Back to Journals » OncoTargets and Therapy » Volume 15
Upregulated Ubiquitin D is a Favorable Prognostic Indicator for Rectal Cancer Patients Undergoing Preoperative Concurrent Chemoradiotherapy
Authors Chou CL, Chen TJ, Li WS, Lee SW, Yang CC, Tian YF, Lin CY, He HL, Wu HC, Shiue YL, Li CF, Kuo YH
Received 28 June 2022
Accepted for publication 23 September 2022
Published 11 October 2022 Volume 2022:15 Pages 1171—1181
DOI https://doi.org/10.2147/OTT.S378666
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Geoffrey Pietersz
Chia-Lin Chou,1,2 Tzu-Ju Chen,1,3– 5 Wan-Shan Li,1,3,5 Sung-Wei Lee,6 Ching-Chieh Yang,7,8 Yu-Feng Tian,2 Cheng-Yi Lin,9 Hong-Lin He,10 Hung-Chang Wu,8,11 Yow-Ling Shiue,5,12 Chien-Feng Li,12– 14 Yu-Hsuan Kuo5,8,11,12
1Department of Medical Technology, Chung Hwa University of Medical Technology, Tainan, 717, Taiwan; 2Division of Colon and Rectal Surgery, Department of Surgery, Chi Mei Medical Center, Tainan, 710, Taiwan; 3Department of Pathology, Chi Mei Medical Center, Tainan, 710, Taiwan; 4Department of Clinical Pathology, Chi Mei Medical Center, Tainan, 710, Taiwan; 5Institute of Biomedical Science, National Sun Yat-Sen University, Kaohsiung, 804, Taiwan; 6Department of Radiation Oncology, Chi Mei Medical Center, Liouying, 736, Taiwan; 7Department of Radiation Oncology, Chi Mei Medical Center, Tainan, 710, Taiwan; 8College of Pharmacy and Science, Chia Nan University, Tainan, Taiwan; 9Division of Gastroenterology and Hepatology, Department of Internal Medicine, Chi Mei Medical Center, Tainan, 710, Taiwan; 10Department of Pathology, E-DA Hospital & E-DA Cancer Hospital, I-Shou University, Kaohsiung, 82445, Taiwan; 11Division of Hematology and Oncology, Department of Internal Medicine, Chi-Mei Medical Center, Tainan, Taiwan; 12Institute of Precision Medicine, National Sun Yat-Sen University, Kaohsiung, 804, Taiwan; 13Department of Medical Research, Chi Mei Medical Center, Tainan, 710, Taiwan; 14National Institute of Cancer Research, National Health Research Institutes, Miaoli County, 35053, Taiwan
Correspondence: Yu-Hsuan Kuo; Chien-Feng Li, No. 901, Zhonghua Road Yongkang Dist, Tainan City, Taiwan, Tel +886-6-2812811, Fax +886-6-2510218 ; Fax +886-6-2510218, Email [email protected]; [email protected]
Purpose: For locally advanced rectal cancer, neoadjuvant concurrent chemoradiotherapy (CCRT) allows tumor downstaging and makes curative radical proctectomy possible. However, we lack a genetic biomarker to predict cancer prognosis or treatment response. We investigated the association between ubiquitin D (UBD) expression and clinical outcomes in rectal cancer patients receiving CCRT.
Patients and Methods: We analyzed the genes associated with the protein modification process (GO:0036211) and identified the UBD gene as the most relevant among the top 7 differentially expressed genes associated with CCRT resistance. We collected tissue specimens from 172 rectal cancer patients who had received CCRT followed by a curative proctectomy. We examine the relationship between UBD expression and patient characteristics, pathological findings, and patient survival, such as metastasis-free survival (MeFS) and disease-specific survival.
Results: Upregulated UBD expression was associated with lower pre-CCRT tumor T stage (P = 0.009), lower post-CCRT tumor T stage (P < 0.001), lower post-CCRT nodal stage (P < 0.001), less vascular invasion (P = 0.015), and better tumor regression (P < 0.001). Using univariate analysis, we found that high UBD expression was correlated with better disease-free survival (DFS) (P < 0.0001), local recurrence-free survival (LRFS) (P < 0.0001) and MeFS (P < 0.0001). Moreover, multivariate analysis demonstrated that high UBD expression was associated with superior DFS (P < 0.001), LRFS (P = 0.01), and MeFS (P = 0.004).
