Back to Journals » OncoTargets and Therapy » Volume 12
Upregulation of circulating miR130a is correlated with development of Barrett’s esophagus and esophageal adenocarcinoma
Authors Wang L, Ji F, Liu G, Wang W, Li Z, Yue Y, Wang Z
Received 15 January 2018
Accepted for publication 21 September 2018
Published 17 December 2018 Volume 2019:12 Pages 1—7
DOI https://doi.org/10.2147/OTT.S162603
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Faris Farassati
Li Wang,1 Feng Ji,1 Gang Liu,2 Wei Wang,3,4 Zhitong Li,1 Yongqiang Yue,1 Zhonggao Wang1,5,6
1Department of Interventional Radiology, First Affiliated Hospital of Zhengzhou University, Zhengzhou 450052, China; 2Department of Center for Clinical Single Cell Biomedicine, People’s Hospital of Zhengzhou University, Henan Provincial People’s Hospital, Zhengzhou 450003, China; 3Department of Clinical Research Center, People’s Hospital of Zhengzhou University, Provincial People’s Hospital, Zhengzhou 450003, China; 4Department of Obstetrics and Gynecology, People’s Hospital of Zhengzhou University, Provincial People’s Hospital, Zhengzhou 450003, China; 5Department of Gastroesophageal Reflux Disease, Rocket Force General Hospital of Chinese People’s Liberation Army, Beijing 100088, China; 6Department of Vascular Surgery, Xuan Wu Hospital, Capital Medical University, Beijing 100053, China
Background: Barrett’s esophagus (BE) is one of the major known risk factors for esophageal adenocarcinoma (EAC). Circulating miRNAs are emerging as predictive biomarkers for early detection of malignancy. However, the potential for circulating miRNAs to be used as biomarkers for BE neoplastic progression to EAC has not been well characterized.
Method: We performed a systematic screening approach to identify spectrum miRNAs in the serum of both BE and EAC patients.
Results: miRNA-array web-based software identified 116 sequences differentially expressed between BE patients and healthy controls. Subsequent study revealed that miR130a was significantly upregulated in serum samples of BE and EAC patients compared to healthy controls. We found an increase in serum miR130a in low-grade and high-grade dysplasia BE patients compared to individuals with metaplasia. We also observed that miR130a expression levels increased gradually from early-stage (I, II) to advanced-stage (III, IV) EAC patients.
Conclusion: Our preliminary results provide evidence that circulating miR130a is correlated with the development of BE and EAC.
Keywords: Barrett’s esophagus, esophageal adenocarcinoma, microRNA, miR130a, biomarker
Introduction
Esophageal adenocarcinoma (EAC) is one of the most invasive malignancies with poor prognosis.1,2 Barrett’s esophagus (BE) is a metaplastic precursor of EAC.3 The rapid increase in incidence and poor prognosis of EAC highlights the urgent need for improving early-detection methods and management.4 A newly uncovered class of small noncoding RNAs that can negatively regulate the protein-coding gene, miRNAs are associated with physiological and pathological processes, including cell apoptosis, metastasis, proliferation, and angiogenesis.5 There have been recent advances in our understanding of the role of miRNAs as novel, specific, and sensitive biomarkers for malignant tumors, especially as blood-based markers, and multiple investigators have studied biomarker signatures that can predict malignant progression of various malignancies.6,7 However, the potential for circulating miRNAs as biomarkers in BE-related EAC has not been well characterized. Therefore, we investigated whether circulating miRNAs have the potential to predict the clinical characteristics of EAC patients. In the present study, we used serum miRNA expression profiles to identify miRNAs that are upregulated in EAC induced by BE disease. Among these, miR130a was identified as a potentially predictive biomarker for BE-mediated EAC.
Methods
Patient blood samples
A total of 130 preoperative blood samples were collected from patients with EAC (n=40) and BE (n=60) at the First Affiliated Hospital of Zhengzhou University and Henan Provincial People’s Hospital, with 30 healthy volunteers as a control group included. The diagnosis of BE and EAC was confirmed by two pathologists separately. EAC patients did not receive chemotherapy or radiotherapy before blood collection. This research was approved by the Life Science Ethics Review Committee of Zhengzhou University, and all patients provided written informed consent.
