Back to Journals » Cancer Management and Research » Volume 11
Upregulation of family with sequence similarity 83 member D expression enhances cell proliferation and motility via activation of Wnt/β-catenin signaling and predicts poor prognosis in gastric cancer
Authors Wang F, Zhang S , Wei Y, Chen H, Jiao Z , Li Y
Received 27 January 2019
Accepted for publication 17 June 2019
Published 22 July 2019 Volume 2019:11 Pages 6775—6791
DOI https://doi.org/10.2147/CMAR.S203082
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Kenan Onel
Furong Wang, 1, 2,* Sigong Zhang, 2, 3,* Yucai Wei, 2,4 Hao Chen, 2,4 Zuoyi Jiao, 2,4 Yumin Li 2,4
1Department of Pathology, The Second Hospital of Lanzhou University, Lanzhou 730000, People’s Republic of China; 2The Key Laboratory of the Digestive System Tumors of Gansu Province, The Second Hospital of Lanzhou University, Lanzhou 730000, People’s Republic of China; 3Department of Rheumatology, The Second Hospital of Lanzhou University, Lanzhou 730000, People’s Republic of China; 4Department of General Surgery, The Second Hospital of Lanzhou University, Lanzhou 730000, People’s Republic of China
*These authors contributed equally to this work
Background/aims: Gastric cancer (GC) is the third most common cause of cancer-related death worldwide. The molecular mechanisms underlying the progression of gastric cancer are still not fully elucidated. In this study, we focused on exploring the role of family with sequence similarity 83, member D (FAM83D) in gastric cancer progression.
Methods: The expression of FAM83D in GC tissues was detected by immunohistochemistry (IHC) staining. FAM83D knockdown or overexpression were constructed in AGS and SGC-7901 cells with two distinct siRNA duplexes and lentivirus infection, respectively, to explore the role of FAM83D in gastric cancer progression. Nude mouse xenograft assay was used to further explore the role of FAM83D in tumorigenesis in vivo.
Results: We found that FAM83D mRNA and protein levels were higher in human GC tumor tissues and in GC cell lines, compared with the adjacent normal tissues and non-malignant gastric epithelial cell lines, respectively, and that higher FAM83D expression was correlated with worse overall survival (p< 0.0001) and disease-free survival (p< 0.0001) in GC patients. Additionally, our results showed that FAM83D overexpression significantly enhanced the proliferation, clonogenicity, and motility of GC cells, whereas FAM83D depletion caused a dramatic increase in the number of cells arrested at the G1 phase of the cell cycle. Consistent with these findings from in vitro experiment, our data also indicated that FAM83D knockdown significantly repressed GC tumor growth in vivo. Furthermore, we demonstrated that FAM83D depletion was associated with reduced Wnt/β-catenin signaling.
Conclusions: This study suggested that FAM83D overexpression enhanced the proliferation, clonogenicity, and motility of GC cells by activating Wnt/β-catenin signaling, and FAM83D may be a promising diagnostic and therapeutic target for human GC.
Keywords: gastric cancer, FAM83D, proliferation, Wnt/β-catenin
Corrigendum for this paper has been published
Introduction
Gastric cancer (GC) is the fifth most common malignancy worldwide and the third most frequent cause of cancer-related death. The risk factors for GC oncogenesis include food storage, dietary patterns, the availability of fresh produce, and the prevalence of Helicobacter pylori infection.1 Regardless of great advances in the early detection of GC and therapeutic strategy, the 5-year overall survival (OS) rate of GC patients remains lower than 30%.2 Furthermore, though numerous researchers have strived to investigate the molecular mechanisms underlying GC progression, the genetic events mediating metastasis have not be thoroughly elucidated yet due to their complexity. Therefore, it remains imperative to further clarify the mechanisms response for GC progression.
Currently, an improvement in the classification of the molecular pathogenesis of GC has been largely achieved.3–5 The crucial drivers responsible for gastric tumor initiation and progression have been identified via genomic analyses. As previous studies demonstrated, multiple oncogenes, such as human epidermal growth factor receptor 2 (HER2, also known as ERBB2), epidermal growth factor receptor (EGFR), and hepatocyte growth factor receptor (c-MET, also known as tyrosine-protein kinase (MET), are overexpressed in GC, whereas several tumor suppressors, such as tumor protein 53, phosphatase and tensin homolog, and SMAD4, are down-regulated.6 Numerous studies have shown that overexpression of HER2 and c-MET could aberrantly activate their downstream signaling pathways in GC, including mitogen-activated protein kinase (MAPK) and AKT/PI3K signaling.7,8 Additionally, EGFR is overexpressed in GC as well, resulting in a poor prognosis.9 Efforts have been made to target these key signaling pathways for treating patients with GC. Nevertheless, none of them always works well, so more efforts ought to be exerted to identify novel biomarkers in GC,10,11 which have the potential of being the new and effective targets.
Family with sequence similarity 83 consisting of 8 members is a superfamily whose members share a conserved N-terminal domain of unknown function (DUF1669), and function as the key intermediates in oncogenic EGFR, MAPK, and PI3K/AKT signaling.12–15 A recent multi-omics analyses suggested that the members of this family, as oncogenes, played prominent roles in the development of 17 different tumor types.16 Here, we focused on the effects of the member D of this family (FAM83D). The FAM83D gene is located on human chromosome 20q17 and was first reported to encode a protein that localizes to spindles during mitosis.18 FAM83D serves as a pivotal interaction partner of the chromokinesin KID22 that is required for the generation of polar ejection forces and chromosome congression during metaphase.19 FAM83D is overexpressed in several malignancies, including breast cancer, colorectal cancer, lung cancer, ovarian cancer, and hepatocellular carcinoma, and its overexpression is associated with poor prognosis.17,20–24 These reports indicate that FAM83D is a potential candidate for diagnostic and prognostic biomarkers in several cancers. However, in human GC, the biological function and prognostic value of FAM83D and its molecular mechanisms have not been elucidated fully.
In this study, we examined the mRNA expression of FAM83D in human GC, and found that its expression was remarkably up-regulated in GC tissues and GC cell lines. We also showed that a high level of FAM83D expression was associated with poor prognosis in patients with GC from our center. Moreover, in in-vitro cell experiment, we demonstrated that FAM83D promoted cell proliferation, clonogenic potential, and migration, as well as invasion mainly by activating the Wnt/β-catenin signaling pathway. We propose that FAM83D could be a diagnostic biomarker and promising molecular target for anticancer therapy in patients with GC.