Conclusion: UBD upregulation was linked to better clinical prognosis, favorable pathological features, and good treatment response in rectal cancer patients undergoing CCRT. These results suggest UBD is a biomarker for rectal cancer.
Keywords: ubiquitin D, UBD, FAT10, rectal cancer, concurrent chemoradiotherapy
Introduction
Colorectal cancer (CRC) is a common disease in the modern world. Although CRC mortality has been progressively declining since 1990,1 it still remains the third most common cause of cancer-related death in the United States in women and the second leading cause of death in men. Rectal cancer accounting for one-third of these cases. In contrast to these trends, the incidence of CRC in men and women under the age of 50 climbed at a rate of 2.1% per year from 1992 to 2012, and has continued to rise since.2 These increases are driven predominantly by left-sided cancers in general and rectal cancer in particular (3.9% per year).3 The Taiwan Cancer Registry shows that male young-onset rectal cancer incidence rates increased from 4.0 per 100,000 in 1995 to 8.3 per 100,000 in 2014. Female young-onset rectal cancer incidence also increased significantly from 3.8 per 100,000 to 6.4 per 100,000 over the same time period.4
Although surgical resection is the cornerstone of curative therapy for patients with potentially resectable rectal cancer, radiotherapy with concurrent chemotherapy (termed concurrent chemoradiotherapy [CCRT]) has emerged as an important component of curative therapy for transmural or node-positive rectal cancers because local recurrences are more common than with colon primaries. Neoadjuvant CCRT before surgery allows tumor downstaging and increases the rate of sphincter conservation. However, only approximately 20% of the patients can achieve pathological complete response (pCR).5 We had known that patients who achieve pCR have better outcomes and less recurrence of their cancer. Moreover, considering the declining age group of patients with rectal cancer, maintaining the long-term quality of life of rectal cancer patients after tumor resection has become very important and urgent. A biomarker predicting pCR could be useful in treatment decisions.
Ubiquitination is a kind of posttranslational modification involved in the regulation of many biological processes, including the cell cycle, differentiation, transcription regulation, signal transmission, and damage repair.6 Ubiquitin D (UBD), also known as Human HLA-F adjacent transcript locus 10 (FAT10), is a protein containing 165 amino acid residues. It belongs to a class of ubiquitin-like proteins and is a member of the ubiquitin-like protein family, whose protein sequence and three-dimensional core structure resembles ubiquitin (UB).7 Binding of the UBD molecule to its target protein will result in the protein degrading due to the proteasome.8 Deregulation of UBD may result in aberrant modifications in apoptosis, cell division, or chromosomal instability, all of which are linked to cancer.9 Increased studies on UBD in malignancies have emerged in recent years, revealing a link between increased UBD expression and disease development in several tumors,10,11 such as hepatocellular carcinoma,12,13 colon cancer,14 gastric cancer,15 oral squamous cell carcinoma,16 breast cancer,17 and glioma.18 However, the role of UBD in rectal cancer had never been explored. This study aimed to investigate the associations between UBD expression and clinical outcomes in nonmetastatic rectal cancer patients receiving neoadjuvant CCRT.
Materials and Methods
Data Mining of a Public Transcriptome Dataset
A public rectal cancer dataset (GSE35452) with 46 patients receiving CCRT followed by curative resection was used for transcriptome profiling to determine the efficacy of preoperative CCRT. Biopsy specimens were taken during a colonoscopy prior to CCRT in this dataset. The raw CEL files from the Affymetrix Human Genome U133 Plus 2.0 microarray platform were loaded into the Nexus Expression 3 statistical program (BioDiscovery, El Segundo, CA, USA) to evaluate all probes without filtering in order to computerize expression levels. The tumor specimens were divided into two groups by tumor regression grade (TRG) based on their response to neoadjuvant CCRT: “responders” and “non-responders”. The statistical significance of each transcript was tested by comparing responders to non-responders. We selected genes with a p-value less than 0.0001 and absolute expression fold change >1 log2 ratio for further analysis among differentially expressed genes associated with the protein modification process (GO:0036211). From the same database, we profiled the expression level of all genes and probes and identified 200 top-ranking genes whose expression is positively and negatively correlated with UBD. These genes of selection were also submitted to perform enrichment ontology analysis on the GENEONTOLOGY website (http://geneontology.org/) and assess their molecular function.