Serum miRNA expression profiling
We used a human serum and plasma 384HC miScript miRNA PCR array (MIHS-3106Z; Qiagen, Venlo, the Netherlands), which profiles the expression of 372 miRNAs detectable in serum and plasma. This high-throughput array also includes 84 miRNAs detectable and differentially expressed in serum, plasma, and other bodily fluids. The miScript array kit was used for miRNA profiling in blood samples. RNA was reverse-transcribed to cDNA with miScript according to the manufacturer’s instructions. Quantitative real-time PCR (qRT-PCR) was performed using the miScript SYBR green PCR kit (Qiagen) with the manufacturer-provided universal primer. Array data were analyzed using free web-based software (http://www.exiqon.com/mirna-pcr-analysis), which automatically performed all ΔΔCt fold-change calculations.
miRNA-specific quantitative real-time reverse-transcription PCR
Total RNA was isolated from blood samples using a Mirvana Paris kit (Ambion AMI1556, Life Technologies, Carlsbad, CA, USA) according to the manufacturer’s instructions. Reverse-transcription qRT-PCR was performed. Synthetic spiked-in Caenorhabditis elegans miR39 was added to the isolated RNA elution during reverse transcription as an internal control. Research has revealed that frequently used reference genes like RNU6B and 5S ribosomal RNA degrade easily in plasma/serum samples.8 Furthermore, large variation in serum U6 levels has been reported.9 We used TaqMan qRT-PCR assays to examine miRNA expression in blood RNA in all samples. RT-PCR was performed using an ABI 7300 system. For the evaluation of the data we used the 2−ΔΔCt method.
Statistical analysis
Statistical analysis of qRT-PCR data was performed by GraphPad Prism 6.0 (GraphPad Software, La Jolla, CA, USA). Receiver-operating characteristic (ROC) curves and area under the curve (AUC) were used to evaluate the specificity and sensitivity of predictive values of using blood miRNAs as diagnostic markers for BE and EAC. Student’s t-test was used to analyze differentially expressed miRNA between two groups. P<0.05 was considered significant.
Results
Expression profiling of circulating miRNAs in BE patients
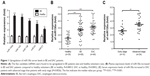
To identify expression profiling of circulating miRNAs in BE patients, an miRNA expression profile was generated using the Qiagen miRNA array web-based software. A total of 116 miRNAs were differentially expressed in BE patients (six) compared to healthy volunteers (six). We selected the top five upregulated miRNAs (miR130a, miR145, miR126, miR93, and miR21) that displayed significant differences in fold change between BE patients and healthy controls to validate reliable and reproducible circulating miRNA biomarkers in the same samples used for the microarrays by qRT-PCR assay. The selected miRNAs are listed in Table 1. Consistent with the miRNA array results, serum expression levels of the five dysregulated miRNAs were significantly upregulated in BE patients compared to healthy individuals (Figure 1A). We observed that miR130a expression levels were most upregulated in BE patients. Based on the miRNA array results by RT-PCR, we chose miR130a as a predictor for BE neoplastic progression to EAC.
  | Table 1 Top five differentially expressed miRNAs between BE patients and healthy individuals |
Candidate miRNA miR130a upregulated in serum of EAC patients
We validated the expression of miR130a in sera from a small cohort of 30 healthy controls, 60 BE patients, and 40 EAC patients. As shown in Figure 1, serum expression levels of miR130a were significantly upregulated in BE and EAC patients compared to healthy volunteers (Figure 1B). Furthermore, serum expression levels of miR130a increased gradually from early-stage (I, II) to advanced-stage (III, IV) EAC patients (Figure 1C). Correlations between miR130a expression and clinical characteristics of BE and EAC patients are summarized in Table 2.
The prediction accuracy of miR130a as a specific and sensitive biomarker in differentiating healthy individuals and BE or EAC patients was evaluated by ROC curve analysis. miR130a levels in 30 healthy controls, 60 BE disease patients, and 40 EAC patients was measured. ROC data revealed that miR130a had an AUC of 0.828 (95% CI 0.739–0.918) in separating healthy controls from BE patients, with a sensitivity and specificity of 75.0% and 80.0%, respectively (Figure 2A). ROC data also showed that miR130a had an AUC of 0.907 (95% CI 0.834–0.980) in separating healthy controls from EAC patients, with a sensitivity and specificity of 88.0% and 76.7%, respectively (Figure 2B).