Materials and methods
Materials
The following commercially available antibodies were used: anti-FAM83D (HPA049333; Sigma-Aldrich, St. Louis, MO), anti-GAPDH (60004-1-Ig; Proteintech, Chicago, IL), anti-GSK-3β (#8213;Cell Signaling Technology, Danvers, MA), anti-cyclinD1 (#3300; Cell Signaling Technology), anti-c-Myc (#9402; Cell Signaling Technology), and anti-β-catenin (#8480; Cell Signaling Technology).
Patient selection and tissue preparation
We obtained paraffin-embedded GC specimens (n=360) for prognostic survival analysis from Lanzhou University Second Hospital. A radical surgical tumor resection was performed on each patient in the Department of Gastrointestinal Surgery. The tissues were cut into 1×1 cm cubes and stored in liquid nitrogen directly for RNA and protein extraction or fixed in 4% paraformaldehyde for immunohistochemistry (IHC).25 This study was approved by the Institute Research Ethics Committee of the Second Hospital of Lanzhou University. Written informed consent was obtained from each patient. The experiments performed on these samples were in accordance with the relevant regulations. All experiments involving patient tissue specimen have been approved by the Ethics Committee of Lanzhou University Second Hospital.
RNA extraction, reverse transcriptional PCR and real-time PCR analyses
Briefly, samples were lysed with Trizol reagent (Invitrogen, Life Technologies), then total RNA was isolated according to the manufacturer’s instructions. The extracted RNA was resuspended in RNA-free water, and 2 μg RNA from each sample was used for cDNA synthesis primed with random hexamers. Quantitative RT-PCR was performed using cDNAs prepared from paired HCC and their corresponding non-cancerous tissues. All qRT–PCR reactions were performed using Light Cycler 480 (Roche). The fold change in expression was calculated using the 2-ΔΔCt method26 with the GAPDH mRNA as an internal control (ΔΔCt= ΔCt (FAM83D-GAPDH) normal-ΔCt (FAM83D-GAPDH) cancer). PCR amplification was performed using the SYBR Green PCR master mix Kit (Promega, USA). Specific primers were 5′-GCCTGGCTCGTTTCCTGAA-3′ (forward) and 5′- GGAAGTGCGTCTCGACACG −3′ (reverse) for FAM83D, and 5′-GGAGCGAGATCCCTCCAAAAT-3′ (forward) and 5′-GGCTGTTGTCATACTTCTCATGG-3′ (reverse) for GAPDH.27
IHC
IHC was performed as described previously.28 Briefly, formalin-fixed, paraffin-embedded GC samples were cut into 4 μm sections on glass slides. The slides were dried overnight at 37 °C, deparaffinized in xylene twice for 10 min, rehydrated through graded alcohol five times for 5 min, and immersed in 3% hydrogen peroxide for 15 min to block endogenous peroxidase activity. The sections were boiled in an electric pressure cooker in ethylenediamine tetraacetic acid buffer (pH 8.0) for 3 min for antigen retrieval. The slides were incubated in10% normal goat serum at room temperature for 30 min to reduce nonspecific reactions. The sections were then incubated overnight with a primary antibody against FAM83D at 4 °C. After washing three times inphosphate-buffered saline (PBS), the sections were incubated with an anti-rabbit/mouse secondary antibody at room temperature for 1 h. The signals were detected in a freshly prepared 3,3ʹ-diaminobenzidine substrate solution at room temperature for 5 min. Finally, the sections were counterstained with Mayer’s hematoxylin, dehydrated, and mounted. Each section was evaluated by three independent pathologists who were blinded to the clinical status of the patients and graded as described below. For each sample, 500 cells from five randomly chosen fields were counted. The staining intensity score was determined as: 0= negative, 1= weak, 2= moderate, and 3= strong. The staining area score was determined as: 0=0%, 1=1–25%, 2=26–50%, 3=51–75%, and 4=76–100%. These scores were added together to produce the final score: 0–1, negative expression; 2–4, weak positive expression; and 5–7, strong positive expression. Given the limited number of patients in the FAM83D-negative expression group, the negative expression group and weak positive expression group were considered to be the low expression group.29 In addition, strong FAM83D expression was considered as the high expression group.
Cell culture
Seven GC cell lines (NCI-N87, BGC-823, AGS, SGC-7901, KATO III, MKN-45 and SNU-1) and two immortalized gastric cell lines (GES-1 and Hs 738. St/Int) were employed in this study, which were purchased from the Chinese Academy of Sciences (Shanghai, China), the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China) and Chi Scientific (Jiangsu, China). NCI-N87 cells were established from a liver metastasis of a gastric carcinoma arising in an American patient and are classified as a well-differentiated GC cell line. SNU-1 cells were established from a Korean patient who had not received prior cytotoxic therapy.30 The human gastric cell line SGC-7901 was first established from a metastatic lymph node of a 56-year-old female patient with gastric adenocarcinoma. The BGC-823 cell line was derived from a 62-year-old male patient with gastric cancer.31 The MKN-45 cell line was established from a poorly differentiated gastric adenocarcinoma that had metastasized to the liver.32 GES-1 cells were isolated from gastric epithelial cells from a 9-month-old human fetus.33 NCI-N87, SNU-1, SGC-7901, BGC-823, MKN-45 and GES-1 cells were cultured in RPMI 1640 medium (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA) supplemented with 10% fetal bovine serum (Invitrogen; Thermo Fisher Scientific, Inc.). AGS cells were established from a single biopsy of an untreated human gastric adenocarcinoma of a 54-year-old female patient, and this cell line was cultured in Ham’s F-12K medium (Invitrogen; Thermo Fisher Scientific, Inc.) with 10% fetal bovine serum.34 KATO-III cells were established in vitro from a pleural effusion of a 55-year-old male patient, and this cell line was cultured in Iscove’s modified Dulbecco’s medium (Invitrogen; Thermo Fisher Scientific, Inc.) with 20% fetal bovine serum.35 Hs 738. St/Int cells were established from an 18-week-gestation fetus, and this cell line was cultured in Dulbecco’s modified Eagle’s medium (Invitrogen; Thermo Fisher Scientific, Inc.) with 10% fetal bovine serum. All cells were cultured with 1% penicillin/streptomycin under standard conditions (5% CO2, 37 °C, and 95% humidity).
