Back to Journals » International Journal of Nanomedicine » Volume 16
Urinary Exosomal CA9 mRNA as a Novel Liquid Biopsy for Molecular Diagnosis of Bladder Cancer
Authors Wen J, Yang T, Mallouk N, Zhang Y, Li H, Lambert C , Li G
Received 31 March 2021
Accepted for publication 1 July 2021
Published 14 July 2021 Volume 2021:16 Pages 4805—4811
DOI https://doi.org/10.2147/IJN.S312322
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 5
Editor who approved publication: Prof. Dr. Anderson Oliveira Lobo
Jin Wen,1,* Tingkai Yang,1,* Nora Mallouk,2 Yang Zhang,3 Hanzhong Li,1 Claude Lambert,4 Guorong Li5
1Department of Urology, Peking Union Medical College Hospital, Beijing, People’s Republic of China; 2Center of Electronic Microscopy, CMES, Faculty of Medicine, University Jean Monnet, Saint-Etienne, France; 3Department of Molecular Biology, Guangzhou HopeTech Biological Technology Co, LTD, Guangzhou, People’s Republic of China; 4Unit of Flow Cytometry, Immunology Laboratory, North Hospital, CHU Saint-Etienne, Saint-Etienne, France; 5Department of Urology, North Hospital, CHU Saint-Etienne, Saint-Etienne, France
*These authors contributed equally to this work
Correspondence: Hanzhong Li
Department of Urology, Peking Union Medical College Hospital, Beijing, People’s Republic of China
Email [email protected]
Guorong Li
Department of Urology, North Hospital, CHU Saint-Etienne, Saint-Etienne, France
Tel +33 477828814
Email [email protected]
Objective: The objective of this study was to assess the possibility of using urinary exosomal CA9 mRNA as a novel liquid biopsy for the molecular diagnosis of bladder cancer.
Patients and Methods: A total of 168 bladder cancer patients and 90 control subjects were included in the study. An isolation kit was used to isolate urinary exosomes. Transmission electron microscopy (TEM) was used to examine the presence of exosomes. Flow cytometry was used to examine the exosomal marker CD63. The expression level of exosomal CA9 mRNA was detected by RT-qPCR. The diagnostic performance of urinary urinary exosomal CA9 mRNA was evaluated.
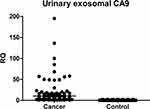
Results: TEM confirmed the enriched exosomes from urinary bladder patients. Flow cytometry indicated a strong positive expression of exosome marker CD63. Successful extraction of RNA was performed from exosome samples. The level of urinary exosomal CA9 mRNA was significantly higher in bladder cancer group than in control group (p< 0.001). The area under the ROC curve was 0.837 (95% CI: 0.743– 0.859) with a sensitivity of 85.18% and a specificity of 83.15% for the diagnosis of bladder cancer.
Conclusion: We found that the urinary exosomes were abundant in the urine of bladder cancer patients. CA9 mRNA could be detectable in urinary exosomes. The urinary exosomal CA9 mRNA may present a new liquid biopsy for the diagnosis of bladder cancer.
Keywords: urinary bladder cancer, urinary exosome, CA9, liquid biopsy
Introduction
Despite of the technique advances, bladder cancer remains a severe health problem. Bladder cancer is the ninth common malignant disease worldwide.1 About 75% of newly diagnosed cases are non-muscle-invasive bladder cancer (NMIBC).2 The treatment of NMIBC involves transurethral resection of bladder tumor. These tumors frequently recur. Approximately 30% of bladder cancer patients have muscle-invasive bladder cancer (MIBC) at diagnosis.2,3 Thirty percent of patients with MIBCs are initially diagnosed with NMIBC. The high rate of recurrence and the high risk of disease progression enforce the regular surveillance to bladder cancer patients.4 All these facts indicate that the search for new and non-invasive methods to detect and monitor bladder cancer is of maximum interest.
