Back to Journals » Journal of Hepatocellular Carcinoma » Volume 10
ULK1 Depletion Protects Mice from Diethylnitrosamine-Induced Hepatocarcinogenesis by Promoting Apoptosis and Inhibiting Autophagy
Authors Duan T, Yang X, Kuang J, Sun W, Li J, Ge J, Zhang M, Cai X , Yu P, Yang J , Zhu X
Received 7 December 2022
Accepted for publication 28 January 2023
Published 27 February 2023 Volume 2023:10 Pages 315—325
DOI https://doi.org/10.2147/JHC.S399855
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 5
Editor who approved publication: Dr David Gerber
Ting Duan,1,2,* Xin Yang,3,* Jingyu Kuang,4 Wenjie Sun,3 Jin Li,3 Juan Ge,3 Mohan Zhang,3 Xiaobo Cai,3 Peilin Yu,3 Jun Yang,5 Xinqiang Zhu6
1School of Pharmacy, Hangzhou Normal University, Hangzhou, 311121, People’s Republic of China; 2Key Laboratory of Elemene Class Anti-Cancer Chinese Medicines, Engineering Laboratory of Development and Application of Traditional Chinese Medicines, Collaborative Innovation Center of Traditional Chinese Medicines of Zhejiang Province, School of Pharmacy, Hangzhou Normal University, Hangzhou, 311121, People’s Republic of China; 3Department of Toxicology, Zhejiang University School of Medicine, Hangzhou, Zhejiang, 310058, People’s Republic of China; 4Department of Biology and Chemistry, College of Sciences, National University of Defense Technology, Changsha, Hunan, 410073, People’s Republic of China; 5Department of Nutrition and Toxicology, Hangzhou Normal University School of Public Health, Hangzhou, Zhejiang, 311121, People’s Republic of China; 6Medical Research Center, The Fourth Affiliated Hospital, School of Medicine, Zhejiang University, Yiwu, 322000, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Xinqiang Zhu; Jun Yang, Email [email protected]; [email protected]
Purpose: The uncoordinated-51 like kinase 1 (ULK1) is an important serine/threonine protein kinase involved in autophagy, especially for the initiation stage. Previous studies have shown that ULK1 could be used as a prognostic marker in predicting poor progression-free survival and a therapeutic target for hepatocellular carcinoma (HCC) when treated with sorafenib; however, its role during hepatocarcinogenesis remains to be elucidated.
Methods: CCK8 and colony formation assay were used to detect cell growth ability. Western blotting was performed to determine expression level of protein. Data from public database were downloaded to analyze expression of ULK1 at mRNA level and predict survival time. RNA-seq was conducted to reveal disturbed gene profile orchestrated by ULK1 depletion. A diethylnitrosamine (DEN)-induced HCC mice model was used to uncover the role of ULK1 in hepatocarcinogenesis.
Results: ULK1 was up-regulated in liver cancer tissues and cell lines, and knockdown of ULK1 promoted apoptosis and suppressed proliferation of liver cancer cells. In in vivo experiments, Ulk1 depletion attenuated starvation-induced autophagy in mice liver, reduced diethylnitrosamine (DEN)-induced hepatic tumor number and size, and prevented tumor progression. Further, RNA-seq analysis revealed a close relationship between Ulk1 and immunity with significant changes in gene sets enriched in the interleukin and interferon pathways.
Conclusion: ULK1 deficiency prevented hepatocarcinogenesis and inhibited hepatic tumor growth, and might be a molecular target for the prevention and treatment of HCC.
Keywords: hepatocellular carcinoma, ULK1, RNA-sequencing, apoptosis, autophagy
Introduction
Hepatocellular carcinoma (HCC) accounts for nearly 85% of primary liver cancer, which is the sixth most prevalent and the third leading lethal malignancy.1 Despite increased global vaccine coverage which has substantially decreased hepatitis virus infection, the global burden from HCC remains challenging, probably due to the increasing population with obesity and diabetes mellitus.2 Most HCC cases were diagnosed at advanced stage with limited therapeutic options, and the 5-year survival rate has been very low (~24.3%).3 Thus, it is of great importance to better understand the underlying molecular mechanisms in hepatocarcinogenesis, which will help improve the current strategies for the prevention, early diagnosis, and treatment of HCC. HCC always arises from the sequence of liver injury, chronic inflammation, fibrosis, and cirrhosis, which can be caused by hepatitis virus infection, aflatoxin, alcohol consumption, obesity, and type II diabetes mellitus.4 During these processes, multiple genes/pathways including Ras/PI3K/mTOR, Wnt/β-catenin, and TP53 are involved,5 many of which have been identified and considered as potential therapeutic targets. Nonetheless, more hepatocarcinogenesis-related genes remain to be identified.