Expression levels of miR130a in BE patients with different clinical status
The malignant development of BE normally follows the sequence of metaplasia, low-grade dysplasia (LGD), high-grade dysplasia (HGD), and adenocarcinoma.10 We next examined the miR130a expression levels in samples from BE patients with different clinical status. miR130a plasma levels in individuals with metaplasia, LGD, and HGD were analyzed. Our data indicated that in LGD and HGD BE patients, miR130a levels were significantly elevated when compared with individuals with metaplasia (Figure 3A). Therefore, an increase in miR130a expression in plasma may correlate with early-stage EAC. Our analysis showed that miR130a had an ROC of 0.794 (95% CI 0.671–0.917) with a sensitivity of 70.5% and a specificity of 62.5% in separating the metaplasia patients from LGD and HGD BE patients (Figure 3B).
We also performed ROC curve analysis to determine the prediction accuracy of miR130a for detection of early-stage BE-mediated EAC progression. ROC data revealed that miR130a had an AUC of 0.899 (95% CI 0.825–0.974) with a sensitivity of 95.5% and a specificity of 66.7% in separating healthy volunteers from healthy LGD and HGD volunteers (Figure 4A). ROC data also showed that miR130a had an AUC of 0.856 (95% CI 0.749–0.964) in separating healthy controls from early-stage EAC patients, with a sensitivity and specificity of 87.5% and 73.3%, respectively (Figure 4B).
Discussion
Most EAC cases are caused by BE, a precursor lesion in which the squamous epithelium of the esophagus is substituted by a metaplastic columnar epithelium.10 Mounting evidence suggests that miRNAs may serve as serum biomarkers for various malignancies and different forms of inflammatory injuries, and thus they provide a new perspective on the tumorigenic process.11,12 Based on observations from human serum specimens, we provide evidence that upregulation of circulating miR130a is associated with BE-induced EAC progression, and miR130a serum level may present a sensitive and specific biomarker for early detection of BE neoplastic progression to EAC.
Studies from different groups have indicated that miR130a has participated in different pathogenesis, including hepatocellular carcinoma (HCC),13,14 serous ovarian cancer,15 gastric carcinoma,16 and breast cancer.17 He et al demonstrated that miR130a directly targeted Dicer mRNA to enhance migration and invasion in cervical cancer cells. Upregulation of miR130a in cervical cancer tissues was significantly associated with poor disease-free survival.18 Dong et al reported that mutant TP53 exerted oncogenic functions and promoted EMT in endometrial cancer by binding directly to the miR130b promoter and inhibiting its transcription.19 Eqawa et al analyzed the functional significance of the miR130 family using a bladder cancer cell line and revealed that the miR130 family of inhibitors suppressed cell migration and invasion by downregulating FAK and Akt phosphorylation. They found that the miR130 family has a crucial role in malignant progression of bladder cancer and thus could be a promising therapeutic target for invasive bladder cancer.20 Hamilton et al integrated the Cancer Genome Atlas pancancer data set with an miRNA target atlas composed of publicly available Ago cross-linking immunoprecipitation data to identify pantumor miRNA drivers of cancer. Combined analyses identified pancancer oncogenic cotargeting of the PI3K, TGFβ, and p53 pathways by the miR17-19-130 superfamily members.21 Wang et al found that miR130a expression could be upregulated by inflammatory factors and was transactivated by NFκB. Their findings established cross talk between inflammation and mTOR signaling that was mediated by miR130a, which might have a pivotal role in the initiation and progression of serous ovarian carcinoma.15 These studies support an important role for miR130a in inflammation-related tumorigenesis. However, its role in EAC is still unclear.
In a study of miRNA profiling, miR130a was highly expressed in esophageal mucosae of patients with achalasia and may be a biomarker of esophageal achalasia.22 Liu et al explored the association of specific miRNAs with the development of esophageal cancer (EC) to identify new molecular markers for EC by analyzing the expression profiles of the miRNA spectrum in EC tissue. They found that miR130a expression was upregulated in EC tissue compared with normal paracancerous tissues. Specific miRNA expression spectra exist in EC tissues and may serve as novel EC molecular markers.23 In these reports, miR130a was not detailed further in deep studies. More intensive investigations are required to verify the potential role and mechanism of miR130a in EAC progression.