Patient Eligibility and Enrollment
This study was approved by the Institutional Review Board of Chi Mei Medical Center (10302014). All patients had signed the informed consent. Our study complies with the Declaration of Helsinki. We collected formalin-fixed, paraffin-embedded Pre-Tx rectal tissue specimens from 172 rectal cancer patients. Imaging tests were used to determine the initial clinical stage, and those who had been diagnosed with distant metastases were ruled out. All patients received 5-fluorouracil-based chemotherapy as well as radiation (45–50 Gy) in 25 fractions over a 5-week period, followed by a curative proctectomy. Those with a nodal status higher than N1 or a pre- or post-CCRT tumor status greater than T3 received adjuvant chemotherapy. After diagnosis, all patients were continuously observed until death or the last follow-up.
Histopathological and Immunohistochemical Evaluations
Two independent pathologists, blinded to the patients’ clinical information, reviewed all tumor specimens to achieve more objective results. The seventh American Joint Committee on Cancer TNM staging system was used to identify the T and N stages. The tumor regression grade (TRG), which is predictive of the tumor response to CCRT, was evaluated in concordance with the description by Dworak et al.19 Immunohistochemical study was performed to assess the expression of UBD. In brief, tissue sections from Pre-Tx rectal tumor biopsies were cut from paraffin-embedded tissue blocks at 3 mm thickness onto precoated glass slides. Slides were then deparaffinized with xylene, rehydrated with ethanol and heated for 7 min by microwave for antigen retrieval in a 10-mM citrate buffer (pH 6). Endogenous peroxidase was blocked by using 3% H2O2. Slides were then washed with Tris-buffered saline for 15 min and then incubated with a primary poly-clonal antibody against UBD (Cat No. NBP2-81752, Novus Biologicals, dilution 1:100). Antibodies were then detected using a DAKO ChemMate EnVision Kit (K5001, Carpinteria, CA, USA). Cell blocks from cell lines known to express UBD were used as positive controls. Sections were processed without the primary anti-UBD antibody as negative controls. The H-score was applied to evaluate UBD immunoreactivity and was quantified with the following equation:
H-score = SPi (i + 1)
where Pi is the percentage of stained tumor cells for each intensity, which ranges from 0% to 100%, and i is the staining intensity (0 to 3+). The H-score, which ranged from 100 to 400, was calculated using a combination of the intensity and proportion of positively stained tumor cells. High UBD expression was defined as H-scores more than or equal to the median of all scored instances.
Statistical Analysis
The associations between clinicopathological characteristics and UBD expression were measured using the chi-square (X2) test. The Kaplan–Meier method was used to plot survival curves, and the Log rank test was employed to quantify and compare the time from surgery to death (or last seen alive) or recurrence (or last seen relapse-free). The Cox proportional hazards model for multivariate analysis includes those parameters clinically significant in the univariate analysis. SPSS software version 22.0 (IBM Corporation, Armonk, NY, USA) was used for all statistical analyses, and two-tailed tests with p < 0.05 were considered statistically significant.
Results
UBD is Recognized as the Most Significant Differentially Expressed Gene Connected with the Protein Modification Process
A published rectal cancer transcriptome dataset (GSE35452) was used for data mining to investigate the potential biomarkers of rectal cancer cells sensitive to preoperative CCRT. The dataset included 46 patients who had neoadjuvant CCRT followed by standardized curative resection. Twenty-four patients (52.2%) were classed as responders, whereas 22 patients (47.8%) were classified as non-responders, based on their response to CCRT. We found seven transcripts focusing on the protein modification process (GO:0036211) (Table 1 and Figure 1). UBD was chosen for further investigation because its expression was significantly lower in CCRT non-responders (p < 0.0001). This discovery prompted us to look into the clinical significance of UBD expression in rectal cancer.