Plasmid construction and RNA interference
The FAM83D coding sequence was amplified and inserted into LV003-IRES-EGFP (Forevergen Biosciences Co., Ltd., Guangzhou, China). Lentiviruses were produced by co-transfecting the constructed plasmid with the packaging plasmids psPAX2 and pMD2.G (Addgene, Watertown, MA) into 293T cells using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) for approximately 72 h. Culture supernatants were collected, filtered, concentrated, and used to infect AGS and SGC-7901 cells. After 48 h of infection, the infected cells were selected by 2 µg/mL puromycin (540411; Merck Millipore, Burlington, MA), and successful establishment was confirmed by Western blotting.
Two small interfering RNA (siRNA) duplexes targeting FAM83D were obtained from RiboB (Lanzhou, China) and gave consistent results. AGS and SGC-7901 cells were transfected with 100 nM siRNA using Lipofectamine RNAiMAX according to the manufacturer’s protocol (Invitrogen). After 48 h, the effect of RNA interference was confirmed using Western blotting.
Western blotting
Western blot analysis was performed according to the standard protocol. Total protein was extracted using radioimmunoprecipitation assay buffer with protease/phosphatase inhibitor cocktail (Roche, Mannheim, Germany). The nuclear protein was extracted using the Subcellular Proteome Extraction Kit (Millipore, USA) according to the manufacturer’s recommendations.36 Protein concentration was measured by a bicinchoninic acid protein assay. Proteins were separated using an 8–12% gradient polyacrylamide gel and transferred onto polyvinylidene difluoride membranes. The membranes were blocked in Tris-buffered saline containing 5% bovine serum albumin at room temperature for 1 h, incubated with the indicated primary antibody at 4 °C overnight, and incubated with a secondary antibody at room temperature for 1 h. Bands were visualized using enhanced chemiluminescence (Pierce, Waltham, MA).
3-(4,5-Dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromidecell viability assay
The cells were seeded in 96-well plates in sextuplicate at a density of 1000 cells/well. Cell proliferation was monitored daily using 3-(4,5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT) (Promega, Madison, WI). In brief, the MTT assay was performed by adding 20 μL MTT (5 mg/mL) for 4 h. The absorbance of the solution was measured at 490 nm on a microplate reader.
Colony formation assay
Five hundred cells were seeded in triplicate in six-well plates. After incubation for 2 weeks, the colonies were fixed, stained with crystal violet (0.1%, w/v in 20 nM 4-morpholinepropanesulfonic acid), and visible colonies were counted to determine the number of cells in each colony.
Cell migration and invasion assays in vitro
Cells (2.5×10e4 for the migration assay; 2.5×10e5 for the invasion assay) were suspended in RPMI 1640 medium and seeded in 8 µm pore Transwell plates with or without pre-coated Matrigel, according to the manufacturer’s instructions (Corning, New York, NY). The Transwell plates were placed in 24-well plates containing complete medium acting as a chemo-attractant. The cells were cultivated for 24 h (migration assay) or 48 h (invasion assay). Non-migrating or non-invading cells on the upper side of the chamber were removed by wiping. Migrating cells and invasive cells were fixed with methanol and stained with crystal violet. Eight randomly selected images of migrating or invading cells were acquired using an inverted microscope. The experiment was repeated three times. The images were analyzed with ImageJ software to determine the number of cells.
Flow cytometry
For cell cycle analysis, the cells were firstly synchronized at the G1/S transition using a double thymidine block and harvested at 16 h after release in fresh medium. The samples were washed twice in PBS and fixed in ice-cold 70% ethanol at −20 °C overnight. The fixed cells were treated with RNase A (R4875; Sigma-Aldrich) for 30 min at room temperature before the addition of 5 μL/mL propidium iodide (P4864; Sigma-Aldrich) for 10 min in the dark and analyzed by flow cytometry.
Nude mouse xenograft assay
Briefly, 10 male BALB/c nude mice (Beijing Slac Laboratory Animal Co., Ltd., Shanghai, China) were injected with cells that had been transfected with either FAM83D siRNA (siFAM83D) or negative control siRNA (siNC). The mice were maintained in microisolator cages. The mice were assigned randomly into two groups, and each received a subcutaneous injection of 150 µL of a viable cell suspension mixture (5.0×106 cells) containing a 90% AGS-siNC or AGS-siFAM83D cell suspension and 10% Matrigel. When the tumors could be palpated, tumor size was measured using calipers every other day. Tumor volume was calculated using the formula L×W2×0.5, where L is the largest dimension and W is the perpendicular diameter. All mice were sacrificed on the third week after injection, and individual tumors were weighed. All procedures related to animal experiment were conducted in accordance with the Guide for the Care and Use of Laboratory Animals (NIH publications Nos. 80–23, revised 1996) and the Institutional Ethical Guidelines for Animal Experiments developed by Lanzhou University Second Hospital.
Bioinformatics analysis
ONCOMINE (http://www.oncomine.com) is a cancer microarray database that integrates and unifies high-throughput cancer profiling data from 715 datasets (version 4.4.4.3 after the Q2 update 2013).37 We compared the expression of FAM83D mRNA from GC datasets containing data from both GC and normal stomach tissues. Five datasets (8 subsets among them) were included in our study: Chen et al,38 Cho et al,39 D’Errico et al,40 Wang et al,41 and Cui et al.42 The differential expression of FAM83D between GC tissues and normal stomach tissues was analyzed, and its fold-change values and statistical significance were determined.
Survival differences were validated at the gene expression level by OncoLnc (www.oncolnc.org) and Kaplan Meier-Plotter (http://kmplot.com/analysis/index.php?p=service&cancer=gastric). OncoLnc is a tool for interactively exploring survival correlations, and it contains survival data for 8,647 patients from 21 cancer studies performed by The Cancer Genome Atlas (TCGA), along with RNA-Seq expression data for mRNAs from TCGA cases. The top third and lowest third were selected as the high and low FAM83D expression groups, respectively, as recommended on the OncoLnc website.43 The Kaplan Meier-Plotter tool is online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in various cancers containing 882 GC patients with survival data. FAM83D was entered as the gene symbol, and the median value of FAM83D expression was selected as the cut-off for the high and low FAM83D expression groups. Kaplan-Meier and log-rank tests were used to estimate and display the outcomes.44
Statistical analysis
An independent t-test was applied to analyze differences between two groups. A chi-squared test was employed to analyze the relationship between FAM83D expression and clinicopathological characteristics. Kaplan-Meier analysis was employed to assess survival. All of the statistical tests were two-sided. Analyses of the differences between the groups in the xenograft assays were achieved using a two-tailed t test. A difference with p<0.05 was considered statistically significant. All statistical tests were performed using SPSS 20.0 statistical software (SPSS, Inc., Chicago, IL).
