Back to Journals » Infection and Drug Resistance » Volume 16
Using Microfluidic Chip and Allele-Specific PCR to Rapidly Identify Drug Resistance-Associated Mutations of Mycobacterium tuberculosis
Authors Chen S, Liu H, Li T, Lai W, Liu L, Xu Y, Qu J
Received 2 March 2023
Accepted for publication 20 June 2023
Published 3 July 2023 Volume 2023:16 Pages 4311—4323
DOI https://doi.org/10.2147/IDR.S410779
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Shan Chen,1 Houming Liu,1 Tianpin Li,1 Wenjie Lai,1 Lei Liu,1 Youchun Xu,2 Jiuxin Qu1
1Department of Clinical Laboratory, Shenzhen Third People’s Hospital, National Clinical Research Center for Infectious Diseases, The Second Affiliated Hospital of Southern University of Science and Technology, Shenzhen, Guangdong Province, 518112, People’s Republic of China; 2Department of Biomedical Engineering, School of Medicine, Tsinghua University, Beijing, 100084, People’s Republic of China
Correspondence: Jiuxin Qu, Department of Clinical Laboratory, Shenzhen Third People’s Hospital, National Clinical Research Center for Infectious Diseases, The Second Affiliated Hospital of Southern University of Science and Technology, Shenzhen, People’s Republic of China, Email [email protected] Youchun Xu, Department of Biomedical Engineering, School of Medicine, Tsinghua University, Beijing, People’s Republic of China, Email [email protected]
Background: The currently used conventional susceptibility testing for drug-resistant Mycobacterium tuberculosis (M.TB) is limited due to being time-consuming and having low efficiency. Herein, we propose the use of a microfluidic-based method to rapidly detect drug-resistant gene mutations using Kompetitive Allele-Specific PCR (KASP).
Methods: A total of 300 clinical samples were collected, and DNA extraction was performed using the “isoChip®” Mycobacterium detection kit. Phenotypic susceptibility testing and Sanger sequencing were performed to sequence the PCR products. Allele-specific primers targeting 37 gene mutation sites were designed, and a microfluidic chip (KASP) was constructed using 112 reaction chambers to simultaneously detect multiple mutations. Chip validation was performed using clinical samples.
Results: Phenotypic susceptibility of clinical isolates revealed 38 rifampicin (RIF)-resistant, 64 isoniazid (INH)-resistant, 48 streptomycin (SM)-resistant and 23 ethambutol (EMB)-resistant strains, as well as 33 multi-drug-resistant TB (MDR-TB) strains and 20 strains fully resistant to all four drugs. Optimization of the chip-based detection system for drug resistance detection showed satisfactory specificity and maximum fluorescence at a DNA concentration of 1× 101 copies/μL. Further analysis revealed that 76.32% of the RIF-resistant strains harbored rpoB gene mutations (sensitivity, 76.32%; specificity 100%), 60.93% of the INH-resistant strains had katG gene mutations (sensitivity, 60.93%; specificity, 100%), 66.66% of the SM-resistant strains carried drug resistance gene mutations (sensitivity, 66.66%; specificity, 99.2%), and 69.56% of the EMB-resistant strains had embB gene mutations (sensitivity, 69.56%; specificity, 100%). Further, the overall agreement between the microfluidic chip and Sanger sequencing was satisfactory, with a turnaround time of the microfluidic chip was approximately 2 hours, much shorter than the conventional DST method.
Conclusion: The proposed microfluidic-based KASP assay provides a cost-effective and convenient method for detecting mutations associated with drug resistance in M. tuberculosis. It represents a promising alternative to the traditional DST method, with satisfactory sensitivity and specificity and a much shorter turnaround time.
Keywords: Mycobacterium tuberculosis, multidrug-resistance, drug resistance, resistance-conferring gene mutations, polymerase chain reaction, microfluidic analytical techniques, Sanger sequencing
Graphical Abstract:
Introduction
Tuberculosis (TB) is one of the most common causes of death and the leading cause of a single infectious agent.1 According to the 2022 Global TB Report of the World Health Organization (WHO), 10.6 million people were infected with TB in 2021, and although the annual number of deaths from TB decreased between 2005 and 2019 worldwide, this trend was reversed in 2020 and 2021.2 The burden of TB varies significantly among countries, with low-income countries experiencing the highest prevalence.3 In some countries, including India, China, the Philippines and South Africa, the incidence of TB is even higher, reaching up to 500/100,000.4–7 Although TB is a treatable and curable disease, the emergence of drug resistance has become a major challenge in its management. Additionally, mismanagement of patients often leads to multidrug-resistant TB (MDR-TB), posing a considerable financial burden on patients and increasing mortality rates. For instance, in 2017 alone, there were 560,000 new cases of TB resistant to rifampicin (RR)-TB, among whom 82% were classified as MDR-TB and among these MDR-TB patients, approximately 8.5% were diagnosed with extensively drug-resistant TB (XDR-TB). Overall, the presence of MDR-TB is particularly concerning as it signifies resistance to the most effective drugs for treating TB. As a result, the remaining treatment options are less effective, have more side effects, are costlier, and are generally associated with higher mortality risks. Despite an annual decline in the incidence of TB by 2% since 2005, drug-resistant TB remains a significant obstacle to effectively managing and eradicating the disease.1,2
Early and accurate identification of drug-resistant Mycobacterium TB (M.TB) is of great significance for the treatment and prevention of the spread of multidrug-resistant disease.8,9 However, current diagnostic methods based on bacterial culture are time-consuming and have low efficiency,10,11 thereby hindering the proper management of TB. Traditional culture-based tests for TB diagnosis typically require a lengthy period of 3 to 6 weeks to deliver results for sensitive cases and even longer for MDR and XDR-TB cases. During this waiting period, clinicians have to rely solely on the patient’s clinical symptoms, imaging assessments, and other available information to develop a treatment plan, which carries the risk of uncertain outcomes. Furthermore, ineffective therapy is considered the driving force for the emergence and spread of MDR-TB and XDR-TB, with the lack of highly accurate and affordable diagnostic techniques remaining one of the causes of the high incidence and prevalence of drug-resistant TB in economically underdeveloped regions.