 |
Table 1 Summary of Differentially Expressed Genes Associated with Protein Modification Process (GO:0036211) in Relation to Response to CCRT in Rectal Carcinoma |
Clinicopathological Features of Patients with Rectal Carcinoma in Our Cohort
We obtained medical records from a total of 172 rectal cancer patients (Table 2). The median age was 63. Most patients were male (n = 108, 62.8%) and less than 70 years old (n = 106, 61.6%). Fifty-three percent of the patients (n = 91) had T3-T4 lesions; 27% of the patients (n = 47) had lymph node metastases before CCRT. After CCRT, 50% of the patients (n = 86) had T3-T4 lesions; 28% of the patients (n = 49) had lymph node metastases. The surgical specimens revealed vascular invasion in 8% of the patients (n =15) and perineural invasion in 3% of the patients (n=5). Tumor regression grade, which was used to evaluate treatment response to CCRT, varied from grade 0 to 1 (n = 37, 22%), grade 2 to 3 (n = 118, 69%) and grade 4 (n = 17, 10%).
 |
Table 2 Associations and Comparisons Between UBD Expression and Clinicopathological Factors in 172 Rectal Cancer Patients Receiving Neoadjuvant CCRT |
Correlation of UBD Immunoexpression with Clinicopathological Parameters
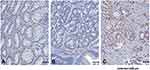
The immunohistochemical stain showed UBD immunoactivity was significantly higher in CCRT-responders than in CCRT non-responders (Figure 2). Table 2 shows the correlation between UBD immunoexpression and clinicopathological parameters. Upregulated UBD expression was associated with lower pre-CCRT tumor T stage (P = 0.009), lower post-CCRT tumor T stage (P < 0.001), lower post-CCRT nodal stage (P < 0.001), less vascular invasion (P = 0.015), and better tumor regression (P < 0.001).
Survival Analyses and Clinical Implications of UBD Expression
Based on univariate analysis, high UBD expression was correlated with better disease-free survival (DFS) (P < 0.0001), local recurrence-free survival (LRFS) (P < 0.0001), and metastases-free survival (MeFS) (P < 0.0001). (Table 3 and Figure 3) Lower post-CCRT tumor T stage and better tumor regression grade were associated with better DFS, LRFS, and MeFS (all P < 0.009). Lower pre-CCRT tumor T stage was associated with better DFS (P = 0.086). Pre-CCRT nodal metastases correlated with worse DFS (P = 0.001) and worse LRFS (P = 0.007). Vascular invasion was correlated with worse DFS (P = 0.0029) and LRFS (P = 0.0028). Based on multivariate analysis (Table 4), high UBD expression was associated with superior DFS (P < 0.001), LRFS (P = 0.01), and MeFS (P = 0.004). Worse tumor regression grade and pre-CCRT nodal metastases were correlated with inferior DFS (P = 0.078 and P = 0.023, respectively).
 |
Table 3 Univariate Log Rank Analysis for Important Clinicopathological Variables and UBD Expression |
 |
Table 4 Multivariate Analysis for Important Clinicopathological Variables and UBD Expression |
 |
Figure 3 Kaplan–Meier analysis showed UBD overexpression was associated with superior disease-specific survival (A), local recurrence-free survival (B) and metastasis-free survival (C). |
UBD Positively and Negatively Correlated Genes
The top 200 genes whose expression is most positively correlated (Table S1) or negatively correlated (Table S2) with UBD in rectal adenocarcinoma from the same transcriptome database (GSE35452) were examined to further predict the biological activities of UBD-interacting networks. The top 10 co-upregulated genes are CXCL11, COL12A1, CTHRC1, COL13A1, PHTF2, EGFL6, CD47, PPT1, BCAT1, and LARP6. The top 10 co-downregulated genes are MOGAT2, STARD5, CD300LG, PTPN18, RASAL1, FAM22A, DNAJC4, GNG13, PHKB, and FAM98C. These genes of selection were also submitted to perform enrichment ontology analysis of their molecular function. We identified the top 200 positively correlated genes are enriched to several pathways affecting molecular functions. Platelet-derived growth factor binding (GO:0048407, fold enrichment:76.4), extracellular matrix (ECM) structural constituent conferring tensile strength (GO:0030020, fold enrichment: 38.4), and IgG binding (GO:0019864, fold enrichment: 38.2) are the top three GO terms associated with UBD upregulation (Table S3A). Meanwhile, enoyl-CoA hydratase activity (GO:0004300, fold enrichment: 41.3) is the only GO term for those molecular functions negatively associated with UBD (Table S3B).
