Back to Journals » Cancer Management and Research » Volume 11
Upregulation of FOXP4 in breast cancer promotes migration and invasion through facilitating EMT
Received 21 October 2018
Accepted for publication 24 January 2019
Published 8 April 2019 Volume 2019:11 Pages 2783—2793
DOI https://doi.org/10.2147/CMAR.S191641
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Chien-Feng Li
Tao Ma,1–4 Jin Zhang1–4
1The Third Department of Breast Cancer, Tianjin Medical University Cancer Institute and Hospital, National Clinical Research Center for Cancer, Tianjin, P.R. China; 2Key Laboratory of Breast Cancer Prevention and Therapy, Tianjin Medical University, Ministry of Education, Tianjin, P.R. China; 3Key Laboratory of Cancer Prevention and Therapy, Tianjin, P.R. China; 4Tianjin’s Clinical Research Center for Cancer, Tianjin, P.R. China
Background: Family of forkhead box transcription factors has been found to play key roles in multiple types of cancer.
Materials and methods: Our study is to decipher the effects of FOXP4 in human breast cancer (BC). Quantitative real-time polymerase chain reaction and Western blot analyses were performed to determine the mRNA and protein expressions of FOXP4 in BC tissue samples and cell lines. The gain and loss of function assay were used to explore the detailed roles of FOXP4 in breast cell lines, including MDA-MB-231 and MCF-7 cells. Its effect on BC growth, migration, and invasion were evaluated by colony formation assay, CCK-8 assay, wound-healing assay, and transwell invasion assay, respectively.
Results: Our findings revealed that FOXP4 promotes cell proliferation, migration, as well as invasion of BC cells. Furthermore, FOXP4 also facilitates epithelial–mesenchymal transition. ChIP, qChIP assay, and dual luciferase reporter assay were used to examine whether Snail is a downstream target of FOXP4. Moreover, overexpression of Snail could partially rescue the effects of FOXP4 inhibition on cancer cell migration and invasion.
Conclusion: Our findings revealed that FOXP4 is a critical regulator in BC.
Keywords: FOXP4, EMT, Snail, migration, breast cancer
Introduction
Breast cancer (BC) is one of the most common malignancy in women with a high morbidity rate.1,2 Incidence of BC indicates an increasing trend and the mortality rate is as high as 36%.3 In P.R. China, onset age of BC is becoming increasingly younger.4 The survival rate of BC is closely associated with the time of diagnosis, the deterioration, metastasis, and drug resistance of BC.5
Forkhead box proteins (FOXP), consisting of FOXP1, FOXP2, FOXP3, and FOXP4, is a family of transcription factors that play important roles in regulating cancer growth, tumorigenesis, and metastasis in multiple types of human cancer.6,7 In BC, FOXP1 was shown to be correlated with calpain II and ER expression, and enhanced tumor migration by repression of NFAT1, predicting a poor outcome.8,9 FOXP2 was found to facilitate tumor proliferation and metastasis through regulation of GRP78 in Triple-negative BC.10 FOXP2 has been found to regulate multiple neurogenesis signaling pathways critical in embryonic development and cell cycle such as Notch, WNT, and Hedgehog pathways.11,12 It is also involved in heart, lung, and central nervous system development. Especially, it plays important functions in human language development. Depletion and mutation in FOXP2 can lead to speech and linguistic impairment, aging, and cancer.13 FOXP3 was demonstrated to serve as a tumor suppressor and repress BC metastasis through downregulation of CD44.14 However, the expression and function of FOXP4 in BC remain little known.
Epithelial–mesenchymal transition (EMT) is a crucial event during metastasis to enable the cancer cells to migration. One of the key features of EMT is downregulation of E-cadherin, a prototypical member of type I classical cadherins.15 Downregulation of EMT caused by multiple mechanisms can result in downregulation of E-cadherin, such as repression of gene transcription and gene mutations.16 Previous reports have found that EMT-associated transcription factors, including SNAIL, SLUG, SIP1, and ZEB1, could lead to transcriptionally repress E-cadherin expression in human malignancy.17 Recent studies have indicated that FOXF1 promotes EMT in lung and BC cells to facilitate invasion and metastasis.18,19 However, exact molecular mechanisms of FOXP4 on EMT remain elusive in BC.