It has been demonstrated that polymerase chain reaction (PCR)-based methods, similar to traditional culture-based tests, can accurately detect drug resistance with high specificity.12,13 However, the risk of carry-over contamination of samples and the ability to detect multiple mutations has not been fully addressed in the present literature. One of the primary concerns in TB diagnosis is the high infectiousness of the disease and the risk of cross-contamination of PCR test amplicons. To address this issue, the GeneXpert MTB/RIF system from Cepheid has been proposed as it can detect TB in a microfluidic cassette, following a sample-in and answer-out approach.14,15 It significantly reduces manual handling, and the PCR product is isolated within the reaction chamber of the cassette. Consequently, the GeneXpert system has gained wide usage in TB diagnosis due to its ability to minimize contamination risks.
Various approaches have been explored to simultaneously detect common mutations in M.TB, such as microarray,16 qualitative DNA strips17 and microfluidic chips.18 While these assays offer rapid detection and the ability to identify a wide range of mutation sites, they are prone to false diagnoses due to issues with multiplex PCR and the open testing environment. Thus, sealing PCR products within an enclosed microfluidic chip has been proposed as a feasible solution to mitigate the risk of contamination.
In this study, we aimed to develop a microfluidic method to rapidly detect gene mutations associated with drug resistance in M.TB with high sensitivity and high specificity while also prioritizing cost-effectiveness and convenience for practical implementation.
Methods
Samples Collection and Preparation
A total of 300 clinical samples comprising 140 sputum samples, 125 alveolar lavage fluid samples, two urine samples, 21 pleural fluid samples and 12 samples of other types of specimens, were obtained from TB patients treated at the Shenzhen Third People’s Hospital between February 2017 to January 2018. The clinical specimens were either used for DNA extraction and drug susceptibility testing (DST), as previously reported.19 DNA extraction from the clinical samples was performed using the “isoChip®” Mycobacterium detection kit (CapitalBio Technology Company, Beijing). After extraction, the concentration of genomic DNA was measured using Nanodrop 1000, and the DNA samples were stored at −20°C. All samples were de-identified, and all participants provided written informed consent. This study was approved by the Institutional Review Board of the Shenzhen Third People’s Hospital (ID: 20180022).
Phenotypic Susceptibility Test
All clinical samples were decontaminated using the NALC-NaOH method with a final NaOH concentration of 1%. After centrifugation at 10,000 rpm for 20 minutes, the precipitate was re-suspended in 1.5 mL of 0.5 M phosphate buffer (pH 6.8) and inoculated on the BACTEC MGIT 960 mycobacteria culture identification system (Becton Dickinson, USA) following the manufacturer’s instructions. The WHO recommended ratio method was used to perform the drug sensitivity test for rifampicin (RIF), isoniazid (INH), streptomycin (SM), and ethambutol (EMB), with a critical concentration of 40 mg/L (RIF), 0.2 mg/L (INH), 4 mg/L (SM), and 2 mg/L (EMB), following which the results were categorized as resistant (R) or sensitive (S).20
Sanger Sequencing
The following loci were extended by PCR for 300 TB clinical isolates, namely rpoB (RIF), katG (INH), embB (EMB), and rpsL and rrs (SM), using the primers listed in Table 1. Sanger sequencing was performed to sequence the PCR products.
 |
Table 1 Primer Sequences for Target Gene Sequencing |
Allele-Specific Primers
Thirty-seven pairs of primers were designed based on the TB resistance-related database and the published literature on the high-frequency resistance mutation sites of rpoB, katG, embB, rrs and rpsL genes.18 Each primer pair contained two allele-specific upstream primers and one shared downstream primer. These primers targeted mutation codons of the most common resistance-related genes and are considered to cover approximately 90% of rpoB mutations in RIF-resistant strains, 87% of katG mutations in INH-resistant strains, 68% of embB mutations in EMB-resistant strains, and 70% of rpsL and rrs mutations in SM-resistant strains. A pair of specific primers targeting the 16S rRNA was designed as the positive control for the PCR reaction. All primers and the master mix of Kompetitive Allele-Specific PCR (KASP) were provided by LGC Corp. (Shanghai, China). The mutation sites are listed in Table 2, and the sequences of primer pairs are shown in Table S1.
 |
Table 2 Resistant Gene Loci and Coverage |
Fabrication of the Microfluidic Chip
A customized microfluidic chip with 112 reaction chambers was used for 37 gene mutation sites responsible for resistance against first-line anti-TB drugs using the KASP principle. Once the samples were infused into the microfluidic chip, the entire process, including sample distribution, reaction chamber sealing, PCR and results detection, could be fully sealed, eliminating potential contamination.