Discussion
UB has been established as a versatile molecule that is involved in both proteolytic and non-proteolytic activities. Ubiquitination is a multi-step process that involves an E1 ubiquitin-activating enzyme, E2 ubiquitin-conjugating enzymes, and E3 ubiquitin ligases to attach ubiquitin to a target protein.20 At least four ubiquitin moieties linked by lysine target a wide range of cytosolic and nucleoplasmic proteins for proteasomal destruction. Polyubiquitylated substrates are transported to the proteasome by various ubiquitin receptors, where ubiquitins are recycled and broken into tiny peptides by proteasomic proteases. UB governs several physiological activities indirectly through proteolysis, including cell cycle progression, differentiation, and development, cell responses to extracellular effectors and stress, induction of inflammatory and immunological responses, and DNA damage repair or tolerance.21,22 The UBD, also called FAT10, is a ubiquitin-like protein and was first discovered in 1996.23 While ubiquitin is recycled from degraded target proteins, UBD degrades with its target, resulting in a relatively short half-life.24,25 It is a protein belonging to the immune system, which can be strongly upregulated by pro-inflammatory cytokines.10 The description of the role of UBD in cancer in the published literature is contradictory. Several clinical studies have shown that UBD could confer malignant characteristics to non-tumorigenic cells and enhance the malignant-related characteristics of cancer cells.26,27 Tumor UBD expression shows some tissue specificity, with transcriptional upregulation observed in liver, uterine cervix, ovarian, pancreatic, gastric and small intestine adenocarcinomas, but not in thyroid, prostate or kidney cancers.28,29 Ji et al demonstrated that overexpression of UBD in gastric cancer is correlated with metastasis and tumor staging, and both UBD mRNA and protein levels were identified as independent prognostic factors for the disease.15 Increased UBD has been also linked to mutant p53 expression, which could activate UBD expression and aid gastric cancer growth indirectly.30 There are also several studies exploring the role of UBD in colon cancer. Yan et al31 demonstrated that increased cytoplasmic UBD immunohistochemical staining was significantly associated with depth of cancer invasion, lymph node metastasis, distant metastasis, tumor histologic grade, advanced clinical stage, and Ki-67 proliferative index. After radical surgery for stage II and III colon cancer, individuals with UBD-positive tumors had a considerably higher disease recurrence rate and shorter survival than patients with UBD-negative tumors. These findings suggest that UBD may contribute to colon carcinogenesis and can be used as a new prognostic indicator for predicting recurrence in stage II and III patients following curative surgery. Another study revealed that UBD-positive expression was a significant, independent predictive high-risk factor for overall survival and DFS in stage IIB–IIC colon cancer patients treated with 5-fluorouracil-based adjuvant chemotherapy.32 However, our study yielded different results. Our study revealed that high UBD expression correlated to better DFS, LRFS, and MeFS in rectal cancer patients receiving CCRT followed by curative resection. The differences between this study and others are that our patients have received radiation therapy while others have not. This is the first study to explore the relationship between UBD and chemoradiotherapy. In a study presented by Yu Wang et al,33 they found that UBD expression was significantly positively correlated with all analyzed immune cells in skin cutaneous melanoma (B cell, CD8+T cell, CD4+T cell, macrophage, neutrophil, and dendritic cell). In this study, high UBD expression predicted better outcomes. Since several studies proposed that tumor-infiltrating immune cells in the tumor microenvironment may improve the response to neoadjuvant CCRT and improve the recurrence-free survival in rectal cancer patients,34–37 the relationships between UBD, tumor-infiltration immune cells and response to neoadjuavant CCRT are worth further exploration. In fact, the conclusion that UBD promotes carcinogenesis is still open to question. Lukasiak et al38 suggested that overexpression of UBD in colon cancer is a consequence of a proinflammatory immune response and so there is no reason to propose that UBD contributes to carcinogenesis. In fact, UBD failed to exert transforming activity in their study. Therefore, it is difficult to conclude whether UBD overexpression in more advanced cancer is the cause or the effect.
In our previous study, we use the same dataset to evaluate the genes associated with maintenance of gastrointestinal epithelium (GO: 0030277) and validate MUC2 as a significant survival biomarker by the same patient cohort.39 Based on this, we believe the pathogenesis of rectal cancer is complex and all biomarkers are different in nature and biology. Searching other possible molecular biomarkers is mandatory. In this study, we use the same transcriptome dataset, where genes related to the protein modification process (GO:0036211) were assessed to better understand the role of UBD in rectal cancer.