Accumulating evidence has indicated that miRNA expression profiling is an effective and reliable biomarker for differential diagnosis.24 miRNAs have been reported as being tumor-specific.25 Circulating cell-free miRNAs are promising candidates as tumor biomarkers, due to their association with malignancy, stability in plasma and serum, and easy and sensitive detection using standard PCR-based methods.5 Ali et al concluded that differentially expressed miRNAs in the serum of hepatitis C virus-infected and HCC patients can be utilized as surrogate and noninvasive biomarkers for segregation of hepatitis C and HCC patients from healthy subjects.26 Evaluation of serum miRNA biomarkers for gastric cancer based on blood- and tissue-pool profiling indicates that the majority of potential serum miRNA biomarkers may originate from tissue other than the primary tumor.27
To date, a few miRNAs have been characterized to possess potential diagnostic or prognostic functions in EC. Serum miR146a was significantly reduced in esophageal squamous-cell cancer (ESCC) patients (n=154) compared to healthy controls (n=154). Both univariate and multivariate analyses identified miR146a expression as an independent prognostic factor in ESCC patients.28 Circulating serum miR367 is aberrantly upregulated in ESCC patients and might serve as a circulating biomarker for ESCC patients.29 Chiam et al found that circulating exosomal miRNAs have potential to discriminate individuals with EAC from healthy controls and nondysplastic BE.30 Herein, we have provided evidence to show that miR130a is expressed at higher levels in sera of EAC patients compared with healthy individuals, suggesting that it may serve as a potential biomarker of EAC disease. We further demonstrated that serum miR130a expression is elevated in LGD and HGD BE patients compared to individuals with metaplasia. Expression in LGD and HGD patients exceeded 95%. However, high expression in metaplasia patients was <44%. Our data suggested that miR130a may serve as a predictive biomarker for BE neoplastic progression to EAC. These results implicate that our findings are clinically relevant. Circulating serum levels of miR130a may be a sensitive and cost-effective biomarker for early detection of EAC. However, the actual source of circulating miRNA biomarkers in EAC has not been adequately evaluated. This has important implications for further studies to elucidate the role and origins of circulating miRNAs to allow early detection of EAC and should be validated by other research groups.
Acknowledgment
This work was supported by the National Natural Science Foundation of China (grant U1204821).
Disclosure
The authors report no conflicts of interest in this work.
References
Shridhar R, Hoffe SE, Meredith KL, Fontaine J, Fulp WJ, Almhanna K. Is it time for quality time? Intensive medical oncology follow-up during treatment for esophageal adenocarcinoma. J Clin Oncol. 2014;32(30 Suppl):29. | ||
Siegel RL, Miller KD, Jemal A, Statistics C. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. | ||
Ashktorab H, Kupfer SS, Brim H, Carethers JM. Racial Disparity in Gastrointestinal Cancer Risk. Gastroenterology. 2017;153(4):910–923. | ||
Kanarek N, Hooker C, Rudin CM, Brock M. Esophageal cancer gender disparity. J Clin Oncol. 2012;30(30 Suppl):47. | ||
Hs Y, Tewari M, Pritchard CC. Alteration of candidate circulating microRNA cancer biomarkers by blood sample processing. J Clin Oncol. 2012;30(30 Suppl):8. | ||
Jacobsen A, Mikkilineni N, Fiegoli B, et al. MicroRNA as novel blood-based biomarkers in clear cell renal cell carcinoma. J Clin Oncol. 2013;31(6 Suppl):375. | ||
Antón Aparicio LM, Medina V, Valladares Ayerbes M, et al. Circulating microRNAs as potential biomarkers in patients with renal tumors. J Clin Oncol. 2012;30(5 Suppl):405. | ||
Peltier HJ, Latham GJ. Normalization of microRNA expression levels in quantitative RT-PCR assays: identification of suitable reference RNA targets in normal and cancerous human solid tissues. RNA. 2008;14(5):844–852. | ||