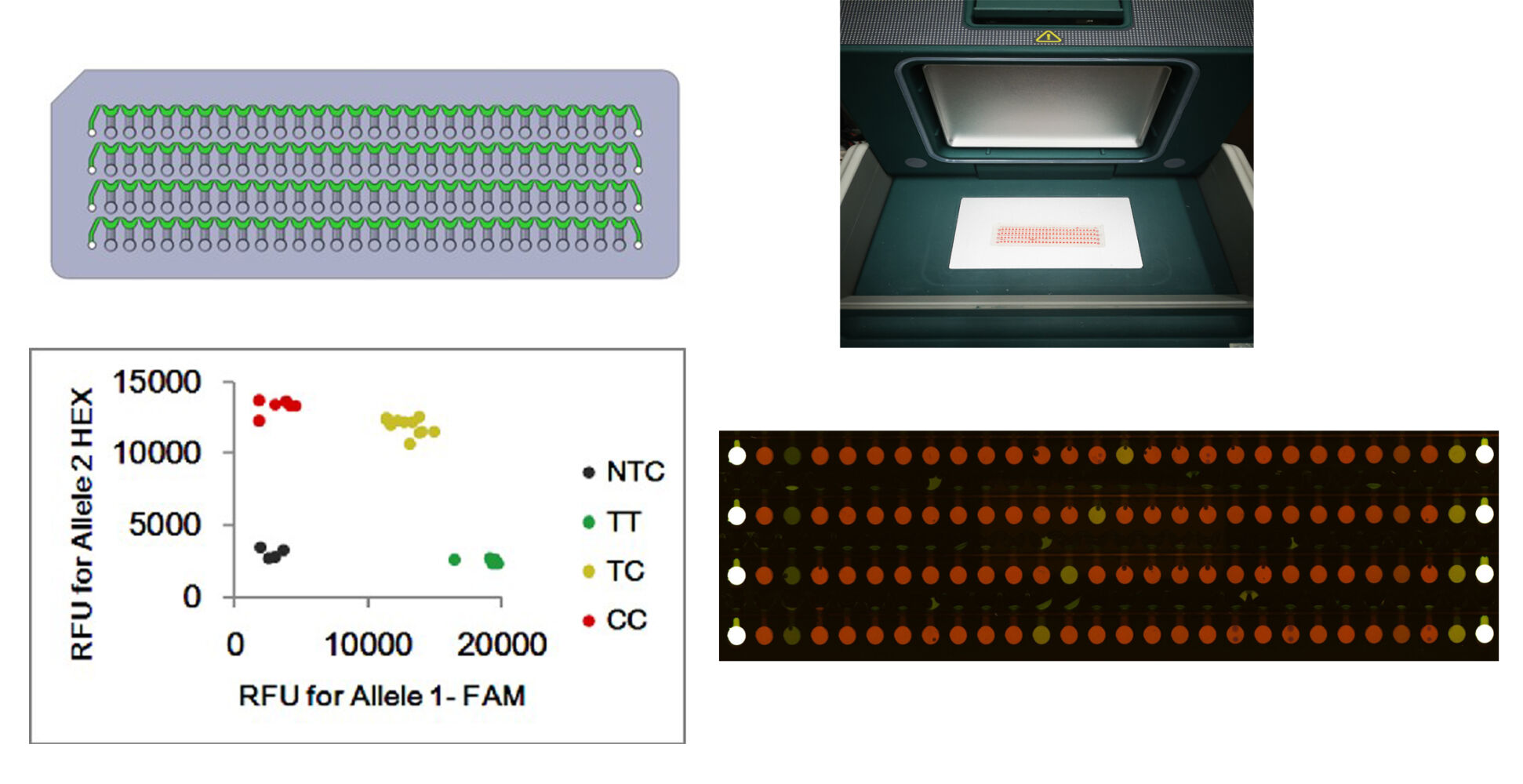
The microfluidic chip was injection-molded with polypropylene of dimensions 75 mm long, 25 mm wide, and 2 mm thick. As shown in Figure S1, four infusing channels, each connected with 28 reaction chambers, could be used to simultaneously detect as many as 112 targets (including control) on a chip. To detect the 37 gene mutation sites of TB, the primer pairs were pre-spotted in the corresponding reaction chambers of the chip (Figure 1). For each target, two specific upstream primers (2.5 μM) and the shared downstream primer (0.5 μM) were loaded in the reaction chamber (final volume: 0.14 μL/chamber) with an automated spotter (PersonalArrayer-16, CapitalBio, Beijing). The primers were air-dried at room temperature, the surface of the chip was then sealed with heat-sealing film (LGC), and the sealed chip was stored at 4°C before use.