Results
FAM83D expression is increased in GC tissues and cell lines
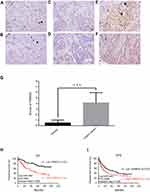
To assess the role of FAM83D in GC, we analyzed 5 microarray datasets from the ONCOMINE database, and found significant overexpression of FAM83D in the majority of GC tissues compared with adjacent non-neoplastic controls. The median rank of FAM83D in the upregulated genes of GC was 300.5 based on a meta-analysis across 8 data subsets using the ONCOMINE algorithms (p=3.59E-5, Figure 1A). We examined whether there was a relationship between FAM83D expression levels and the prognosis of GC patients by using online bioinformatics analysis tools. High FAM83D expression correlated with a poorer clinical outcome in GC patients (Figure 1B, log-rank, p=0.035). We used the online Kaplan Meier-Plotter tool to assess the prognostic value of FAM83D in 882 GC samples. Log-rank and Kaplan-Meier tests were used to compare the survival of patients, and the results indicated that high FAM83D expression was associated with a poorer clinical outcome in GC (Figure 1C, log-rank, p=3.3e-7). To validate further the relationship between FAM83D expression status and GC, we detected the mRNA levels of FAM83Din tumor tissues compared with paired non-cancerous tissues using quantitative PCR. The data confirmed that FAM83D mRNA levels were significantly elevated in tumor tissues (25/33, 75.8%, Figure 1D). To explore further the potential role of FAM83D in GC tumorigenesis, we detected FAM83D protein expression in the normal gastric cell lines GES-1 and Hs 738.St/Int and in 7 different GC cell lines using Western blotting. Compared with normal gastric cells, FAM83D was overexpressed in all of the examined GC cell lines (Figure 1E). Taken together, these results demonstrated that FAM83D expression was increased in GC tumor tissues and cells and suggested that the upregulation of FAM83D in GC might play a significant role in tumor development.
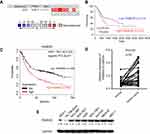 |
Figure 1 FAM83D is overexpressed in human GC. (A) FAM83D mRNA expression in liver cancer tissues is higher than normal tissues from ONCOMINE database. Dataset 1, Diffuse Gastric Adenocarcinoma vs Normal, Chen Gastric; Dataset 2, Gastric Intestinal Type Adenocarcinoma vs Normal, Chen Gastric; Dataset 3, Diffuse Gastric Adenocarcinoma vs Normal, Cho Gastric; Dataset 4, Gastric Intestinal Type Adenocarcinoma vs Normal, Cho Gastric; Dataset 5, Gastric Mixed Adenocarcinoma vs Normal Cho Gastric; Dataset 6, Gastric Cancer vs Normal, Cui Gastric; Dataset 7, Gastric Intestinal Type Adenocarcinoma vs Normal, DErrico Gastric; Dataset 8, Gastric Cancer vs Normal, Wang Gastric. Cell color and the number above the cell in the lower panel indicate the best gene rank percentile for the analysis. Red, upregulated; blue, downregulated. (B) The prognostic value of FAM83D mRNA expression in gastric cancer (www.oncolnc.org). (C) The prognostic value of FAM83D mRNA expression in gastric cancer (www.kmplot.com). (D) The mRNA levels of FAM83D from 33 patients were tested by quantitative PCR using paired t-test. (E) FAM83D protein expressions in seven GC cell lines (NCL-N87, BGC-823, AGS, SGC-7901, KATO III, MKN-45, SNU-1), one immortalized gastric cell line (GES-1, Hs 738.St/Int) were examined by Western blotting. |
Upregulation of FAM83D is associated with poor prognosis in patients with GC
To investigate further FAM83D expression in fresh GC tissues, we employed IHC staining with a total of 360 GC samples from patients at the Second Hospital of Lanzhou University after gastric section who were followed up for 100 months. Among the patients, there were 47 women (13.1%) and 313 men (86.9%) with an age range of 20 to 78 years. Most patients (83.6%) had a single tumor, and the majority were >4 cm in size (216/360 cases, 60.0%). With regard to histological classification, 54 cases (15.0%) were classified as tumor-node-metastasis (TNM) stage I, 184 cases (51.1%) as TNM stage II, 111cases (30.8%) as TNM stage III, and 11 cases (3.1%) as TNM stage IV. FAM83D expression was specifically detected in the cell nucleus in cancer tissues of the vast majority of GC samples (309/360 cases, 85.8%), while the other samples presented with negative expression (51/360cases, 14.2%). No FAM83D signals were found in any of the corresponding adjacent normal tissues. According to the IHC results, 207 cases were defined as low FAM83D expression, while the other 153 cases were identified as high expression (Figure 2A–F). Significantly higher FAM83D expression was observed in gastric tumor tissues compared with normal tissues (Figure 2G, p<0.001). Further investigations of the relationship between FAM83D protein expression and the clinicopathological features are summarized in Table 1. The results indicated that the levels of FAM83D protein expression were markedly correlated with age (p=0.011), tumor size (p<0.001), tumor location (p=0.026) and survival (p<0.001).