The uncoordinated-51 like kinase 1 (ULK1) is the mammalian orthologue of yeast Atg1, a serine/threonine protein kinase that is important for autophagy.6 There are five mammalian ULK homologues (ULK1, ULK2, ULK3, ULK4, and STK36 (serine/threonine kinase 36)), among which ULK1 and ULK2 are reported to be involved in conventional autophagy signaling while ULK3 participates in stress-induced autophagy.7 ULK1 functions in a complex with autophagy-related 13 (ATG13), focal adhesion kinase family-interacting protein of 200 kDa (FIP200), and ATG101 to initiate autophagy in response to upstream signals such as mTORC1 and AMPK.8 Though ULK2 has a higher degree of homology with ULK1 than others, it is ULK1 but not ULK2 that acts as the predominant isoform involved in inducing autophagy.9 Besides the canonical role in autophagy, ULK1 also participates in other physiological processes. For example, ULK1 sustains glucose metabolic fluxes by directly phosphorylating key glycolytic enzymes during deprivation of amino acid and growth factors,10 and promotes cell death via regulating the activity of PARP1 under oxidative stress.11
Accumulating evidences have also uncovered the relationship between ULK1 and cancer; however, the role of ULK1 in cancer remains to be carefully examined as it can either promote or suppress tumor growth depending on the type of cancer investigated. For example, ULK1 was significantly down-regulated in breast cancer,12 and ULK1 inhibited breast cancer metastasis.13 On the other hand, ULK1 inhibition could suppress cell growth in lung cancer, colon cancer, and ovarian cancer.14–16 For HCC, it has been reported that ULK1 was overexpressed in clinical samples, silencing ULK1 inhibited liver cancer cell growth while increasing the therapeutic effects of sorafenib,17 and it was suggested that ULK1 could be used as a potential prognostic biomarker for HCC.18 However, the exact role and function of ULK1 in hepatocarcinogenesis remains to be elucidated.
Therefore, in the current study, we first examined the expression of ULK1 in liver cancer cell lines and human HCC samples, as well as clinical tissues from public databases. Then, using an Ulk1-knockout (Ulk1KO) mouse strain, the development of tumor was evaluated in the diethylnitrosamine (DEN)-induced HCC model. As reported here, we found ULK1 was overexpressed in liver cancer cell lines and clinical samples, and Ulk1 deficiency inhibited DEN-induced hepatocarcinogenesis, probably through the induction of apoptosis along with the inhibition of autophagy.
Materials and Methods
Database Search
A gene expression profile of 424 LIHC (liver hepatocellular carcinoma) patients was downloaded from UCSC Xena datahub (https://gdc-hub.s3.us-east-1.amazonaws.com/download/TCGA-LIHC.htseq_fpkm.tsv.gz). Then, normalized mRNA expression of ULK1 in liver cancer and normal liver tissues was analyzed by GraphPad Prism 7.0 (GraphPad Prism Software, La Jolla, CA). Comparison of ULK1 expression among cancer cell lines was performed using the online tool Cancer Cell Line Encyclopedia (https://sites.broadinstitute.org/ccle/). Tumor tissues from patients diagnosed with HCC accompanied by hepatitis B virus infection and paired adjacent non-tumor tissues were used to detect ULK1 protein expression in liver cancer.
Cell Lines
The human HCC cell lines Huh-7, Hep3B, and HCCLM3, as well as the normal human liver cell line HL7702, were obtained from The Cell Bank of the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). The cells were maintained in DMEM medium (Cienry, China) supplemented with 10% fetal bovine serum (Gibco, USA) at 37°C with 5% CO2 in a humid atmosphere.
Animals and DEN Treatment
The Ulk1KO mice and wild-type C57BL/6J mice were bred by the Laboratory Animal Center, Zhejiang Chinese Medical University and maintained in a specific pathogen-free environment. Animals were housed in individual ventilated cages (no more than five animals per cage) under standard laboratory conditions (temperature 24 ± 0.5°C, relative humidity 55 ± 5%, and a 12-h dark/light cycle).
The Ulk1KO mouse strain was originally generated as previously described19 and was a generous gift from Dr. Toshifumi Tomoda (University of Toronto) by way of Dr. Hanming Shen (National University of Singapore). And the use of Ulk1KO mouse strain was approved by Ethics Committee of Laboratory Animal Care and Welfare, Zhejiang University School of Medicine.