To ensure a more comprehensive coverage of resistance-associated mutation sites, we designed 37 pairs of specific primers that can adequately target the essential gene segments related to drug resistance (Figure 2A). Each site was amplified in the presence of three primers, yielding two bands with 3′-end allele-specific primers. One of the bands possessed a nucleotide sequence bound to the FAM probe, and the other had a nucleotide sequence bound to the HEX probe at the 5′-end. After amplification, the different fluorescence of the product was analyzed using scanning software. Homozygous mutants generated only green fluorescence, homozygous wild-type genes yielded only red fluorescence, while heterozygotes produced mixed red and green fluorescence.
Multiple tests were performed to establish the optimal measurement conditions of the detection system and to determine the acceptable range of variation for the standard protocol. First, varying concentrations and ratios of different types of primers were analyzed to establish optimal conditions ensuring the highest specificity and signal-to-noise ratio. Second, the sensitivity of the method was assessed by testing a range of DNA concentrations in the samples. The concentration of DNA was 1×10° copies/µL to 1×106 copies/µL, with each concentration pro-spotted in three chambers for PCR amplification.
Microfluidic System for Detecting Mutations
The template, including 5 μL of the extracted genomic DNA, 22.5 μL of 2× KASP PCR master mix, and 17.5 μL of ddH2O, was injected into the infusing channel of the chip. Then, the chip was sealed with single-sided tape and centrifuged at 4000 rpm for 2 minutes to distribute the template into the reaction chambers with primer pairs. Afterward, the chip was thermally sealed to prevent the medium from evaporating following PCR. The PCR cycles were conducted as follows: an initial step of incubation at 37°C for 5 minutes, followed by denaturation at 95°C for 15 minutes, then cycles consisting of denaturation at 95°C for 20 seconds, annealing at 61°C for 60 seconds and a gradual stepwise cooling from 61°C to 55°C, decreasing the temperature by 0.6°C with each cycle, followed by denaturation at 95°C for 20 seconds and annealing at 55°C for 60 seconds; for a total of 29 PCR cycles. Strain H37Rv was used as a wild-type control, and the plasmid containing mutated DNA was used as a positive control. Once the PCR was completed, the chip was scanned using a LuxScan 10K-D microarray scanner (CapitalBio, Beijing), and the fluorescence data were analyzed and normalized using the instrument’s software.
Data Analysis
Genotyping was performed based on the following principles: 1) Median fluorescence intensity of the reaction chamber was calculated using the following formula: (6-carboxyfluorescein [FAM] fluorescence median - median background signal) / (5-hexachlorofluorescein [HEX] fluorescence median - median background signal); 2) The signal was considered to be positive if the signal intensity of the reaction chamber was more than three times higher than that of the negative control; 3) For determining the mutation type, the value of (FAM fluorescence median / HEX fluorescence median) was assessed, whereby a value greater than 0.5 and less than 2.0 indicated a heterozygous mutation, while any other value was interpreted as a homozygous mutation.
Results
Phenotypic Susceptibility Test
Among the 300 TB clinical isolates, 38 strains were identified as RIF-resistant, 64 were INH-resistant, 48 were SM-resistant, 23 were EMB-resistant, 33 were MDR-TB and 20 were fully four-drug-resistant (Table 3).
 |
Table 3 Phenotypic Drug Susceptibility Testing of 300 Clinical TB Isolates |
Optimization and Analysis of the Drug Resistance Detection System
As shown in Figure 2B, the reaction chambers pre-spotted with the primer pair for rpsL43Arg in the chip for detecting sample No. 49,701 showed a significantly different color (green) than other samples (red), indicating homozygous mutation. Moreover, DNA sequencing results showed that it existed 43th codon homozygous mutation of rpsL gene (Figure 2C). The results also showed that the 2:2:5 ratio of the two allele-specific primers and the reverse primer provided adequate reaction specificity and maximum fluorescence value. Figure S2 shows that a DNA concentration of less than 1×101 copies/µL did not produce fluorescence signals.