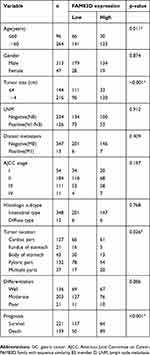 |
Table 1 Association between FAM83D expression and clinicopathologic factors in 360 GC specimens |
According to Kaplan-Meier survival analysis, the median survival time of all patients was 45.1 months, while patients with higher FAM83D expression had decreased OS (p<0.0001) and disease-free survival (DFS, p=0.0068, Figure 2H and I). Multivariate Cox regression analysis showed that FAM83D expression (hazard ratio [HR] =2.674, p<0.001), gender (HR =0.522, p=0.032), and TNM stage (HR =1.522, p<0.001) were independent predictors of survival in GC patients (Table 2). These data indicated that FAM83D expression was associated with certain clinicopathological factors and could be a prognostic marker for both early and late stage GC.
 |
Table 2 Univariate and multivariate analysis of overall survival in 360 GC specimens |
FAM83D promotes the proliferation and clonogenicity of GC cells
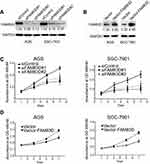
To address the cellular mechanism by which FAM83D promoted tumor progression, FAM83D knockdown or overexpression GC cell models were constructed in AGS and SGC-7901 cells with two distinct siRNA duplexes and lentivirus infection, respectively. Protein levels were determined to confirm FAM83D silencing and overexpression in the cell lines. FAM83D expression was almost eliminated in the knockdown cell model (Figure 3A) and increased in the overexpression cell model, indicating their successful establishment (Figure 3B). An MTT assay was performed to assess cell viability at the indicated times. The data showed that FAM83D inhibition markedly reduced the viability of GC cells (Figure 3C). On the contrary, cellular proliferation was greatly increased after FAM83D overexpression (Figure 3D). A colony formation assay showed that the size and number of siFAM83D transfectants were dramatically decreased compared with the siNC cells (Figure 4A). On the other hand, cell size and number were significantly increased in FAM83D overexpression cells (Figure 4B). Taken together, these results indicated that FAM83D played an important role in GC proliferation and clonogenicity.
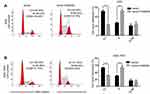
FAM83D facilitates migration and invasion in vitro
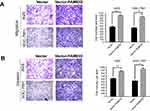
As FAM83D expression was correlated with lymph node metastasis, Transwell assays were performed to explore the effects of FAM83D on cell motility. The data showed that inhibition of endogenous FAM83D decreased the number of cancer cells dramatically during migration (Figure 5A) or invasion (Figure 5B) through the Transwell membranes. Conversely, the ectopic expression of FAM83D significantly increased migration (Figure 6A) and invasion (Figure 6B). Thus, FAM83D is considered as a stimulator of tumor migration and invasion.
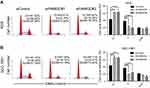
FAM83D accelerates cell cycle progression in GC cells
Having found an effect of FAM83D on the proliferation and motility of GC cell lines, we examined its impact on the cell cycle. AGS and SGC-7901 cells transfected with FAM83D siRNA showed a decreased percentage of cells in the S phase and a marked accumulation in the number of cells in the G0/G1 phase (Figure 7). On the contrary, using the FAM83D overexpression cell model, we found an increase in the percentage of cells in the S phase and a decrease in the percentage of cells in theG0/G1 phase (Figure 8). Taken together, we can summarize that FAM83D promotes cell proliferation by controlling the G1 and S checkpoints to promote cell cycle progression.
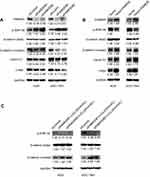
FAM83D activates Wnt/β-catenin signaling
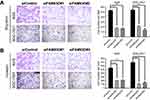
Recently, a study showed that FAM83G (PAWS1) is overexpressed in xenopus laevis embryonic cells, which could activate Wnt/beta-catenin pathway and caused complete axis duplication.45 Similarly, Yang M. et al reported that the FAM83H mutation could weaken the mineralization in ameloblasts by activating Wnt/β-catenin signaling pathway.46 To determine whether the observed phenotypic changes in the malignant behavior of GC cells as a result of FAM83D overexpression were related to epithelial-mesenchymal transition (EMT), the expression of markers associated with EMT was quantified in the GC cell lines by Western blot analysis. We silenced FAM83D with siRNA in AGS and SGC-7901 cells. Effective knockdown of FAM83D led to a significant decrease of β-catenin expression in GC cells; GSK-3β was significantly up-regulated, while the expression of the downstream proteins β-catenin, cyclin D1, and c-Myc was down-regulated (Figure 9A). Conversely, the opposite results were obtained following FAM83D overexpression in GC cells (Figure 9B). These results suggested that FAM83D may promote β-catenin expression via the activation of Wnt/β-catenin signaling.
Lithium chloride (Licl) is a typical activator of Wnt/β-catenin signaling pathway which can inhibit the activity of GSK3-β and thereby stabilize the β-catenin in cytoplasm.47 To explore further FAM83D-regulated Wnt/β-catenin signaling in GC cells, 20 mmol/L LiCl was used on the siNC and siFAM83D groups of AGS and SGC-7901 cells to examine whether the activation of Wnt/β-catenin signaling could restore the effects of FAM83D inGC cells. We found that LiCl administration recovered the level of β-catenin protein expression after FAM83D knockdown (Figure 9C). Thus, these results suggest that FAM83D declines the expression of GSK-3β and consequently activates the Wnt/β-catenin signaling pathway in the nucleus.
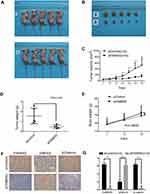
FAM83D knockdown inhibits GC tumorigenesis in nude mice
To explore further the role of FAM83D in tumorigenesis in vivo, AGS cells that were transfected with siFAM83D were injected subcutaneously into 5-week-old BALB/c male mice. Tumor formation was assessed after 7 days. FAM83D repression significantly reduced xenograft volume (Figure 10A–C) and weight (Figure 10D, P=0.0031). In addition, there was no significant difference in body weight between both groups (Figure 10E). The siFAM83D treated tumors showed a significant reduction in FAM83D and β-catenin staining and increase in GSK3-β when compared to the siControl treated group (Figure 10F and C). In summary, our data indicate that FAM83D knockdown repressed tumor growth in nude mice.
Discussion
GC is a heterogeneous disease involving numerous genetic and epigenetic alterations. In recent years, there has been growing interest in identifying novel genes that have pivotal roles in cancer initiation and progression. Thus, we used 5 datasets and the ONCOMINE algorithms to preliminarily screen differentially expressed genes between GC and normal tissues. As a result, we found that there was a significant difference in FAM83D expression, signifying that it might be a candidate oncogene that facilitates the tumor cell growth, invasion and metastasis in GC.