Exosomes are membrane-bound nanoparticles secreted by cells into the extracellular space.5 It has been proved that exosomes play a significant role in physiological and pathological processes.6 Numerous studies have shown that exosomes participate in cancer oncogenesis and cancer progression.7,8 Exosomes have become recently a hot topic in the research area of biomarkers because exosomes carry specific proteins, lipids and nucleic acids of their origins. More importantly, exosomes provide a convenient source of potential disease biomarkers because exosomes are abundant in accessible body fluids.9–11 In fact, exosome is generally considered as a major source of liquid biopsy. Urine contains a large number of exosomes. Carbonic anhydrase 9 (CA9) is a transmembrane member of the carbonic anhydrase family. It catalyses the reversible hydration of carbon dioxide into bicarbonate and a proton, thus enabling tumor cells to keep a neutral pH despite of an acidic microenvironment. CA9 is overexpressed in urinary bladder cancer and is considered as a new promising marker for urinary bladder cancer.12 We hypothesize that urine from bladder cancer patients may contain tumoral exosomes that carry CA9 marker. These exosomes may serve as a new source of liquid biopsy for the detection of bladder cancer.
Patients and Methods
Patients
Inclusion criteria included patients with the suspicion of bladder cancer due to the occurrence of hematuria or on the basis of initial findings during cystoscopy or during imaging examinations. In total, 168 patients undergoing surgical resection were included. Histopathological examination of the resected bladder specimens served as the final diagnosis. The patients’ characteristics were summarized in Table 1. Urine samples from patients without urinary bladder cancer were used as controls. Ninety urine samples were obtained as controls. There was no significant difference in average ages between the two groups of cancer patients and control subjects. The diagnosis of bladder cancer was based on pathology or biopsy analysis. The tumor stage and grade were based respectively according to the tumor-node metastasis (TNM) staging system and the WHO 2004 grading system. This study was reviewed and approved by Peking Union Medical College Hospital review board/ethics committee (G20190001645). All participants got necessary information and a written informed consent form was given to each participant. The written informed consent was obtained from all participants. This study was conducted in accordance with the Declaration of Helsinki.
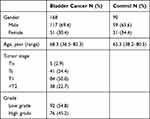 |
Table 1 The Patients’ Characteristics |
Isolation of Urinary Exosomes
The urine was collected before surgery. A total of about 50–100 mL morning urine was collected in a sterile cup. Urine samples were put in a refrigerator and processed within 12 hours. Centrifugation was performed at 3000 rpm at 4°C for 15 minutes to remove cells and large debris. Twenty-milliliter urine was used to isolate the urinary exosomes by using a urinary exosome isolation solution (Hope Tech Biotechnology, Guangzhou). Briefly, 4 mL isolation solution was added into urine. The mixture of solution was vortexed. Then the mixture was put in the refrigerator overnight. The mixture was centrifuged at 3000 rpm at 4°C for 30 minutes to recover the urinary exosomes.
Transmission Electron Microscopy (TEM) of Exosomes
TEM was performed on the pellet of exosomes suspended in 50 µL PBS. Ten microliters of exosome solution was placed on a 200 mesh-copper grid and left to adhere for 20 minutes. The grids were washed three times in distilled water. The exosomes were negatively stained with uranyl acetate and observed at an accelerating voltage value of 100 kV using a Hitachi electron microscopy.
Flow Cytometry of Exosome Marker
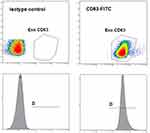
CD63 is frequently used as an exosomal marker.13 We used flow cytometry to study urinary exosomal CD63. We added 1 µL of Aldehyde/sulphate latex beads (A37304, ThermoFisher) to 100 µL of exosome suspension, and mixed well. Nine hundred microliters of PBS was then added, and the mixture was left to incubate for 2 hours at room temperature on a rotary wheel. One hundred microliters of 100 mM glycine was then added and the mixture incubated at room temperature for 30 minutes to block binding. The beads were then pelleted by centrifugation at 2650 g for 5 minutes, and the supernatant removed. The pellet was washed by addition of 1mL PBS and centrifugation at 2650 g for 5 minutes. Finally, the pellet was resuspended in PBS and the antibody of anti-human CD63-FITC FITC (IM1165U, Beckman Coulter) or the antibody of isotype control (A07795, Beckman Coulter) was added according to standard flow cytometry protocol. The antibody was incubated for 30 minute at dark. After twice washing with PBS, the staining was observed by using a conventional flow cytometry. Data were analyzed by using the NaviosTM software (Beckman-Coulter, CA).