To further predict the molecular functions of UBD, the top 200 genes co-upregulated (Supplementary Table 1) or co-downregulated (Supplementary Table 2) with UBD in colorectal adenocarcinoma from the transcriptomic database were evaluated. We found that the genes negatively associated with UBD expression are significantly enriched to enoyl-CoA hydratase activity (GO:0004300, Fold enrichment:41.3) (Supplementary Table 3A). Moreover, among the top 10 pathways to which the positively correlated genes are enriched (Supplementary Table 3B), ECM-related pathways accounted for four places: ECM structural constituent conferring tensile strength (GO:0030020, fold enrichment: 38.4), ECM structural constituent (GO:0005201, fold enrichment: 19.4), ECM structural constituent conferring compression resistance (GO:0030021, fold enrichment: 19.1), and ECM binding (GO:0050840, fold enrichment: 13.1). The ECM proteins confer distinct biochemical and biophysical properties that influence cell phenotype. The ECM’s composition and organization are spatiotemporally regulated to control cell behavior and differentiation, but dysregulation of ECM dynamics may lead to the development of cancer.40 ECM does not only influence malignancy and growth of the tumor but also its response to therapy.41 The exact mechanisms to link UBD and the ECM-related pathway need further exploration and analysis. There are still several limitations to the existing studies. First, this is a single institution’s retrospective analysis of rectal cancer patients who received preoperative CCRT. Second, this study lacks a validation cohort. Third, the role of UBD function in cancer progression is generally indirect. Fourth, if the ideal cutoff for high UBD expression is not established, clinical assessment will be useless. And finally, there were not enough samples. Therefore, more multicenter investigations should be conducted to confirm the significance of UBD expression.
Conclusion
In conclusion, our study demonstrated that high UBD expression has been linked to less advanced rectal cancer characteristics and is a unique predictive indicator of better patient outcomes for rectal cancer patients undergoing neoadjuvant CCRT.
Abbreviations
CCRT, concurrent chemoradiotherapy; CRC, colorectal cancer; DFS, disease-free survival; ECM, extracellular matrix; FAT10, human HLA-F adjacent transcript locus 10; LRFS, local recurrence-free survival; MeFS, metastasis-free survival; pCR, pathological complete response; UB, ubiquitin; UBD, ubiquitin D.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Islami F, Ward EM, Sung H, et al. Annual report to the nation on the status of cancer, part 1: National cancer statistics. J Natl Cancer Inst. 2021;113(12):1648–1669. doi:10.1093/jnci/djab131
2. Stoffel EM, Murphy CC. Epidemiology and mechanisms of the increasing incidence of colon and rectal cancers in young adults. Gastroenterology. 2020;158(2):341–353. doi:10.1053/j.gastro.2019.07.055
3. Ahnen DJ, Wade SW, Jones WF, et al. The increasing incidence of young-onset colorectal cancer: a call to action. Mayo Clin Proc. 2014;89(2):216–224. doi:10.1016/j.mayocp.2013.09.006
4. Sung JJY, Chiu HM, Jung KW, et al. Increasing trend in young-onset colorectal cancer in Asia: more cancers in men and more rectal cancers. Am J Gastroenterol. 2019;114(2):322–329. doi:10.14309/ajg.0000000000000133
5. Martin ST, Heneghan HM, Winter DC. Systematic review and meta-analysis of outcomes following pathological complete response to neoadjuvant chemoradiotherapy for rectal cancer. Br J Surg. 2012;99(7):918–928. doi:10.1002/bjs.8702