Our results showed that FAM83D was overexpressed in GC tissues and cell lines, compared with non-tumorous gastric tissues and normal cell lines, respectively, indicating it could be a potential prognostic biomarker in GC. Consistently, we found that high FAM83D expression was significantly correlated with large tumor size, distant metastasis, poor tumor differentiation, and poor overall survival (OS). We then used the in vitro GC cell models to explore the molecular mechanisms by which FAM83D promoted gastric malignant transformation and tumor progression. We observed the decreased motility of GC cell lines when FAM83D gene was silenced, but the opposite results were observed when FAM83D gene was upregulated, implying that FAM83D may play a pivotal role in GC metastasis. Additionally, using a loss or gain of function study, we demonstrated that FAM83D induced cells in the G0/G1 phase to pass into the S phase by promoting mitotic progression. Furthermore, we found that the upregulation of FAM83D expression had the capability of inducing these malignant phenotypes via the activation of the Wnt/β-catenin signaling. In line with the results of in-vitro experiments, we observed that FAM83D knockdown significantly inhibited GC tumorigenesis in nude mice as well. Overall, this present study presents a key role for FAM83D in promoting cellular growth and progression in GC.
Several studies have investigated the potential mechanisms underlying the roles of FAM83D in cancer initiation and progression. It has been reported that FAM83D could promote cell cycle progression of hepatocellular carcinoma cells through activating the MEK/extracellular signal-regulated kinase signaling pathway.48 FAM83D has also been supposed to physically interact with FBXW7, a tumor suppressor protein, and meanwhile downregulate FBXW7 expression, which enhanced cell proliferation and motility in breast cancer.17 Additionally, it was reported FAM83D downregulation could restrain tumor growth and metastasis via the inhibition of CD44 expression, which was induced by inactivating the MAPK, transforming growth factor-β, and Hippo pathways.24 In accordance with these previous studies, herein our present study also showed that FAM83D facilitated GC progression by promoting cell proliferation, migration, and invasion.
The aberrant activation of Wnt/β-catenin signaling is involved in tumorigenesis in GC.49 Particularly, it has been reported that FAM83A overexpression could activate the Wnt/β-catenin signaling pathway in other malignancies.50 Similarly, in the current study, we found that FAM83D knockdown increased GSK-3β expression, but decreased the expression of several Wnt/β-catenin target genes, including β-catenin, cyclin D1, and c-Myc, whereas the aberrant activation of Wnt/β-catenin signaling occurred with FAM83D gene overexpressed in GC cell lines. GSK-3β reduces the nuclear accumulation of β-catenin and Wnt target gene expression.51 Cyclin D1, as a regulator of cell cycle progression through the G1/S checkpoint, is upregulated in several types of cancer.52,53 c-Myc protein promotes cell growth and proliferation and inhibits apoptosis by regulating its target genes.54 Thus, the phenotypes induced by FAM83D depletion and overexpression may be partly attributed to the subsequent upregulation of cyclin D1 and c-Myc. The Wnt/β-catenin pathway is of decisive importance in epithelial-mesenchymal transition (EMT), a key step in oncogenic transformation.55,56 β-Catenin is a key component of the Wnt/β-catenin pathway, and its free cytoplasmic levels lead to the reduction of E-cadherin.57 The loss of E-cadherin on the membrane, coupled with N-cadherin overexpression, is one of the hallmarks of EMT.58 Thus, the upregulation of β-catenin plays an important role in enhancing the invasion and metastasis of several types of cancer by inducing EMT.59,60 A previous study reported that FAM83D could induce several EMT-associated factors in breast cancer.17 Analogously, in the present study, we also found that FAM83D may be associated with the nuclear translocation of β-catenin.
In general, the present study suggested that FAM83D may be a useful prognostic biomarker in GC, and promoted cell cycle progression, migration, and invasion mainly by activating the Wnt/β-catenin signaling pathway. Actually, there are many factors that induce dysregulation of the Wnt/β-catenin pathway, such as ubiquitination, microRNA dysregulation, and DNA methylation.61,62 Nevertheless, in this study, we did not explore the precise mechanisms underlying how FAM83D interacted with or affected β-catenin in GC cells, which requires clarification in the future studies.
Conclusion
This study demonstrated that FAM83D was overexpressed in GC tissues and cell lines and its overexpression was closely correlated with advanced clinicopathological characteristics and poor prognosis. Additionally, FAM83D could promote cell cycle progression, migration, and invasion mainly by activating the Wnt/β-catenin signaling pathway. Therefore, FAM83D may serve as a prognostic biomarker and a potential target for GC therapy.
Acknowledgments
This work was supported by Cuiyin Scienticfic and Technology Innotation Program of Lanzhou University Second hospital (CY2018-MS15), China Postdoctoral Science Foundation (2016M600826), the Provincial Natural Science Foundation of Gansu (No.18JR3RA306, No.1606RJZA213), and the grants from National Natural Science Foundation of China (31770537, 31270532).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi:10.3322/caac.21262
2. Gupta GP, Massague J. Cancer metastasis: building a framework. Cell. 2006;127(4):679–695. doi:10.1016/j.cell.2006.11.001
3. Chen D, Lin X, Zhang C, et al. Activated Wnt signaling promotes growth and progression of AFP-producing gastric cancer in preclinical models. Cancer Manag Res. 2019;11:1349–1362. doi:10.2147/CMAR.S187219