In this work, we detected the expression levels of FOXP4 in BC cell lines and tissues from BC patients. Then, we overexpressed or knocked down FOXP4 in BC cell line MDA-MB-231 and MCF-7 cells to evaluate its effect on BC growth, migration as well as invasion. Moreover, we found that FOXP4 promotes EMT in BC cells. Additionally, we examined whether FOXP4 promotes EMT through regulation of EMT-associated transcription factor. Our results provide that FOXP4 serves as an oncogene and might be as a target for molecular therapy in BC.
Materials and methods
Cell lines
Human BC cell lines, including MDA-MB-231 and MCF-7, and human normal breast epithelial cell line, such as MCF-10A, were purchased from American Type Culture Collection (ATCC, Rockville, MD, USA). MDA-MB-231 and MCF-7 cells were cultured in DMEM (Fisher Scientific International, Inc., Hampton, NH, USA) supplemented with 100 µg/mL penicillin/streptomycin (Fisher Scientific International, Inc.) and 10% FBS (Fisher Scientific International, Inc.) and cultured at 37°C in a humidified atmosphere with 5% CO2. MCF-10A cells were cultured in DMEM/F12 medium supplemented with insulin (10 µg/mL), cholera toxin (0.1 µg/mL), EGF (10 ng/mL), heat-inactivated horse serum (5%), and hydrocortisone (1 µg/mL) at 37°C in a humidified atmosphere with 5% CO2.
Tissue samples
Total of 134 tissue samples were collected from patients with BC which was diagnosed at the Tianjin Medical University Cancer Institute and Hospital from December 2006 to March 2009. None of the patients underwent therapy prior to specimen collection, including chemotherapy and radiotherapy. Tissue samples were conducted in accordance with the Declaration of Helsinki. We had obtained the approval of the ethics committee of the Tianjin Medical University Cancer Institute and Hospital before this study. The informed consent has been signed by all patients who participated in this study.
Transfection
The vectors (pcDNA3.1), pcDNA3.1-FOXP4 and pcDNA3.1-SNAI1, were purchased from Vigenbio (Shandong, P.R. China). Scramble siRNA (SCR) and FOXP4 siRNA (siFOXP4) were purchased from GenePharma Corporation (Shanghai, P.R. China). For transfection, a total of 2×105 cells were placed into 6-well plates, and transfected with 2.5 µg plasmid or 40 nM siRNA using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. After transfection for 48 hours, the cells were collected and used for further experiments.
Western blotting assay
The whole protein was isolated from tissue samples and cells using RIPA lysis buffer (Beyotime Biotechnology, Beijing, P.R. China) with protease inhibitor (Roche, P.R. China). BCA protein assays (Fisher Scientific International, Inc.) was used to measure the protein concentration according to manufacturer’s method, and 40 µg protein samples were separated by using 10% SDS-PAGE. Subsequently, proteins were transferred onto polyvinylidene fluoride membrane (Millipore, Bedford, MA, USA). After blocking membranes with 5% skim milk in Tris-buffered saline and 1% Tween 20 (Sigma-Aldrich, St. Louis, MO, USA), then membranes were incubated with indicated primary antibodies at 4°C overnight followed by incubation with HRP-conjugated secondary antibody at room temperature for 1 hour. The signals were captured by using a Pierce™ ECL Western Blotting Substrate kit (Fisher Scientific International, Inc.). β-actin was used as an internal control.