Comparison of Chip Scanning, Phenotypic Drug Sensitivity, and Sequencing Results
RIF-Resistant Strains
The results of microarray scanning indicated that among the 38 isolates with the RIF-resistant phenotype, 29 strains contained mutations in the rpoB gene. Notably, none of the 262 strains with the RIF-sensitive phenotype contained rpoB gene mutations. The chip scanning results were consistent with the sequencing data. Among the 29 strains harboring a mutated rpoB gene, the most frequent mutation was rpoB531 (TCG-TTG) detected in 24 (63.15%) strains, followed by rpoB526 (CAC-CGC) mutation detected in 3 strains (7.89%), and rpoB522 (TCG-CAG) and rpoB526 (CAC-TAC) mutations detected in 1 strain (2.63%) each. The sensitivity and specificity of the chip system for detecting RIF resistance were 76.32% and 100%, respectively (Tables 4–7).
 |
Table 4 Evaluation Between Genetic and Phenotypic Characteristics of 20 XDR-TB Isolates and 127 Pan-Sensitive Isolates |
 |
Table 5 Correlation Between Genotypic and Phenotypic Drug Susceptibility Testing |
 |
Table 6 Evaluation Between Phenotypic and Genetic Characteristics of 300 Clinical Isolates* |
 |
Table 7 Comparison of the Chip Results, Phenotypic DST and Sanger-Sequencing Results |
INH-Resistant Strains
The results of microarray scanning indicated that among the 64 isolates with the INH-resistant phenotype, katG gene mutations were detected in 39 isolates. No mutations were detected in INH-sensitive strains. Further, the chip scan results were consistent with the sequencing results. Among the katG mutant strains, there were 33 strains (84.61%) with the katG315 (AGC-ACC) mutation, 3 isolates (4.68%) with the katG315 (AGC-AAC) mutation, 2 strains (3.12%) with the katG315 (AGC-GGC) mutation, and 1 strain (1.56%) with the katG315 (AGC-ATC) mutation. The sensitivity and specificity of the chip system for detecting INH resistance were 60.93% and 100%, respectively (Tables 4–7).
SM-Resistant Strains
According to the chip scan results, among the 48 strains with the SM-resistant phenotype, a mutated drug resistance gene was present in 32 isolates. The rpsL43 (AAG-AGG) mutation was detected in 2 SM-sensitive strains. The chip scanning results were consistent with the sequencing results. Among the SM-resistant strains, 26 (54.16%) carried the rpsL43 (AAG-AGG) mutation, 3 (6.25%) the rpsL88 (AAG-AGG) mutation, and 3 (6.25%) the rrs513 (AGC-AGT) mutation. The sensitivity and specificity of the chip system for detecting SM resistance were 66.66% and 99.2%, respectively (Tables 4–7).
EMB Resistant Strains
The results of microarray scanning indicated that among the 23 isolates with the EMB-resistant phenotype, embB gene mutations were detected in 16 strains. No mutations of the embB gene were detected in EMB-sensitive strains. The results of microarray scanning were consistent with the sequencing results. Among the embB mutants, 14 isolates (60.86%) had embB306 (ATG-ATA) mutation, and 2 (8.69%) had embB306 (ATG-CTA) mutation. The sensitivity and specificity of the chip system for detecting EMB resistance were 69.56% and 100%, respectively (Tables 4–7).
Thus, our results indicated that the proposed chip detected all drug-resistant mutations found by Sanger sequencing, maintaining 100% sensitivity and specificity (Table 4). It was also consistent with DST, both in terms of detecting the presence of resistance and the types of mutations present (Table 6 and Table 7). In regard to detection accuracy, the chip and Sanger sequencing had similar accuracies in comparison to phenotypic DST, ranging from 92% to 98% across all drugs (Table 6). In addition, our proposed chip exhibited high positive predictive value (PPV) and negative predictive value (NPV), indicating high accuracy in predicting resistance (or a lack of resistance), and as demonstrated in Table 5, the PPV was 100% for all drugs, while the NPV ranged from 90.42% to 97.54%. Lastly, unlike phenotypic DST, which only provides information regarding overall resistance, both our proposed chip and Sanger sequencing provided detailed mutation information, which could be particularly valuable for tracking the spread of specific resistance alleles and developing targeted treatments (Table 7).
Discussion
In this present study, we demonstrated the effectiveness of a microfluidic chip and allele-specific PCR assay for rapidly detecting drug resistance-associated mutations in M.TB by assessing different mutations and using different methods compared to previous related studies. Our results indicated that the proposed chip could perform at a similar level to Sanger sequencing but provides results more quickly and at a potentially lower cost, making it a promising alternative for drug susceptibility testing of M.TB. Additionally, compared to phenotypic DST, the chip also provides molecular-level information, which is useful for understanding the genetic basis of drug resistance.
Previously, Mohajeri et al investigated the distribution and drug resistance of the M.TB Beijing genotype in western Iran,21 using PCR-based methods to detect the Beijing genotype and performed phenotypic drug susceptibility testing to evaluate drug resistance. They reported that their results significantly differed from previously reported mutation frequencies for codon 526 (CAC to GAC) among Italian isolates (40.1%) and Greek isolates (17.6%). However, the reported mutations differed from ours, making direct comparison difficult. Further, Sadri et al assessed the frequency of mutations associated with isoniazid and rifampicin resistance in M.TB strains isolated from patients in western Iran.22 They also used PCR-based methods to detect mutations in the katG, inhA and rpoB genes associated with drug resistance. Although some of the mutations they evaluated were similar to those examined in our current study, it is important to note that the methods employed in our research differed. Specifically, our engineered microfluidic chip detection system displayed significant potential with its high sensitivity and specificity. It demonstrated superior accuracy in detecting drug resistance compared to traditional assays and sequencing methods that depend on bacterial culture, thereby highlighting the potential of this innovative detection system as a more precise and efficient method for evaluating drug sensitivity in TB cases.