4. Fang J, Wang H, Fang X, et al. Low STYK1 expression indicates poor prognosis in gastric cancer. Cancer Manag Res. 2018;10:6669–6676. doi:10.2147/CMAR.S181910
5. Koper-Lenkiewicz OM, Kaminska J, Gawronska B, Matowicka-Karna J. The role and diagnostic potential of gastrokine 1 in gastric cancer. Cancer Manag Res. 2019;11:1921–1931. doi:10.2147/CMAR.S194949
6. Strand MS, Lockhart AC, Fields RC. Genetics of gastric cancer. Surg Clin North Am. 2017;97(2):345–370. doi:10.1016/j.suc.2016.11.009
7. Comoglio PM, Giordano S, Trusolino L. Drug development of MET inhibitors: targeting oncogene addiction and expedience. Nat Rev Drug Discov. 2008;7(6):504–516. doi:10.1038/nrd2530
8. Menard S, Pupa SM, Campiglio M, Tagliabue E. Biologic and therapeutic role of HER2 in cancer. Oncogene. 2003;22(42):6570–6578. doi:10.1038/sj.onc.1206779
9. Kim MA, Lee HS, Lee HE, Jeon YK, Yang HK, Kim WH. EGFR in gastric carcinomas: prognostic significance of protein overexpression and high gene copy number. Histopathology. 2008;52(6):738–746. doi:10.1111/j.1365-2559.2008.03021.x
10. Jiang X, Wu M, Xu Z, et al. HJC0152, a novel STAT3 inhibitor with promising anti-tumor effect in gastric cancer. Cancer Manag Res. 2018;10:6857–6867. doi:10.2147/CMAR.S188364
11. Sun MY, Zhang H, Tao J, Ni ZH, Wu QX, Tang QF. Expression and biological function of rhotekin in gastric cancer through regulating p53 pathway. Cancer Manag Res. 2019;11:1069–1080. doi:10.2147/CMAR.S185345
12. Cipriano R, Miskimen KL, Bryson BL, Foy CR, Bartel CA, Jackson MW. Conserved oncogenic behavior of the FAM83 family regulates MAPK signaling in human cancer. Mol Cancer Res. 2014;12(8):1156–1165. doi:10.1158/1541-7786.MCR-13-0289
13. Cipriano R, Graham J, Miskimen KL, et al. FAM83B mediates EGFR- and RAS-driven oncogenic transformation. J Clin Invest. 2012;122(9):3197–3210. doi:10.1172/JCI60517
14. Cipriano R, Miskimen KL, Bryson BL, Foy CR, Bartel CA, Jackson MW. FAM83B-mediated activation of PI3K/AKT and MAPK signaling cooperates to promote epithelial cell transformation and resistance to targeted therapies. Oncotarget. 2013;4(5):729–738. doi:10.18632/oncotarget.1027
15. Lee SY, Meier R, Furuta S, et al. FAM83A confers EGFR-TKI resistance in breast cancer cells and in mice. J Clin Invest. 2012;122(9):3211–3220. doi:10.1172/JCI60498
16. Snijders AM, Lee SY, Hang B, Hao W, Bissell MJ, Mao JH. FAM83 family oncogenes are broadly involved in human cancers: an integrative multi-omics approach. Mol Oncol. 2017;11(2):167. doi:10.1002/1878-0261.12016
17. Wang Z, Liu Y, Zhang P, et al. FAM83D promotes cell proliferation and motility by downregulating tumor suppressor gene FBXW7. Oncotarget. 2013;4(12):2476–2486. doi:10.18632/oncotarget.1581
18. Sauer G, Korner R, Hanisch A, Ries A, Nigg EA, Sillje HH. Proteome analysis of the human mitotic spindle. Mol Cell Proteomics. 2005;4(1):35–43. doi:10.1074/mcp.M400158-MCP200
19. Santamaria A, Nagel S, Sillje HH, Nigg EA. The spindle protein CHICA mediates localization of the chromokinesin kid to the mitotic spindle. Curr Biol. 2008;18(10):723–729. doi:10.1016/j.cub.2008.04.041
20. Liao W, Liu W, Liu X, et al. Upregulation of FAM83D affects the proliferation and invasion of hepatocellular carcinoma. Oncotarget. 2015;6(27):24132–24147. doi:10.18632/oncotarget.4432
21. Hashimoto M, Kobayashi T, Tashiro H, Arihiro K, Kikuchi A, Ohdan H. h-Prune is associated with poor prognosis and epithelial-mesenchymal transition in patients with colorectal liver metastases. Int J Cancer Manag. 2016;139(4):812–823. doi:10.1002/ijc.30118
22. Mu Y, Zou H, Chen B, Fan Y, Luo S. FAM83D knockdown regulates proliferation, migration and invasion of colorectal cancer through inhibiting FBXW7/Notch-1 signalling pathway. Biomed Pharmacother. 2017;90:548–554. doi:10.1016/j.biopha.2017.03.073
23. Perez-Pena J, Alcaraz-Sanabria A, Nieto-Jimenez C, et al. Mitotic read-out genes confer poor outcome in luminal A breast cancer tumors. Oncotarget. 2017;8(13):21733–21740. doi:10.18632/oncotarget.15562
24. Lin B, Chen T, Zhang Q, et al. FAM83D associates with high tumor recurrence after liver transplantation involving expansion of CD44+ carcinoma stem cells. Oncotarget. 2016;7(47):77495–77507. doi:10.18632/oncotarget.12715
25. Liao Y, Chen J, Ma J, Mao Q, Wei R, Zheng J. Notch-regulated ankyrin-repeat protein is a novel tissue biomarker that predicts poor prognosis in non-small cell lung cancer. Oncol Lett. 2018;16(2):1885–1891. doi:10.3892/ol.2018.8826
26. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods (San Diego, Calif). 2001;25(4):402–408. doi:10.1006/meth.2001.1262
27. Olesen I, Thorsen L, Jespersen L. Relative transcription of listeria monocytogenes virulence genes in liver pates with varying NaCl content. Int J Food Microbiol. 2010;141(Suppl 1):S60–S68. doi:10.1016/j.ijfoodmicro.2010.01.042
28. Zu LD, Peng XC, Zeng Z, et al. Gastrin inhibits gastric cancer progression through activating the ERK-P65-miR23a/27a/24 axis. J Exp Clin Cancer Res. 2018;37(1):115. doi:10.1186/s13046-018-0782-7
29. Mungan MU, Gurel D, Canda AE, Tuna B, Yorukoglu K, Kirkali Z. Expression of COX-2 in normal and pyelonephritic kidney, renal intraepithelial neoplasia, and renal cell carcinoma. Eur Urol. 2006;50(1):
30. Park JG, Frucht H, LaRocca RV, et al. Characteristics of cell lines established from human gastric carcinoma. Cancer Res. 1990;50(9):2773–2780.
31. Li Y, Ling B, Xiang C-P, Zhang Y, Li -Y-Y, Wu X-L. Characterization of gastric cancer models from different cell lines orthotopically constructed using improved implantation techniques. World J Gastroenterol. 2012;18(2):136–143. doi:10.3748/wjg.v18.i2.136
32. Takahashi N, Okumura T, Motomura W, Fujimoto Y, Kawabata I, Kohgo Y. Activation of PPARgamma inhibits cell growth and induces apoptosis in human gastric cancer cells. FEBS Lett. 1999;455(1–2):135–139.
33. Ke Y, Ning T, Wang B. [Establishment and characterization of a SV40 transformed human fetal gastric epithelial cell line-GES-1]. Chin J Oncol. 1994;16(1):7.