RNA extraction and quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was extracted from cells and tissue samples by using a Trizol reagent (Takara, Otsu, Japan) according to the manufacturer’s protocol. A 2 µg RNA was used to reversely transcribe into cDNA by using TransScript Fly First-Strand cDNA Synthesis SuperMix kit (Transgen Biotech Co., Ltd., Beijing, P.R. China). The thermocycling conditions were as follows: 5 minutes at 65°C, 5 minutes at 42°C, and 1 minute at 85°C. The qRT-PCR was performed by using SYBR Green PCR Kit (Takara). The primers sequences were as following: FOXP4 forward: 5′-CGACATGATGGTGGAATCTG-3′ and reverse: 5′-TGTTTGCTGTCATTGTTCCC-3′; E-cadherin forward: 5′-TGCCCAGAAAATGAAAAAGG-3′ and reverse: 5′-GTGTATGTGGCAATGCGTTC-3′; N-cadherin forward: 5′-GTCATCACAGTGACAGATGTC-3′ and reverse: 5′-TTCAAAGTCGATTGGTTTGACC-3′; Vimentin forward: 5′-GAGAACTTTGCCGTTGAAGC-3′ and reverse: 5′-CTCAATGTCAAGGGCCATCT-3′; Snail forward, 5′-TCTAATCCAGAGTTTACCTTCCAG-3′ and reverse, 5′-TGAAGTAGAGGAGAAGGACGA-3′; Slug forward, 5′-ACACATACAGTGATTATTTCCC-3′ and reverse, 5′-ACTGTAGTCTTTCCTCTTCAT-3′; ZEB1 forward: 5′-AAGCAAACGATTCTGATTCCC3′ and reverse, 5′-CTTGTCTTTCATCCTGATTTCC-3′; GAPDH forward: 5′-ACGGATTTGGTCGTATTGGGCG-3′ and reverse: 5′-CTCCTGGAAGATGGTGATGG-3′. An initial pre-denaturation step of 10 minutes at 95°C, followed by 32 cycles of denaturation at 94°C for 20 seconds and annealing at 55°C for 34 seconds were performed. GADPH acted as an internal control. The relative expression levels were counted using the 2−∆∆Cq method.20
Transwell invasion assay
The invasion ability of MDA-MB-231 cells was detected by using Transwell chambers (8 mm pore filter, Corning Incorporated, Corning, NY, USA) which were coated with matrigel. In brief, a total of 5×104 transfected MDA-MB-231 cells were resuspended in 500 µL serum-free DMEM and placed into the upper chambers. Subsequently, the lower chambers were filled with 500 µL DMEM supplemented with 10% FBS. After incubation at 37°C for 24 hours, the invaded cells were fixed with methanol at room temperature for 10 minutes and stained with 0.5% crystal violet at room temperature for 5 minutes (Amresco Co., Solon, OH, USA). Finally, the cells adhering to the lower membrane of the inserts were counted and imaged with an IX71 inverted microscope (Olympus Co., Tokyo, Japan).
ChIP and qChIP analyses
ChIP and qChIP analyses were performed by using a EZ-ChIP Kit (Millipore, Billerica, MA, USA) according to the manufacturer’s protocol. In brief, MCF-7 and MDA-MB-231 cells were cultured to 90%–100% confluence, and the chromatin was cross-linked by 1% formaldehyde at 37°C for 20 minutes, subsequently, cross-linked chromatin was sonicated (1 seconds on, 1 seconds off, 3×15 times) at 4°C to generate 200–1,000 bp fragments. Next, 5 µg anti-IgG antibodies or anti-FOXP4 were used to immunoprecipitate hromatin fragments at 4°C overnight. IgG antibody was used as control. After purifying the antibody-interact DNA, qRT-PCR was conducted to analyze the precipitated chromatin DNA. The primer sequences were as following: Snail forward, 5′-GTTCTGCCCTTCAGGTTGGT-3′ and reverse 5′-AGGCTGTAACACGGCTCCAT-3′. The qPCR conditions were as following: 5 minutes at 98°C, denaturation at 98°C for 30 seconds, annealing at 56°C for 30 seconds, and extension at 72°C for 20 seconds, performed for 32 cycles.
Luciferase reporter assay
The promoter region (−2,000 to +200) of Snail was cloned into pGL3 plasmid. A 2 µg pGL3-Snail-luc and 2 ng renilla were co-transfected with 2 µg pcDNA3.1 (vector) or 0.5 µg, 1 µg, 2 µg FOXP4 into MCF-7 or MDA-MB-231 cells. After transfection for 24 hours, it was used to detect the luciferase activities which were measured by using a dual-luciferase reporter assay system (Promega, Madison WI, USA) according to the manufacturer’s instructions. The firefly luciferase activity was normalized to the renilla luciferase activity.
Colony formation assay
Approximately 5 103 transfected cells were plated in 6-well plates and cultured for 12 days at 37°C. The cells were fixed with methanol and stained with 0.1% crystal violate. The number of clones (cell number >50 cells/colony) was counted under a light microscope.
Cell-counting kit 8 (CCK-8) assay
CCK-8 assay (Beyotime Institute of Biotechnology, Jiangsu, P.R. China) was used to determine the effect of FOXP4 on cell proliferation in BC. In brief, 3×103 transfected MCF-7 and MDA-MB-231 cells were placed in 96-well plates with 200 µL DMEM and incubated at 37°C. When cells were incubated at 24 hours, 48 hours, and 72 hours, 20 µL CCK-8 solution was added to each well. After incubation at 37°C for 30 minutes, the absorbance at 450 nm was measured by using the microplate reader (EL340; Bio-Tek Instruments, Hopkinton, MA, USA).
