 2023;27(1):85–102. doi:10.1016/j.cld.2022.08.004
5. Stępniak J, Krawczyk-Lipiec J, Lewiński A, Karbownik-Lewińska M. Sorafenib versus Lenvatinib Causes Stronger Oxidative Damage to Membrane Lipids in Noncancerous Tissues of the Thyroid, Liver, and Kidney: effective Protection by Melatonin and Indole-3-Propionic Acid. Biomedicines. 2022;10(11):2890. doi:10.3390/biomedicines10112890
6. Guo N, Miao Y, Sun M. Transcatheter hepatic arterial chemoembolization plus cinobufotalin injection adjuvant therapy for advanced hepatocellular carcinoma: a meta-analysis of 27 trials involving 2079 patients. Onco Targets Ther. 2018;11:8835–8853. doi:10.2147/ott.S182840
7. Wei J, Liu Z, He J, et al. Traditional Chinese medicine reverses cancer multidrug resistance and its mechanism. Clin Translational Oncol. 2022;24(3):471–482. doi:10.1007/s12094-021-02716-4
8. Wang Z, Qi F, Cui Y, et al. An update on Chinese herbal medicines as adjuvant treatment of anticancer therapeutics. Biosci Trends. 2018;12(3):220–239. doi:10.5582/bst.2018.01144
9. Pharmacopoeia CC. Pharmacopoeia of the People’s Republic of China. 2010.
10. Song GP. Effect of Cinobufagin Injection on Prevention of Recurrence of Primary Liver Cancer with Radical Resection. Chin J Exp Traditional Med Formulae. 2012.
11. Zhang P, Zhang D, Zhou W, et al. Network pharmacology: towards the artificial intelligence-based precision traditional Chinese medicine. Briefings Bioinf. 2023;25(1):675.
12. Safran M, Rosen N, Twik M, et al. The GeneCards Suite. In: Abugessaisa I, Kasukawa T, editors. Practical Guide to Life Science Databases. Springer Nature Singapore; 2021. 27–56.
13. Lee L, Wang K, Li G, et al. Liverome: a curated database of liver cancer-related gene signatures with self-contained context information. BMC Genomics. 2011;12 Suppl 3(Suppl 3):S3. doi:10.1186/1471-2164-12-s3-s3
14. Tang G, Cho M, Wang X. OncoDB: an interactive online database for analysis of gene expression and viral infection in cancer. Nucleic Acids Res. 2022;50(D1):D1334–d1339. doi:10.1093/nar/gkab970
15. Fang S, Dong L, Liu L, et al. HERB: a high-throughput experiment- and reference-guided database of traditional Chinese medicine. Nucleic Acids Res. 2021;49(D1):D1197–d1206. doi:10.1093/nar/gkaa1063
16. Daina A, Michielin O, Zoete V. SwissTargetPrediction: updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 2019;47(W1):W357–w364. doi:10.1093/nar/gkz382
17. Daina A, Michielin O, Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep. 2017;7:42717. doi:10.1038/srep42717
18. Otasek D, Morris JH, Bouças J, Pico AR, Demchak B. Cytoscape Automation: empowering workflow-based network analysis. Genome Biol. 2019;20(1):185. doi:10.1186/s13059-019-1758-4
19. Szklarczyk D, Kirsch R, Koutrouli M, et al. The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023;51(D1):D638–d646. doi:10.1093/nar/gkac1000
20. Chin CH, Chen SH, Wu HH, Ho CW, Ko MT, Lin CY. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 2014;8 Suppl 4(Suppl 4):S11. doi:10.1186/1752-0509-8-s4-s11
21. Sherman BT, Hao M, Qiu J, et al. DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022;50(W1):W216–w221. doi:10.1093/nar/gkac194
22. Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47(W1):W556–w560. doi:10.1093/nar/gkz430
23. Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: a Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia. 2017;19(8):649–658. doi:10.1016/j.neo.2017.05.002
24. Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31(2):455–461. doi:10.1002/jcc.21334
25. Ibrahim AA, Kareem MM, Al-Noor TH, et al. Pt(II)-Thiocarbohydrazone Complex as Cytotoxic Agent and Apoptosis Inducer in Caov-3 and HT-29 Cells through the P53 and Caspase-8 Pathways. Pharmaceuticals. 2021;14:6.
26. Sun S, Zheng G, Zhou D, et al. Emodin Interferes With Nitroglycerin-Induced Migraine in Rats Through CGMP-PKG Pathway. Front Pharmacol. 2021;12:758026. doi:10.3389/fphar.2021.758026
27. Dwivedi AK, Mallawaarachchi I, Alvarado LA. Analysis of small sample size studies using nonparametric bootstrap test with pooled resampling method. Stat Med. 2017;36(14):2187–2205. doi:10.1002/sim.7263
28. Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100(1):57–70. doi:10.1016/s0092-8674(00)81683-9
29. Hsin KY, Ghosh S, Kitano H. Combining machine learning systems and multiple docking simulation packages to improve docking prediction reliability for network pharmacology. PLoS One. 2013;8(12):e83922. doi:10.1371/journal.pone.0083922
30. Shen JJ. The clinical effect of Cinobufagin injection by transcatheter arterial chemoembolization(TACE) combined with intravenous on treating primary liver cancer(PLC). J Clin Hepatol. 2009;207–209.