6. Dikic I, Wakatsuki S, Walters KJ. Ubiquitin-binding domains — from structures to functions. Nat Rev Mol Cell Biol. 2009;10(10):659–671. doi:10.1038/nrm2767
7. Cappadocia L, Lima CD. Ubiquitin-like protein conjugation: structures, chemistry, and mechanism. Chem Rev. 2018;118(3):889–918. doi:10.1021/acs.chemrev.6b00737
8. Hofmann K. Ubiquitin-binding domains and their role in the DNA damage response. DNA Repair (Amst). 2009;8(4):544–556. doi:10.1016/j.dnarep.2009.01.003
9. Adler M, Muller K, Rached E, Dekant W, Mally A. Modulation of key regulators of mitosis linked to chromosomal instability is an early event in ochratoxin A carcinogenicity. Carcinogenesis. 2009;30(4):711–719. doi:10.1093/carcin/bgp049
10. Aichem A, Groettrup M. The ubiquitin-like modifier FAT10 in cancer development. Int J Biochem Cell Biol. 2016;79:451–461. doi:10.1016/j.biocel.2016.07.001
11. Xiang S, Shao X, Cao J, Yang B, He Q, Ying M. FAT10: function and relationship with Cancer. Curr Mol Pharmacol. 2020;13(3):182–191. doi:10.2174/1874467212666191113130312
12. Luo C, Xiong H, Chen L, et al. GRP78 Promotes hepatocellular carcinoma proliferation by increasing FAT10 expression through the NF-kappaB pathway. Exp Cell Res. 2018;365(1):1–11. doi:10.1016/j.yexcr.2018.02.007
13. Yuan R, Wang K, Hu J, et al. Ubiquitin-like protein FAT10 promotes the invasion and metastasis of hepatocellular carcinoma by modifying beta-catenin degradation. Cancer Res. 2014;74(18):5287–5300. doi:10.1158/0008-5472.CAN-14-0284
14. Qing X, French BA, Oliva J, French SW. Increased expression of FAT10 in colon benign, premalignant and malignant epithelial neoplasms. Exp Mol Pathol. 2011;90(1):51–54. doi:10.1016/j.yexmp.2010.09.005
15. Ji F, Jin X, Jiao CH, Xu QW, Wang ZW, Chen YL. FAT10 level in human gastric cancer and its relation with mutant p53 level, lymph node metastasis and TNM staging. World J Gastroenterol. 2009;15(18):2228–2233. doi:10.3748/wjg.15.2228
16. Song A, Wang Y, Jiang F, et al. Ubiquitin D Promotes Progression of Oral Squamous Cell Carcinoma via NF-Kappa B Signaling. Mol Cells. 2021;44(7):468–480. doi:10.14348/molcells.2021.2229
17. Han T, Liu Z, Li H, et al. High expression of UBD correlates with epirubicin resistance and indicates poor prognosis in triple-negative breast cancer. Onco Targets Ther. 2015;8:1643–1649. doi:10.2147/OTT.S81214
18. Yuan J, Tu Y, Mao X, et al. Increased expression of FAT10 is correlated with progression and prognosis of human glioma. Pathol Oncol Res. 2012;18(4):833–839. doi:10.1007/s12253-012-9511-2
19. Bozzetti F, Andreola S, Bertario L. Pathological features of rectal cancer after preoperative radiochemotherapy. Int J Colorectal Dis. 1998;13(1):54–55. doi:10.1007/s003840050134
20. Hoeller D, Dikic I. Targeting the ubiquitin system in cancer therapy. Nature. 2009;458(7237):438–444. doi:10.1038/nature07960
21. Ciechanover A, Orian A, Schwartz AL. Ubiquitin-mediated proteolysis: biological regulation via destruction. Bioessays. 2000;22(5):442–451. doi:10.1002/(SICI)1521-1878(200005)22:5<442::AID-BIES6>3.0.CO;2-Q
22. Elsasser S, Finley D. Delivery of ubiquitinated substrates to protein-unfolding machines. Nat Cell Biol. 2005;7(8):742–749. doi:10.1038/ncb0805-742
23. Fan W, Cai W, Parimoo S, Schwarz DC, Lennon GG, Weissman SM. Identification of seven new human MHC class I region genes around the HLA-F locus. Immunogenetics. 1996;44(2):97–103. doi:10.1007/BF02660056
24. Hipp MS, Kalveram B, Raasi S, Groettrup M, Schmidtke G. FAT10, a ubiquitin-independent signal for proteasomal degradation. Mol Cell Biol. 2005;25(9):3483–3491. doi:10.1128/MCB.25.9.3483-3491.2005
25. Liu S, Jin Y, Zhang D, Wang J, Wang G, Lee CGL. Investigating the Promoter of FAT10 Gene in HCC Patients. Genes. 2018;9(7):319. doi:10.3390/genes9070319