Statistical analysis
All statistical analyses were analyzed by GraphPad Prism 5.0 package (GraphPad Software Inc., La Jolla, CA, USA). Data were shown as the mean ± SD. Two or multiple groups were analyzed by Student’s t-test or one-way ANOVA followed by Tukey’s test, respectively. The expression of FOXP4 and clinicopathological parameters was analyzed using a chi-squared test. The correlation between patient survival and FOXP4 expression was determined by the Kaplan–Meier method. All experiments were performed three times. *P<0.05 was considered statistically significant.
Results
FOXP4 is highly expressed in BC tissues and cell lines
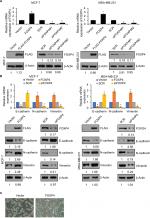
To explore the role of FOXP4 in BC, we examined FOXP4 expression in 134 BC tissues and adjacent non-neoplastic breast tissues using qRT-PCR and Western blotting analyses. As shown in Figure 1A, we found that both mRNA and protein levels of FOXP4 were significantly higher than that in non-neoplastic tissues (Figure 1A). Additionally, we also detected FOXP4 expression in BC cell lines, including MDA-MB-231 and MCF-7 cell lines; MCF-10A cell line was used as control. Consisted with tissue samples, the expression of FOXP4 in BC cell lines was markedly higher than that in MCF-10A cells (Figure 1B). Moreover, compared with the weakly metastatic cell lines MCF-7, highly metastatic cell lines MDA-MB-231 had a higher expression level of FOXP4 (Figure 1B). To investigate whether high FOXP4 expression plays a role in BC, we determined the association between pathological characteristics of BC patients and FOXP4 expression. As shown in Table 1, high expression of FOXP4 was associated with bigger tumor size, pathological grade, as well as lymph node metastasis (Table 1). Moreover, Kaplan–Meier followed by log rank suggested that high expression of FOXP4 predicted a poor prognosis (Figure 1C). Our findings strongly support that FOXP4 plays a key role in BC.
  | Table 1 Clinicopathological variables in the 134 BC patients Abbreviation: BC, breast cancer. |
FOXP4 promotes EMT in BC cells
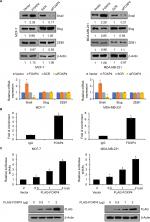
To further investigate the roles of FOXP4 in BC, we overexpressed or knocked down FOXP4 in MDA-MB-231 and MCF-7 cell lines; the efficiency of transfection was determined using qRT-PCR and Western blot analysis (Figure 2A). FOXP4 siRNA#1 was more efficient than FOXP4 siRNA#2, so FOXP4 siRNA#1 was used in the further experiments. EMT is an essential process in BC development. Subsequently, we measured the expression of EMT markers, including E-cadherin, N-cadherin, and Vimentin by using qRT-PCR and Western blotting analyses. Ectopic expression of FOXP4 increased the mRNA and protein levels of N-cadherin and Vimentin and decreased the mRNA and protein levels of E-cadherin (Figure 2B). In contrast, FOXP4 inhibition decreased the mRNA and protein levels of N-cadherin and Vimentin and increased the mRNA and protein levels of E-cadherin (Figure 2B). In addition, the morphological alterations of the cells were examined by phase-contrast microscopy. Notably, FOXP4 overexpressing MCF-7 cells displayed loss of cell–cell contacts, scattering, and a spindle-like, fibroblastic morphology compared with the highly organized cell–cell adhesion, cell polarity, and cobblestone-like appearance of control MCF-7 cells (Figure 2C). In all, our findings indicate that FOXP4 promotes EMT in BC cells.
FOXP4 transcriptionally activates Snail expression
To further investigate the role of FOXP4 in EMT, we assessed whether FOXP4 regulates EMT-associated transcription factors expression, such as ZEB1, Slug, and Snail. As shown in Figure 3A, we found that overexpression of FOXP4 led to an increase in Snail expression both in MCF-7 and MDA-MB-231 cells, whereas siRNA-induced FOXP4 silencing caused an obvious decrease in Snail expression (Figure 3A). However, the expression of FOXP4 had little effect on Slug and ZEB1 expression (Figure 3A). Consistently, qRT-PCR analysis revealed that mRNA of Snail was elevated in the FOXP4 group and reduced in the siFOXP4 group (Figure 3A), suggesting that FOXP4 positively regulated Snail expression at transcriptional level. We first assessed whether FOXP4 could immunoprecipitate the promoter region of Snail using ChIP and qChIP assays. As expected, we found that FOXP4 could bind to Snail promoter (Figure 3B). Moreover, dual luciferase reporter assay demonstrated that FOXP4 transcriptionally activated Snail (Figure 3C). Therefore, FOXP4 could bind to the promoter of Snail to promote the expression of Snail.