In the samples included in the present study, the mutation rate of the rpoB gene in the RIF-resistant strain was approximately 76%, and the high-frequency mutation sites in rpoB531 and rpoB526 were consistent with those previously reported.23 Specifically, the presence of high-frequency mutation sites in rpoB531 and rpoB526, which are commonly associated with RIF resistance, further validates the reliability of our proposed chip assay in identifying these mutations. Further, mutations in the katG gene, which is associated with INH resistance, were found to be linked to high levels of resistance to this important anti-TB drug. The proposed chip assay demonstrated a specificity of 100% in detecting INH-resistant TB cases, while the sensitivity was observed to be 60.93%, indicating room for improvement in accurately detecting INH resistance. In addition, it should be noted that the frequency of INH-resistant TB is influenced by the specific location of mutations within the katG gene, suggesting that the lower sensitivity observed in the chip assay for INH resistance detection in this study could be attributed to the particular distribution of mutations within the gene, which may not have been fully captured by our proposed assay design.24 Thus, while these findings underscore the relevance of the microfluidic chip assay in identifying drug resistance mutations in TB, with rpoB mutations associated with RIF resistance being detected with high sensitivity and specificity, the sensitivity for detecting INH resistance was relatively lower, and the role of mutation distribution within katG should be further investigated to improve the sensitivity of the chip assay in further improving its accuracy, especially in regard to INH-resistant TB cases.
The high prevalence of drug-resistant mutations in this study might have been attributed to the high levels of drug-resistant TB within the region from which the isolates were collected. Importantly, our department specializes in treating challenging cases, usually referred from lower-tier hospitals that struggle to effectively manage drug-resistant TB, thereby, to a certain extent, contributed to the higher prevalence of drug-resistant mutations observed in the study population. In addition, several other factors might have also contributed to the high levels of drug-resistant TB, such as poor treatment adherence (can lead to the development of drug resistance), inappropriate treatment regimens (either due to suboptimal drug choices or inadequate treatment duration, can also contribute to the emergence of drug-resistant strains), and the circulation of drug-resistant strains within the community, which might have contributed to the overall prevalence of drug-resistant mutations. Thus, to gain a more comprehensive understanding of the dynamics and underlying factors contributing to these observations, further studies are needed into factors such as treatment practices, patient adherence, healthcare infrastructure, and the epidemiology of drug-resistant TB in the region, which might aid in devising targeted interventions to effectively manage and control drug-resistant TB in the affected population.
In China, the resistance rate of SM is very high and is mostly related to its long-standing and frequent use as a first-line anti-TB drug. The TB database shows that the two-point mutations rpsL43 and rpsL 88 are responsible for 75% of cases of SM-resistant TB, and the sensitivity of the chip developed in this study for detecting SM resistance was 66.66%. EMB is typically used in combination with other first-line anti-TB drugs such as RIF, INH or pyrazinamide, and the resistance rate for EMB is generally lower compared to other first-line drugs. Although the embB306 codon is a mutation hotspot for M.TB resistant to EMB, the reliability of embB306 as a marker for identifying EMB resistance remains controversial. It was reported that this mutation was only present in 28–68% of EMB-resistant TB cases, with the sensitivity of molecular diagnostic methods for detecting EMB resistance ranging from 60–80% and specificity ranging from 93–100%.25–27 These values, characterized by high specificity but relatively weaker sensitivity, are consistent with the results obtained from our developed chip assay in our present study.28 Collectively, these findings highlight the specific challenges and characteristics of resistance rates for different drugs, especially in China. Streptomycin resistance is prevalent due to its historical usage, while EMB resistance, despite having embB306 as a potential marker, presents controversies and limitations in its detection. Our proposed chip assay demonstrated consistency with previous reports in terms of sensitivity and specificity for detecting these drug resistances, indicating its suitability and alignment with existing diagnostic methods.
A rapid, sensitive and specific genetic method for identifying resistance to TB medication is important to improve the treatment outcomes of TB patients and inhibit the development of acquired drug resistance.15 This study focused on designing and developing a microfluidic chip-based detection system for common drug resistance-related mutations of M.TB. Our proposed method demonstrated an average accuracy of 95% (range: 92–98%) for detecting resistance-linked mutations, a result superior to that of tests based on liquid culture. It should be noted that the variations in performance observed in detecting different drug-resistance genes may not solely be attributed to limitations in the methodology used. These discrepancies may also arise from a lack of correlation between drug-resistance phenotype and genotype, and comparing the assay results with data obtained through Sanger sequencing can provide further evidence to support this possibility. Further, although this study did not include specific cost information for each test method, the potential cost-effectiveness of the chip method compared to Sanger sequencing can be inferred based on the current nature of these technologies and their typical usage. For instance, our proposed chip method seems capable of testing multiple samples simultaneously, increasing its throughput compared to Sanger sequencing, which generally processes one sample at a time; thereby leading to greater testing efficiency, allowing more tests to be conducted in a shorter time frame and potentially lowering the cost per test. Also, the microfluidic chip detection system appears to be simpler to operate and faster in producing results than Sanger sequencing, reducing the person-hours required to process and interpret results, hence reducing labor costs. Lastly, given the high degree of sensitivity, specificity and accuracy of the chip method, there might be less need for additional confirmatory testing, which is often required when results are ambiguous, thereby resulting in further cost savings. However, it is important to note that the real cost-effectiveness would need to be evaluated in a detailed cost-analysis study comprising all the direct and indirect costs associated with both methods (ie, reagents, equipment, labor, etc.) and that the cost-effectiveness could vary depending on the specific settings and scale of use.