Extraction of RNA
Total RNA extraction was performed by using miRNeasy Micro Kit according to the manufacturer indications with some modifications (Qiagen S.A.). Briefly, 500 µL Qiazol solution (GITC-containing buffer) was used to dissolve the urinary exosomes. One hundred-microliter Chloroform was added. The mixture was vortexed and centrifuged at 14,000 rpm at 4°C for 15 minutes. The supernatant was obtained and 1.5 volume of 100% ethanol was added. The mixture was centrifuged by the extraction column (Qiagen S.A.). The column was washed respectively by RWT buffer and by REP buffer. The RNA was eluted in 20 µL RNase free water and then quantified by using Nanodrop. The RNA specimens were stored in –80°C until RT-PCR.
RT-qPCR
Complementary DNA (cDNA) was synthesized from total RNA using TaqMan Reverse Transcription Kit (Invitrogen). For RT reactions, 100–500 ng of RNA extract, 4 µL of RT Buffer and 1 µL of reverse transcription mix were used. Reaction mixture was incubated at 37°C for 60 minutes, at 95°C for 5 minutes and then held at 4°C. The cDNA specimens were stored at −20°C until PCR. For PCR detection, we used the TaqMan technique. PCR was performed in 25 µL reaction mixture containing 2 µL cDNA product, 1 µL of sense and antisense primers and 12.5 µL PCR Master MIX (Applied Biosystems). After a denaturing temperature at 95°C for 10 minutes, 40 cycles were performed with denaturing temperature at 95°C for 15 seconds, annealing temperature at 60°C for 20 seconds and extension at 72°C for 34 seconds. A final dissociation cycle was performed at 95°C for 15 seconds, 60°C for 1 minute and 95°C for 15 seconds. A positive and negative (PCR mix without cDNA) controls were included in each round of PCR. To check the quality of extracted RNA, we examined the GAPDH gene expression. We designed GAPDH primers as follows: sense primer 5ʹ- CACATGGCCTCCAAGGAGTAA −3ʹ and antisense primer 5ʹ- TGAGGGTCTCTCTCTTCCTCTTGT −3ʹ. The TaqMan probe of GAPDH was 5ʹ-FAM-CTGGACCACCAGCCCCAGCAAG-3ʹ-TAMRA. The primers for CA9 were as follows: sense primer 5ʹ- CATCCTAGCCCTGGTTTTTGG −3ʹ and antisense primer 5ʹ- CCTTCTGTGCTGCCTTCTCAT −3ʹ. The TaqMan probe of CA9 was 5ʹ-FAM- CTGTCACCAGCGTCGCGTTCCTT-3ʹ-TAMRA. The primers were designed to span the exons to avoid the genomic contamination. The cycle threshold (Ct) value was established by using the detection software SDS v2.0.1 (Applied Biosystems). GAPDH was used as the housekeeping gene and the relative quantification of urinary exosomal CA9 mRNA was calculated by comparative Ct method (2−Δct).
Statistical Analysis
Statistical analyses were based on SPSS v13 software package. The Mann–Whitney U-test was used to check the difference of urinary exosomal CA9 mRNA levels between the bladder cancer group and control group. The chi-square test was used to study the differences of positive numbers between the different tumor grades and tumor stages. Significant difference was established when p<0.05. ROC curve and the area under the ROC curve (AUC) were used to evaluate the diagnostic value.
Results
Analysis of Exosomes by TEM and Flow Cytometry
The isolation kit was effective for the isolation of urinary exosomes. TEM analysis confirmed that the isolated exosomes were within the expected size range (30 to 150 nm) and presented with a general exosome-like morphology (Figure 1). Typical cup-shaped exosomes were observed, along with aggregated exosomal vesicles. The lipid bilayer membrane was clearly visible. Three experiments were performed with similar results. As shown in Figure 2, flow cytometry indicated that the exosomes showed a strong positive for exosome marker CD63.