31. Wei J, Li M, Xue C, et al. Understanding the roles and regulation patterns of circRNA on its host gene in tumorigenesis and tumor progression. J Exp Clin Cancer Res. 2023;42(1):86. doi:10.1186/s13046-023-02657-6
32. Li M, Wei Y, Liu Y, et al. BRD7 inhibits enhancer activity and expression of BIRC2 to suppress tumor growth and metastasis in nasopharyngeal carcinoma. Cell Death Dis. 2023;14(2):121. doi:10.1038/s41419-023-05632-3
33. Liu K, Zheng M, Lu R, et al. The role of CDC25C in cell cycle regulation and clinical cancer therapy: a systematic review. Can Cell Inter. 2020;20:213. doi:10.1186/s12935-020-01304-w
34. Islam B, Yu HY, Duan TQ, et al. Cell cycle kinases (AUKA, CDK1, PLK1) are prognostic biomarkers and correlated with tumor-infiltrating leukocytes in HBV related HCC. J Biomol Struct Dyn. 2023;41(21):11845–11861. doi:10.1080/07391102.2022.2164056
35. de Groot CO, Hsia JE, Anzola JV, et al. A Cell Biologist’s Field Guide to Aurora Kinase Inhibitors. Front Oncol. 2015;5:285. doi:10.3389/fonc.2015.00285
36. Infante JR, Cassier PA, Gerecitano JF, et al. A Phase I Study of the Cyclin-Dependent Kinase 4/6 Inhibitor Ribociclib (LEE011) in Patients with Advanced Solid Tumors and Lymphomas. Clin Cancer Res. 2016;22(23):5696–5705. doi:10.1158/1078-0432.Ccr-16-1248
37. Rolf MG, Curwen JO, Veldman-Jones M, et al. In vitro pharmacological profiling of R406 identifies molecular targets underlying the clinical effects of fostamatinib. Pharmacol Res Perspectives. 2015;3(5):e00175. doi:10.1002/prp2.175
38. Jin X, Wang J, Zou S, et al. Cinobufagin Triggers Defects in Spindle Formation and Cap-Dependent Translation in Liver Cancer Cells by Inhibiting the AURKA-mTOR-eIF4E Axis. Am J Chin Med. 2020;48(3):651–678. doi:10.1142/s0192415x20500330
39. Liu JH, Yang HL, Deng ST, et al. The small molecule chemical compound cinobufotalin attenuates resistance to DDP by inducing ENKUR expression to suppress MYH9-mediated c-Myc deubiquitination in lung adenocarcinoma. Acta Pharmacol Sin. 2022;43(10):2687–2695. doi:10.1038/s41401-022-00890-x
40. Meng H, Shen M, Li J, et al. Novel SREBP1 inhibitor cinobufotalin suppresses proliferation of hepatocellular carcinoma by targeting lipogenesis. Eur J. Pharmacol. 2021;906:174280. doi:10.1016/j.ejphar.2021.174280
41. Han Q, Zhang C, Zhang Y, Li Y, Wu L, Sun X. Bufarenogin induces intrinsic apoptosis via Bax and ANT cooperation. Pharmacol Res Perspectives. 2021;9(1):e00694. doi:10.1002/prp2.694
42. Nero TL, Morton CJ, Holien JK, Wielens J, Parker MW. Oncogenic protein interfaces: small molecules, big challenges. Nat Rev Cancer. 2014;14(4):248–262. doi:10.1038/nrc3690
43. Weigelt J, McBroom-Cerajewski LD, Schapira M, Zhao Y, Arrowsmith CH. Structural genomics and drug discovery: all in the family. Curr Opin Chem Biol. 2008;12(1):32–39. doi:10.1016/j.cbpa.2008.01.045
44. Arkin MR, Tang Y, Wells JA. Small-molecule inhibitors of protein-protein interactions: progressing toward the reality. Chem Biol. 2014;21(9):1102–1114. doi:10.1016/j.chembiol.2014.09.001
45. Zhang G, Zhang J, Gao Y, Li Y. Strategies for targeting undruggable targets. Expert Opin Drug Discov. 2022;17(1):55–69. doi:10.1080/17460441.2021.1969359
46. Li Y, Zhang J, Gao W, et al. Insights on Structural Characteristics and Ligand Binding Mechanisms of CDK2. Int J Mol Sci. 2015;16(5):9314–9340. doi:10.3390/ijms16059314
47. Hardcastle IR, Golding BT, Griffin RJ. Designing inhibitors of cyclin-dependent kinases. Annu Rev Pharmacol Toxicol. 2002;42:325–348. doi:10.1146/annurev.pharmtox.42.090601.125940
48. Zhang MS, Cui JD, Lee D, et al. Hypoxia-induced macropinocytosis represents a metabolic route for liver cancer. Nat Commun. 2022;13(1):954. doi:10.1038/s41467-022-28618-9
49. Liu W, Zhou X, Yao Q, et al. In situ expansion and reprogramming of Kupffer cells elicit potent tumoricidal immunity against liver metastasis. J Clin Invest. 2023;133(8).
50. Arifin WN, Zahiruddin WM. Sample Size Calculation in Animal Studies Using Resource Equation Approach. Malaysian j med sci. 2017;24(5):101–105. doi:10.21315/mjms2017.24.5.11
51. Dai CL, Zhang RJ, An P, Deng YQ, Rahman K, Zhang H. Cinobufagin: a promising therapeutic agent for cancer. J Pharm Pharmacol. 2023;75(9):1141–1153. doi:10.1093/jpp/rgad059
 © 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.