SNAI1 rescues FOXP4-induced tumor progression in vitro
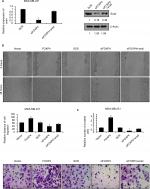
To reinforce the link between snail and FOXP4, we overexpressed Snail in FOXP4-depleted cells. The expression of Snail was detected by using qRT-PCR and Western blotting analyses (Figure 4A). Subsequently, we performed wound-healing assay and transwell invasion assay to assess the effect of FOXP4 on cell migration and invasion ability. The wound-healing assay indicated that FOXP4 inhibition significantly inhibited cell migration, compared to control, and simultaneous overexpression of Snail could rescue this effect (Figure 4B). Similarly, in transwell invasion assay, compared with the SCR group, invasive abilities were reduced upon FOXP4 inhibition, which was rescued by Snail overexpression (Figure 4C). Together, Snail can rescue FOXP4-induced tumor progression in vitro, suggesting that FOXP4 facilitates cell migration and invasion through regulation of Snail.
FOXP4 facilitates cell proliferation in BC
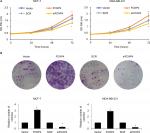
To investigate the involvement of FOXP4 in tumor proliferation, we performed CCK-8 and colony formation assay. The results of CCK-8 assay showed that ectopic expression of FOXP4 obviously improved the proliferation rate of MCF-7 compared with vector group (Figure 5A). Conversely, inhibition of FOXP4 decreased the proliferation rate of MCF-7 compared with SCR group (Figure 5A). The similar results were found in MDA-MB-231 cells. Consistently, overexpression of FOXP4 increased colony formation in both MDA-MB-231 and MCF-7 cell lines compared with the vector group, while the colony formation of these cells was suppressed by inhibition of FOXP4 (Figure 5B). These results suggest that FOXP4 facilitates cell proliferation in BC.
Discussion
Here, we found that FOXP4 is highly expressed in BC tumor tissues than adjacent normal tissues. BC cell lines showed similar result compared with normal breast epithelial cell line MCF-10A. Additionally, upregulation of FOXP4 is positively associated with many clinicopathologic, such as tumor size, pathological grade, and metastasis. These evidences remind us that FOXP4 play a key role in BC.
Several FOX transcription factors, such as FOXK1, FOXC1, FOXC2, FOXQ1, and FOXM1, have been proven to induce or suppress EMT.6,18,19,21 Our findings revealed that the upregulation of FOXP4 could promote migration and invasion in BC. Numerous EMT-induced signaling pathways as well as genes activation, including the TGF-β signaling pathways, the Snail/Slug family of transcription factors, play key role in invasive cancers.22 Our study demonstrated that FOXP4 promoted EMT in BC by decreasing the expression of epithelial markers, such as E-cadherin, and increasing the levels of mesenchymal markers, such as N-cadherin and Vimentin.
Previous work has identified that Snail serves as a crucial transcription regulator of EMT.23 It can recruit other factors, such as PRC2 to induce histone deacetylation and hypermethylation,24 finally suppresses epithelial genes expression, such as E-cadherin.25,26 Our work suggested that FOXP4 could transcriptionally activate Snail expression. By targeting on Snail, FOXP4 promotes the EMT to facilitate the migration and invasion of BC in vitro. In conclusion, we reveal a new mechanism of inducing EMT by targeting Snail in BC.
Overall, our findings suggest that FOXP4 expression is upregulated in BC tissues and cell lines, and its high expression is associated with tumor size and the advanced progression, which predicts a poor diagnosis of BC. Moreover, FOXP4 facilitates migration and invasion by promoting EMT in BC progression by transcriptionally regulating Snail. Our findings highlight the FOXP4–Snail axis as a potential therapeutic target in BC.
Acknowledgment
This study was supported by the National Natural Scientific Foundation (81672623).
Disclosure
The authors report no conflicts of interest in this work.