34. Barranco SC, Townsend CM, Casartelli C, et al. Establishment and characterization of an in vitro model system for human adenocarcinoma of the stomach. Cancer Res. 1983;43(4):1703–1709.
35. Sekiguchi M, Sakakibara K, Fujii G. Establishment of cultured cell lines derived from a human gastric carcinoma. Jpn J Exp Med. 1978;48(1):61–68.
36. Börner A, Warnken U, Schnölzer M, et al. Subcellular protein extraction from human pancreatic cancer tissues. Biotechniques. 2009;46(4):297–304. doi:10.2144/000113090
37. Rhodes DR, Yu J, Shanker K, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6(1):1–6. doi:10.1016/s1476-5586(04)80047-2
38. Chen X, Leung SY, Yuen ST, et al. Variation in gene expression patterns in human gastric cancers. Mol Biol Cell. 2003;14(8):3208–3215. doi:10.1091/mbc.e02-12-0833
39. Cho JY, Lim JY, Cheong JH, et al. Gene expression signature-based prognostic risk score in gastric cancer. Clin Cancer Res. 2011;17(7):1850–1857. doi:10.1158/1078-0432.CCR-10-2180
40. D’Errico M, de Rinaldis E, Blasi MF, et al. Genome-wide expression profile of sporadic gastric cancers with microsatellite instability. Eur J Cancer. 2009;45(3):461–469. doi:10.1016/j.ejca.2008.10.032
41. Wang Q, Wen YG, Li DP, et al. Upregulated INHBA expression is associated with poor survival in gastric cancer. Med Oncol. 2012;29(1):77–83. doi:10.1007/s12032-010-9766-y
42. Cui J, Chen Y, Chou WC, et al. An integrated transcriptomic and computational analysis for biomarker identification in gastric cancer. Nucleic Acids Res. 2011;39(4):1197–1207. doi:10.1093/nar/gkq960
43. Jordan A. OncoLnc: linking TCGA survival data to mRNAs, miRNAs, and lncRNAs. PeerJ Comput Sci. 2016;2(2):e67.
44. Szász AM, Lánczky A, Á N, et al. Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients. Oncotarget. 2016;7(31):49322–49333. doi:10.18632/oncotarget.10337
45. Bozatzi P, Dingwell KS, Wu KZ, et al. PAWS1 controls Wnt signalling through association with casein kinase 1alpha. EMBO Rep. 2018;19:4. doi:10.15252/embr.201744807
46. Yang M, Huang W, Yang F, Zhang T, Wang C, Song Y. Fam83h mutation inhibits the mineralization in ameloblasts by activating Wnt/beta-catenin signaling pathway. Biochem Biophys Res Commun. 2018;501(1):206–211. doi:10.1016/j.bbrc.2018.04.216
47. Xia MY, Zhao XY, Huang QL, et al. Activation of Wnt/beta-catenin signaling by lithium chloride attenuates d-galactose-induced neurodegeneration in the auditory cortex of a rat model of aging. FEBS Open Bio. 2017;7(6):759–776. doi:10.1002/2211-5463.12220
48. Wang D, Han S, Peng R, et al. FAM83D activates the MEK/ERK signaling pathway and promotes cell proliferation in hepatocellular carcinoma. Biochem Biophys Res Commun. 2015;458(2):313–320. doi:10.1016/j.bbrc.2015.01.108
49. Chiurillo MA. Role of the Wnt/beta-catenin pathway in gastric cancer: an in-depth literature review. World J Exp Med. 2015;5(2):84–102. doi:10.5493/wjem.v5.i2.84
50. Chen S, Huang J, Liu Z, Liang Q, Zhang N, Jin Y. FAM83A is amplified and promotes cancer stem cell-like traits and chemoresistance in pancreatic cancer. Oncogenesis. 2017;6(3):e300. doi:10.1038/oncsis.2017.3
51. Radulescu S, Ridgway RA, Cordero J, et al. Acute WNT signalling activation perturbs differentiation within the adult stomach and rapidly leads to tumour formation. Oncogene. 2013;32(16):2048–2057. doi:10.1038/onc.2012.224
52. Knudsen KE, Diehl JA, Haiman CA, Knudsen ES. Cyclin D1: polymorphism, aberrant splicing and cancer risk. Oncogene. 2006;25(11):1620–1628. doi:10.1038/sj.onc.1209371
53. Kishimoto I, Mitomi H, Ohkura Y, Kanazawa H, Fukui N, Watanabe M. Abnormal expression of p16(INK4a), cyclin D1, cyclin-dependent kinase 4 and retinoblastoma protein in gastric carcinomas. J Surg Oncol. 2008;98(1):60–66. doi:10.1002/jso.21087
54. Nie Z, Hu G, Wei G, et al. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell. 2012;151(1):68–79. doi:10.1016/j.cell.2012.08.033
55. Howard S, Deroo T, Fujita Y, Itasaki N. A positive role of cadherin in Wnt/beta-catenin signalling during epithelial-mesenchymal transition. PLoS One. 2011;6(8):e23899. doi:10.1371/journal.pone.0023899
56. Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119(6):1420–1428. doi:10.1172/JCI39104
57. Heuberger J, Birchmeier W. Interplay of cadherin-mediated cell adhesion and canonical Wnt signaling. Cold Spring Harb Perspect Biol. 2010;2(2):a002915. doi:10.1101/cshperspect.a002915
58. Zeisberg M, Neilson EG. Biomarkers for epithelial-mesenchymal transitions. J Clin Invest. 2009;119(6):1429–1437. doi:10.1172/JCI36183
59. Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139(5):871–890. doi:10.1016/j.cell.2009.11.007
60. Valenta T, Hausmann G, Basler K. The many faces and functions of beta-catenin. Embo J. 2012;31(12):2714–2736. doi:10.1038/emboj.2012.150
61. Liu Y, Huang T, Zhao X, Cheng L. MicroRNAs modulate the Wnt signaling pathway through targeting its inhibitors. Biochem Biophys Res Commun. 2011;408(2):259–264. doi:10.1016/j.bbrc.2011.04.009
62. Ying Y, Tao Q. Epigenetic disruption of the WNT/beta-catenin signaling pathway in human cancers. Epigenetics. 2009;4(5):307–312. doi:10.4161/epi.4.5.9371
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.