Several limitations of the newly developed microfluidic chip detection system should be acknowledged. First, the system requires specialized equipment, including a flatbed PCR instrument, heat sealer, blocker and scanner compatible with the chip. Additionally, manual processing of scan data is necessary. Second, the fixed 3′-end of the competitive allele-specific primer limits the flexibility in selecting optimal PCR primers. Third, reliance on high-quality M.TB DNA may introduce errors in complex samples containing a mixture of TB and non-TB mycobacteria. Lastly, a comparison of the efficacy and applicability of the proposed method with other widely used existing methods (ie, GeneXpert, LPA, etc.) was not performed and should be evaluated in future studies. Collectively, the preliminary efforts in this study focused on enhancing the usability of the chip, streamlining chip processing steps, optimizing reaction systems to reduce reliance on sample quality and validating its potential clinical applicability.
In conclusion, the microfluidic chip detection system outperformed traditional drug sensitivity assays and sequencing methods based on bacterial culture due to its higher accuracy in detecting TB drug sensitivity, especially in screening mutations related to the resistance of first-line anti-TB drugs. Our proposed detection system offers a range of advantages, including flexibility in the number of detection sites or samples, ease of operation, and protection against aerosol contamination. Consequently, it represents a more convenient and efficient detection tool for the clinical diagnosis of TB drug resistance.
Acknowledgments
This study was supported by the State Key Laboratory of Infectious Disease Prevention and Control (2019SKLID302), the Science and Technology Program of Shenzhen, China (JCYJ20170307095303424), the Special Support Funds of Shenzhen for Introduced High-Level Medical Team, China (SZSM201412005), the National Key R&D Program of China (2017YFC0909900).
Disclosure
The authors report no conflicts of interest in this work.
References
1. World Health Organization. Global tuberculosis report 2021. Geneva, Switzerland: World Health Organization; 2021. Available from: https://www.who.int/publications/i/item/9789240037021.
2. World Health Organization. Global tuberculosis report 2022. Geneva, Switzerland: World Health Organization; 2021. Available from: https://www.who.int/teams/global-tuberculosis-programme/tb-reports/global-tuberculosis-report-2022.
3. Reid MJA, Arinaminpathy N, Bloom A, et al. Building a tuberculosis-free world: the lancet commission on tuberculosis. Lancet. 2019;393(10178):1331–1384. doi:10.1016/S0140-6736(19)30024-8
4. Subbaraman R, Nathavitharana RR, Satyanarayana S, et al. The tuberculosis cascade of care in india’s public sector: a systematic review and meta-analysis. PLoS Med. 2016;13(10):e1002149. doi:10.1371/journal.pmed.1002149
5. Sylvia S, Xue H, Zhou C, et al. Tuberculosis detection and the challenges of integrated care in rural China: a cross-sectional standardized patient study. PLoS Med. 2017;14(10):e1002405. doi:10.1371/journal.pmed.1002405
6. Zignol M, Cabibbe AM, Dean AS, et al. Genetic sequencing for surveillance of drug resistance in tuberculosis in highly endemic countries: a multi-country population-based surveillance study. Lancet Infect Dis. 2018;18(6):675–683. doi:10.1016/S1473-3099(18)30073-2
7. Naidoo P, Theron G, Rangaka MX, et al. The south African tuberculosis care cascade: estimated losses and methodological challenges. J Infect Dis. 2017;216(suppl_7):S702–S713. doi:10.1093/infdis/jix335
8. Oxlade O, Piatek A, Vincent C, Menzies D. Modeling the impact of tuberculosis interventions on epidemiologic outcomes and health system costs. BMC Public Health. 2015;15:141. doi:10.1186/s12889-015-1480-4
9. Arinaminpathy N, Dowdy D. Understanding the incremental value of novel diagnostic tests for tuberculosis. Nature. 2015;528(7580):S60–S67. doi:10.1038/nature16045
10. Simons SO, van Soolingen D. Drug susceptibility testing for optimizing tuberculosis treatment. Curr Pharm Des. 2011;17(27):2863–2874. doi:10.2174/138161211797470255