RT-qPCR of Exosomal CA9 mRNA
Figure 3 shows the distribution of relative expression levels of urinary exosomal CA9 mRNA. The average level of urinary exosomal CA9 mRNA was significantly higher in bladder cancer group (8.35, 0.04–186.6) than in control group (0.32, 0.03–3.45) (p<0.001). Among the patients with Ta and T1 of NMIBCs, the positive percentage of urinary exosomal CA9 was greater in low-grade cancers (89.2%) than in high-grade cancers (83.3%), but without statistical significance (p>0.05). Among those with detectable urinary exosomal CA9, the positive percentage was higher in NMIBCs than in MIBCs (86.2% versus 81.6%, p>0.05).
To evaluate the diagnostic value of urinary exosomal CA9 mRNA, we performed the ROC curve analysis (Figure 4). The AUC of ROC curve was 0.837 (95% CI: 0.743–0.859). With an optimal cutoff of 0.418, the sensitivity was 85.18% and the specificity was 83.15% for the diagnosis of bladder cancer.
Discussion
Currently, cystoscopy and cytology are considered as the standards for the diagnosis and surveillance of bladder cancer.2,3 Cystoscopy is an invasive examination which can cause the discomfort and possible complications for the patients such as haematuria or urinary tract infections. Cytology is a non-invasive examination with a high specificity,14 but the major problem with cytology is its low sensitivity because urine samples often lack the tumoral cells. Therefore, the search for urine markers has always been a research subject.15
It is now believed that exosomes play important roles in normal and pathological conditions and can serve as useful biomarkers for cancer diagnosis.16 Exosomes from bladder tumor cells accumulate in the urine, where they are stable and easily accessible. Urine can be non-invasively accessible, making it the most attractive source of liquid biopsy. Many mRNA markers have been found to be useful.17–20 However, the urine markers depend on urinary tumor cells. This hinders the sensitivity of urine markers. The supernatant of urine is usually discarded. In this study, we found that abundant exosomal RNAs existed in urinary supernatant.
We used an isolation kit that is easily implanted into clinical laboratory. The study of electron microscopy indicated the success of isolation of urinary exosomes. Recently, the mRNA markers in urinary exosomes are successfully used as a source of liquid biopsy of prostate cancer.21 In this study, we tested if CA9 mRNA markers can be detected in urinary exosomes to serve as a new source of liquid biopsy for bladder cancer. Compared to the control group, the average level of urinary exosomal CA9 was significantly elevated in bladder cancer patients. Urinary cellular CA9 was recently studied as a urine marker for the diagnosis of bladder cancer.12 In that study, the sensitivity and specificity for bladder cancer detection were respectively 86.2% and 95.1%. However, the informative urine specimen rate was 85% in that study. About 15% urine samples were without urinary cells and were excluded from that study when urinary cellular CA9 was studied.12 In the present study, we showed the feasibility of the detection of tumoral CA9 in urinary exosomes. The urinary exosomal CA9 mRNA may produce an acceptable sensitivity and specificity for the detection of bladder cancer. To our knowledge, it is the first article to deal with urinary exosomal CA9 mRNA for the diagnosis of bladder cancer. This strategy can enlarge the use of urinary mRNA markers such as CA9 for the detection or surveillance of bladder cancer, which depends solely on urinary cells. For example, when tumor cells cannot be found in the urine of many cancer patients, the urinary exosomal CA9 may be useful. Now we proved that urinary supernatant contained abundant tumor exosomes. We anticipate that urinary exosomes from the tumor cells may be abundant even at the early stage. We believe that urinary exosomes will become a new source of liquid biopsy for bladder cancer. We admit that it is a pilot study. The next step is to evaluate its clinical use. It will be interesting to study how this technique can improve the urinary cytology or urinary cellular mRNA markers as a diagnostic and surveillance method for bladder cancer.
In conclusion, we found that the urinary exosomes were abundant in the urine of bladder cancer patients. Tumoral CA9 mRNA can be detectable in urinary exosomes. Urinary exosomal CA9 mRNA may provide a new liquid biopsy for the detection of bladder cancer.
Disclosure
Yang Zhang is an employee of Guangzhou HopeTech Biological Technology Co, LTD. The authors reported no other potential conflicts of interest for this work.