Qi R, Weiland M, Gao XH, Zhou L, Mi QS. Identification of endogenous normalizers for serum microRNAs by microarray profiling: U6 small nuclear RNA is not a reliable normalizer. Hepatology. 2012;55(5):1640–1642. | ||
Coleman HG, Xie SH, Lagergren J. The Epidemiology of Esophageal Adenocarcinoma. Gastroenterology. 2018;154(2):390–405. | ||
Yuan Z, Baker K, Redman MW, et al. Dynamic plasma microRNAs are biomarkers for prognosis and early detection of recurrence in colorectal cancer. Br J Cancer. 2017;117(8):1202–1210. | ||
Schwarzenbach H. Clinical Relevance of Circulating, Cell-Free and Exosomal microRNAs in Plasma and Serum of Breast Cancer Patients. Oncol Res Treat. 2017;40(7–8):423–429. | ||
Wang H, Huo X, Yang XR, et al. STAT3-mediated upregulation of lncRNA HOXD-AS1 as a ceRNA facilitates liver cancer metastasis by regulating SOX4. Mol Cancer. 2017;16(1):136. | ||
Liu Y, Li Y, Wang R, et al. MiR-130a-3p regulates cell migration and invasion via inhibition of Smad4 in gemcitabine resistant hepatoma cells. J Exp Clin Cancer Res. 2016;35:19. | ||
Wang Y, Zhang X, Tang W, et al. miR-130a upregulates mTOR pathway by targeting TSC1 and is transactivated by NF-kappaB in high-grade serous ovarian carcinoma. 2017;24(12):2089–2100. | ||
Lee SH, Jung YD, Choi YS, Lee YM. Targeting of RUNX3 by miR-130a and miR-495 cooperatively increases cell proliferation and tumor angiogenesis in gastric cancer cells. Oncotarget. 2015;6(32):33269–33278. | ||
Wu Q, Guo L, Jiang F, Li L, Li Z, Chen F. Analysis of the miRNA-mRNA-lncRNA networks in ER+ and ER− breast cancer cell lines. J Cell Mol Med. 2015;19(12):2874–2887. | ||
He L, Wang HY, Zhang L, et al. Prognostic significance of low DICER expression regulated by miR-130a in cervical cancer. Cell Death Dis. 2014;5:e1205. | ||
Dong P, Karaayvaz M, Jia N, et al. Mutant p53 gain-of-function induces epithelial-mesenchymal transition through modulation of the miR-130b-ZEB1 axis. Oncogene. 2013;32(27):3286–3295. | ||
Egawa H, Jingushi K, Hirono T, et al. The miR-130 family promotes cell migration and invasion in bladder cancer through FAK and Akt phosphorylation by regulating PTEN. Sci Rep. 2016;6:20574. | ||
Hamilton MP, Rajapakshe K, Hartig SM, et al. Identification of a pan-cancer oncogenic microRNA superfamily anchored by a central core seed motif. Nat Commun. 2013;4:2730. | ||
Shoji H, Isomoto H, Yoshida A, et al. MicroRNA-130a is highly expressed in the esophageal mucosa of achalasia patients. Exp Ther Med. 2017;14(2):898–904. | ||
Liu SG, Qin XG, Zhao BS, et al. Differential expression of miRNAs in esophageal cancer tissue. Oncol Lett. 2013;5(5):1639–1642. | ||
Cappellesso R, Galasso M, Nicolè L, Dabrilli P, Volinia S, Fassina A. miR-130A as a diagnostic marker to differentiate malignant mesothelioma from lung adenocarcinoma in pleural effusion cytology. Cancer Cytopathol. 2017;125(8):635–643. | ||
Garzon R, Calin GA, Croce CM. MicroRNAs in Cancer. Annu Rev Med. 2009;60:167–179. | ||
Ali HEA, Abdel Hameed R, Effat H, et al. Circulating microRNAs panel as a diagnostic tool for discrimination of HCV-associated hepatocellular carcinoma. Clin Res Hepatol Gastroenterol. 2017;41(4):e51–e62. | ||
Sierzega M, Kaczor M, Kolodziejczyk P, Kulig J, Sanak M, Richter P. Evaluation of serum microRNA biomarkers for gastric cancer based on blood and tissue pools profiling: the importance of miR-21 and miR-331. Br J Cancer. 2017;117(2):266–273. | ||
Wang C, Guan S, Liu F, et al. Prognostic and diagnostic potential of miR-146a in oesophageal squamous cell carcinoma. Br J Cancer. 2016;114(3):290–297. | ||
Sun J, Song K, Feng X, Gao S. MicroRNA-367 is a potential diagnostic biomarker for patients with esophageal squamous cell carcinoma. Biochem Biophys Res Commun. 2016;473(2):363–369. | ||
Chiam K, Wang T, Watson DI, et al. Circulating Serum Exosomal miRNAs as Potential Biomarkers for Esophageal Adenocarcinoma. J Gastrointest Surg. 2015;19(7):1208–1215. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.