11. Minion J, Leung E, Menzies D, Pai M. Microscopic-observation drug susceptibility and thin layer agar assays for the detection of drug resistant tuberculosis: a systematic review and meta-analysis. Lancet Infect Dis. 2010;10(10):688–698. doi:10.1016/S1473-3099(10)70165-1
12. Igarashi Y, Chikamatsu K, Aono A, et al. Laboratory evaluation of the anyplex II MTB/MDR and MTB/XDR tests based on multiplex real-time PCR and melting-temperature analysis to identify Mycobacterium tuberculosis and drug resistance. Diagn Microbiol Infect Dis. 2017;89(4):276–281. doi:10.1016/j.diagmicrobio.2017.08.016
13. Chikamatsu K, Aono A, Kato T, et al. COBAS(R) TaqMan(R) MTB, smear positivity grade and MGIT culture; correlation analyses of three methods for bacillary quantification. J Infect Chemother. 2016;22(1):19–23. doi:10.1016/j.jiac.2015.09.005
14. Denkinger CM, Schumacher SG, Boehme CC, Dendukuri N, Pai M, Steingart KR. Xpert MTB/RIF assay for the diagnosis of extrapulmonary tuberculosis: a systematic review and meta-analysis. Eur Respir J. 2014;44(2):435–446. doi:10.1183/09031936.00007814
15. Gu Y, Wang G, Dong W, et al. Xpert MTB/RIF and GenoType MTBDRplus assays for the rapid diagnosis of bone and joint tuberculosis. Int J Infect Dis. 2015;36:27–30. doi:10.1016/j.ijid.2015.05.014
16. Lyu J, Wu W, Cheng P, et al. A chip for detecting tuberculosis drug resistance based on Polymerase Chain Reaction (PCR)-magnetic bead molecule platform. Front Microbiol. 2018;9:2106. doi:10.3389/fmicb.2018.02106
17. Stephen S, Muzhizhizhi D, Dhibi N, et al. Validation of the GenoType((R)) MTBDRplus Ver 2.0 assay for detection of rifampicin and isoniazid resistance in Mycobacterium tuberculosis complex isolates at UZCHS-CTRC TB research laboratory. Int J Mycobacteriol. 2019;8(1):83–88. doi:10.4103/ijmy.ijmy_170_18
18. Pholwat S, Liu J, Stroup S, et al. Integrated microfluidic card with TaqMan probes and high-resolution melt analysis to detect tuberculosis drug resistance mutations across 10 genes. mBio. 2015;6(2):e02273. doi:10.1128/mBio.02273-14
19. Woods GL, Brown-Elliott BA, Conville PS, et al. Susceptibility Testing of Mycobacteria, Nocardiae, and Other Aerobic Actinomycetes.
20. World Health Organization. Consolidated Guidelines on Tuberculosis: Module 3: Diagnosis – Tests for Tuberculosis Infection. Geneva: World Health Organization; 2022.
21. Mohajeri P, Sadri H, Farahani A, Norozi B, Atashi S. Frequency of mutations associated with rifampicin resistance in Mycobacterium tuberculosis strains isolated from patients in west of Iran. Microb Drug Resist. 2015;21(3):315–319. doi:10.1089/mdr.2014.0075
22. Sadri H, Farahani A, Mohajeri P. Frequency of mutations associated with isoniazid-resistant in clinical Mycobacterium tuberculosis strains by low-cost and density (LCD) DNA microarrays. Ann Trop Med Public Health. 2016;9:307–311. doi:10.4103/1755-6783.190166
23. Zaw MT, Emran NA, Lin Z. Mutations inside rifampicin-resistance determining region of rpoB gene associated with rifampicin-resistance in Mycobacterium tuberculosis. J Infect Public Health. 2018;11(5):605–610. doi:10.1016/j.jiph.2018.04.005
24. Zhao LL, Sun Q, Zeng CY, et al. Molecular characterisation of extensively drug-resistant Mycobacterium tuberculosis isolates in China. Int J Antimicrob Agents. 2015;45(2):137–143. doi:10.1016/j.ijantimicag.2014.09.018
25. Ahmad S, Jaber AA, Mokaddas E. Frequency of embB codon 306 mutations in ethambutol-susceptible and -resistant clinical Mycobacterium tuberculosis isolates in Kuwait. Tuberculosis. 2007;87(2):123–129. doi:10.1016/j.tube.2006.05.004
26. Shi D, Li L, Zhao Y, et al. Characteristics of embB mutations in multidrug-resistant Mycobacterium tuberculosis isolates in Henan, China. J Antimicrob Chemother. 2011;66(10):2240–2247. doi:10.1093/jac/dkr284
27. Gupta A, Singh SK, Anupurba S. Mutations at embB306 codon and their association with multidrug resistant M. tuberculosis clinical isolates. Indian J Med Microbiol. 2015;33(3):387–392. doi:10.4103/0255-0857.158560
28. Huang WL, Chi TL, Wu MH, Jou R. Performance assessment of the GenoType MTBDRsl test and DNA sequencing for detection of second-line and ethambutol drug resistance among patients infected with multidrug-resistant Mycobacterium tuberculosis. J Clin Microbiol. 2011;49(7):2502–2508. doi:10.1128/JCM.00197-11
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.