References
Hu Z, Mao JH, Curtis C, et al. Genome co-amplification upregulates a mitotic gene network activity that predicts outcome and response to mitotic protein inhibitors in breast cancer. Breast Cancer Res. 2016;18(1):70. | ||
Wang J, Xu H, Wang Q, et al. CIAPIN1 targets Na+/H+ exchanger 1 to mediate MDA-MB-231 cells’ metastasis through regulation of MMPs via ERK1/2 signaling pathway. Exp Cell Res. 2015;333(1):60–72. | ||
Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62(1):10–29. | ||
Chen WQ, Zeng HM, Zheng RS, Zhang SW, He J. Cancer incidence and mortality in China, 2007. Chin J Cancer Res. 2012;24(1):1–8. | ||
Macià F, Porta M, Murta-Nascimento C, et al. Factors affecting 5- and 10-year survival of women with breast cancer: an analysis based on a public General Hospital in Barcelona. Cancer Epidemiol. 2012;36(6):554–559. | ||
Myatt SS, Lam EW. The emerging roles of forkhead box (Fox) proteins in cancer. Nat Rev Cancer. 2007;7(11):847–859. | ||
Katoh M, Katoh M. Human Fox gene family (Review). Int J Oncol. 2004;25(5):1495–1500. | ||
Yu BH, Li BZ, Zhou XY, Shi DR, Yang WT. Cytoplasmic FOXP1 expression is correlated with ER and calpain II expression and predicts a poor outcome in breast cancer. Diagn Pathol. 2018;13(1):36. | ||
Oskay Halacli S. FOXP1 enhances tumor cell migration by repression of NFAT1 transcriptional activity in MDA-MB-231 cells. Cell Biol Int. 2017;41(1):102–110. | ||
Wu J, Liu P, Tang H, et al. FOXP2 promotes tumor proliferation and metastasis by targeting GRP78 in triple-negative breast cancer. Curr Cancer Drug Targets. 2018;18(4):382–389. | ||
Chiu YC, Li MY, Liu YH, Ding JY, Yu JY, Wang TW. FOXP2 regulates neuronal differentiation and neuronal subtype specification. Dev Neurobiol. 2014;74(7):723–738. | ||
Tsui D, Vessey JP, Tomita H, Kaplan DR, Miller FD. FOXP2 regulates neurogenesis during embryonic cortical development. J Neurosci. 2013;33(1):244–258. | ||
Fisher SE, Scharff C. FOXP2 as a molecular window into speech and language. Trends Genet. 2009;25(4):166–177. | ||
Douglass S, Meeson AP, Overbeck-Zubrzycka D, et al. Breast cancer metastasis: demonstration that FOXP3 regulates CXCR4 expression and the response to CXCL12. J Pathol. 2014;234(1):74–85. | ||
Zheng H, Kang Y. Multilayer control of the EMT master regulators. Oncogene. 2014;33(14):1755–1763. | ||
Li C, Wu W, Sang M, et al. Comparative RNA-sequencing analysis of mthl1 functions and signal transductions in Tribolium castaneum. Gene. 2014;547(2):310–318. | ||
Gall TM, Frampton AE. Gene of the month: E-cadherin (CDH1). J Clin Pathol. 2013;66(11):928–932. | ||
Nilsson J, Helou K, Kovács A, et al. Nuclear Janus-activated kinase 2/nuclear factor 1-C2 suppresses tumorigenesis and epithelial-to-mesenchymal transition by repressing Forkhead box F1. Cancer Res. 2010;70(5):2020–2029. | ||
Wei HJ, Nickoloff JA, Chen WH, et al. FOXF1 mediates mesenchymal stem cell fusion-induced reprogramming of lung cancer cells. Oncotarget. 2014;5(19):9514–9529. | ||
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402–408. | ||
Xia L, Huang W, Tian D, et al. Forkhead box Q1 promotes hepatocellular carcinoma metastasis by transactivating ZEB2 and VersicanV1 expression. Hepatology. 2014;59(3):958–973. | ||
Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15(3):178–196. | ||
Barrallo-Gimeno A, Nieto MA. The snail genes as inducers of cell movement and survival: implications in development and cancer. Development. 2005;132(14):3151–3161. | ||
Herranz N, Pasini D, Díaz VM, et al. Polycomb complex 2 is required for E-cadherin repression by the Snail1 transcription factor. Mol Cell Biol. 2008;28(15):4772–4781. | ||
Cano A, Pérez-Moreno MA, Rodrigo I, et al. The transcription factor Snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat Cell Biol. 2000;2(2):76–83. | ||
Bolós V, Peinado H, Pérez-Moreno MA, Fraga MF, Esteller M, Cano A. The transcription factor slug represses E-cadherin expression and induces epithelial to mesenchymal transitions: a comparison with snail and E47 repressors. J Cell Sci. 2003;116(Pt 3):499–511. |
Supplementary material
  | Figure S1 Analysis of Enerly Yakhini Breast GSE19536 dataset. |
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.