References
1. Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–E386. doi:10.1002/ijc.29210
2. Babjuk M, Bohle A, Burger M, et al. EAU Guidelines on non–muscle-invasive urothelial carcinoma of the bladder: update 2016. Eur Urol. 2017;71:447–461. doi:10.1016/j.eururo.2016.05.041
3. Kamat AM, Hahn NM, Efstathiou JA, et al. Bladder cancer. Lancet. 2016;388(10061):2796–2810. doi:10.1016/S0140-6736(16)30512-8
4. Soukup V, Babjuk M, Bellmunt J, et al. Follow-up after surgical treatment of bladder cancer: a critical analysis of the literature. Eur Urol. 2012;62(2):290–302. doi:10.1016/j.eururo.2012.05.008
5. Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200(4):373–383. doi:10.1083/jcb.201211138
6. Skog J, Wurdinger T, van Rijn S, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470–1476. doi:10.1038/ncb1800
7. Tominaga N, Katsuda T, Ochiya T. Micromanaging of tumor metastasis by extracellular vesicles. Semin Cell Dev Biol. 2015;40:52–59. doi:10.1016/j.semcdb.2015.02.016
8. Skog J, Würdinger T, van Rijn S, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470–1476.
9. Franzen CA, Blackwell RH, Foreman KE, Kuo PC, Flanigan RC, Gupta GN. Urinary exosomes: the potential for biomarker utility, intercellular signaling and therapeutics in urological malignancy. J Urol. 2016;195:1331–1339. doi:10.1016/j.juro.2015.08.115
10. Dear JW, Street JM, Bailey MA. Urinary exosomes: a reservoir for biomarker discovery and potential mediators of intrarenal signalling. Proteomics. 2013;13(10–11):1572–1580. doi:10.1002/pmic.201200285
11. Blackwell RH, Franzen CA, Flanigan RC, Kuo PC, Gupta GN. The untapped potential of urine shed bladder cancer exosomes: biomarkers, signalling and therapeutics. Bladder. 2014;1:e7. doi:10.14440/bladder.2014.38
12. de Martino M, Lucca I, Mbeutcha A, et al. Carbonic anhydrase IX as a diagnostic urinary marker for urothelial bladder cancer. Eur Urol. 2015;60:552–554. doi:10.1016/j.eururo.2015.06.015
13. Hiltbrunner S, Mints M, Eldh M, et al. Urinary exosomes from bladder cancer patients show a residual cancer phenotype despite complete pathological downstaging. Sci Rep. 2020;10(1):5960. doi:10.1038/s41598-020-62753-x
14. Tilki D, Burger M, Dalbagni G, et al. Urine markers for detection and surveillance of non-muscle-invasive bladder cancer. Eur Urol. 2011;60:484–492. doi:10.1016/j.eururo.2011.05.053
15. Lokeshwar VB, Habuchi T, Grossman HB, et al. Bladder tumor markers beyond cytology: international consensus panel on bladder tumor markers. Urology. 2005;66:35–63. doi:10.1016/j.urology.2005.08.064
16. Smalley DM, Sheman NE, Nelson K, Theodorescu D. Isolation and identification of potential urinary microparticle biomarkers of bladder cancer. J Proteome Res. 2008;7:2088–2096. doi:10.1021/pr700775x
17. O’Sullivan P, Sharples K, Dalphin M. A multigene urine test for the detection and stratification of bladder cancer in patients presenting with hematuria. J Urol. 2012;188:741–747. doi:10.1016/j.juro.2012.05.003
18. Holyoake A, O’Sullivan P, Pollock R, et al. Development of a multiplex RNA urine test for the detection and stratification of transitional cell carcinoma of the bladder. Clin Cancer Res. 2008;14:742–749. doi:10.1158/1078-0432.CCR-07-1672
19. Wallace E, Higuchi R, Satya M, et al. Development of a 90-minute integrated noninvasive urinary assay for bladder cancer detection. J Urol. 2018;199:655–666. doi:10.1016/j.juro.2017.09.141
20. Pichler R, Fritz J, Tulchiner G, Klinglmair G, Soleiman A, Horninger W. Increased accuracy of a novel mRNA-based urine test for bladder cancer surveillance. BJU Int. 2018;121:29–37. doi:10.1111/bju.14019
21. McKiernan J, Donovan MJ, O’Neill V, et al. A Novel urine exosome gene expression assay to predict high-grade prostate cancer at initial biopsy. JAMA Oncol. 2015;2:882–889. doi:10.1001/jamaoncol.2016.0097
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.