26. Gao Y, Theng SS, Zhuo J, Teo WB, Ren J, Lee CG. FAT10, an ubiquitin-like protein, confers malignant properties in non-tumorigenic and tumorigenic cells. Carcinogenesis. 2014;35(4):923–934. doi:10.1093/carcin/bgt407
27. Zhang K, Chen L, Zhang Z, Cao J, He L, Li L. Ubiquitin-like protein FAT10: a potential cardioprotective factor and novel therapeutic target in cancer. Clin Chim Acta. 2020;510:802–811. doi:10.1016/j.cca.2020.09.016
28. Lee CG, Ren J, Cheong IS, et al. Expression of the FAT10 gene is highly upregulated in hepatocellular carcinoma and other gastrointestinal and gynecological cancers. Oncogene. 2003;22(17):2592–2603. doi:10.1038/sj.onc.1206337
29. Lukasiak S, Breuhahn K, Schiller C, Schmidtke G, Groettrup M. Quantitative analysis of gene expression relative to 18S rRNA in carcinoma samples using the LightCycler instrument and a SYBR GreenI-based assay: determining FAT10 mRNA levels in hepatocellular carcinoma. Methods Mol Biol. 2008;429:59–72. doi:10.1007/978-1-60327-040-3_5
30. Zhang DW, Jeang KT, Lee CG. p53 negatively regulates the expression of FAT10, a gene upregulated in various cancers. Oncogene. 2006;25(16):2318–2327. doi:10.1038/sj.onc.1209220
31. Yan DW, Li DW, Yang YX, et al. Ubiquitin D is correlated with colon cancer progression and predicts recurrence for stage II-III disease after curative surgery. Br J Cancer. 2010;103(7):961–969. doi:10.1038/sj.bjc.6605870
32. Zhao S, Jiang T, Tang H, et al. Ubiquitin D is an independent prognostic marker for survival in stage IIB-IIC colon cancer patients treated with 5-fluorouracil-based adjuvant chemotherapy. J Gastroenterol Hepatol. 2015;30(4):680–688. doi:10.1111/jgh.12784
33. Wang Y, Zhang H. FAT10 is a prognostic biomarker and correlated with immune infiltrates in skin cutaneous melanoma. Front Mol Biosci. 2022;9:805887. doi:10.3389/fmolb.2022.805887
34. Yang Y, Tian W, Su L, et al. Tumor-infiltrating cytotoxic t cells and tumor-associated macrophages correlate with the outcomes of neoadjuvant chemoradiotherapy for locally advanced rectal cancer. Front Oncol. 2021;11:743540. doi:10.3389/fonc.2021.743540
35. Chen CC, Wu ML, Huang KC, Huang IP, Chung YL. The effects of neoadjuvant treatment on the tumor microenvironment in rectal cancer: implications for immune activation and therapy response. Clin Colorectal Cancer. 2020;19(4):e164–e180. doi:10.1016/j.clcc.2020.04.002
36. Mirjolet C, Charon-Barra C, Ladoire S, et al. Tumor lymphocyte immune response to preoperative radiotherapy in locally advanced rectal cancer: the LYMPHOREC study. Oncoimmunology. 2018;7(3):e1396402. doi:10.1080/2162402X.2017.1396402
37. Matsutani S, Shibutani M, Maeda K, et al. Significance of tumor-infiltrating lymphocytes before and after neoadjuvant therapy for rectal cancer. Cancer Sci. 2018;109(4):966–979. doi:10.1111/cas.13542
38. Lukasiak S, Schiller C, Oehlschlaeger P, et al. Proinflammatory cytokines cause FAT10 upregulation in cancers of liver and colon. Oncogene. 2008;27(46):6068–6074. doi:10.1038/onc.2008.201
39. Chou CL, Chen TJ, Tian YF, et al. Upregulated MUC2 Is an unfavorable prognostic indicator for rectal cancer patients undergoing preoperative CCRT. J Clin Med. 2021;10(14):3030. doi:10.3390/jcm10143030
40. Walker C, Mojares E, Del Rio Hernandez A. Role of extracellular matrix in development and cancer progression. Int J Mol Sci. 2018;19(10):3028. doi:10.3390/ijms19103028
41. Henke E, Nandigama R, Ergun S. Extracellular matrix in the tumor microenvironment and its impact on cancer therapy. Front Mol Biosci. 2019;6:160. doi:10.3389/fmolb.2019.00160
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.