Both Ulk1KO and wild-type male mice were divided randomly into control (n=5 (WT); n=7 (Ulk1KO)), and DEN groups (n=7 (WT); n=12 (Ulk1KO)), respectively. Mice in DEN groups were injected intraperitoneally with 25 mg/kg body weight of DEN (Sigma-Aldrich, Cat# N0756), while mice of control groups received equal volume of saline at the age of 2 weeks and were allowed to grow for 30 weeks. All experiments were performed under the Guide for the Care and Use of Laboratory Animals (The National Academy Press, 2011) and approved by both Ethics Committee of Laboratory Animal Care and Welfare, Zhejiang University School of Medicine and Animal Committee of Zhejiang Chinese Medical University.
Genotyping of Ulk1KO Mice
Mice were genotyped by using tail DNA and PCR as described below: an initial denaturation at 94°C for 3 min; followed by 40 cycles of denaturation at 94°C for 40 s, annealing at 61°C for 40 s, and extension at 72°C for 40 s; ending with a final extension at 72°C for 5 min. A common primer (5’–CCT TCC CAT GCA GGC AAC ATA TAA GC–3’) and a wild-type specific primer (5’–AAG CAC GAC CTG GAG GTG GC–3’) amplified a 500 bp fragment from the wild-type (WT) mice. The common primer (5’–CCT TCC CAT GCA GGC AAC ATA TAA GC–3’) and a mutation-specific primer (5’–AGT TCG AGT TCT CTC GCA AGG AC–3’) amplified a 340 bp fragment from Ulk1KO mice. The PCR products were subjected to agarose gel electrophoresis or Sanger sequencing to identify the genotypes of mice (Figure S1).
Liver Preparation
Mice were killed by anesthesia (pentobarbital sodium, 150 mg/kg (i.p.)) after overnight starvation. Unbroken livers were removed, weighed, and photographed with measuring scale. Sections from left lateral lobe were fixed in 10% formalin for hematoxylin-eosin (HE) staining, and the remaining tissue was snap frozen in liquid nitrogen and then stored at −80°C for further analysis.
For in vivo detection of autophagy in mice liver, WT and Ulk1KO mice were injected intraperitoneally with 50 mg/kg body weight of chloroquine or equal volume of saline, then the mice were subjected to fasting for 24 h. Finally, the mice were killed as mentioned above, and the liver tissues were obtained for Western blot to determine autophagic markers.
Analysis of Liver Tumor
All visible nodules on liver surface were counted for each mouse. Tumor diameters per mouse were measured by vernier caliper or calibrated software according to HE staining. Liver histopathologic lesions were classified according to the standardized and internationally accepted nomenclature for classification of rodent tumors20 by two experienced pathologists independently in a blind fashion.
Cell Transfection
The siRNA oligonucleotides were synthesized by Genepharma (Shanghai, China): siULK1-1: 5’-UCA UCA CCC UUU CCU CGA UTT-3’; siULK1-2: 5’-CCU GUG ACA CAG ACG ACU UTT-3’. Lipofectamine RNAi MAX reagent (Invitrogen) was used for transient knockdown by siRNA following the standard procedure.
Western Blot Analysis
Cells were lysed in RIPA (Beyotime) for 30 mins on ice. Cell lysates were then centrifuged (14,000g for 10 min at 4°C), and protein concentration of the supernatant was determined by BCA protein assay kit (Beyotime). Cell lysates were separated by SDS-PAGE and electro-blotted onto a PVDF membrane (Millipore). In detail, 70 μg of cell lysates was separated on 8% acrylamide gel to detect ULK1 expression in liver tissues and liver cancer cells, 70 μg of cell lysates was separated on 12% acrylamide gel to determine proteins related to apoptosis, and 50 μg of cell lysates was separated on 12% acrylamide gel to measure autophagic markers. The membranes were blocked with 5% BSA for 2 h at room temperature and incubated with different primary antibodies: ULK1 (#8054, Cell Signaling Technology, USA), Bcl-2 (sc-7382, Santa Cruz Biotechnology), Bcl-XL (ab32370, Abcam), caspase-3 (#9662, Cell Signaling Technology), cleaved caspase3 (#9664, Cell Signaling Technology), tubulin (#2128, Cell Signaling Technology), and β-actin (A5316, Sigma). This was followed by incubation with the appropriate secondary antibodies (Anti-mouse IgG #7076, Anti-rabbit IgG#7074, Cell Signaling Technology) for 1 h at room temperature. Protein bands were visualized using Bio-Rad Chemidoc touch. Band quantification was measured in optical density units using Image Lab software (Bio-Rad, CA, USA) and normalized to the corresponding β-actin.
Cell Proliferation and Colony Formation Assay
The HCC cell line Huh-7 was treated with siULK1-1, siULK1-2, or a scramble control; 12 h later, the cells were digested and seeded (4000 cells/well) into 96-well plates. The cells were allowed to grow for 24, 48, 72, and 96 h. Then, into each well was added 10 μL CCK-8 (Beyotime) solution suspended in 100 μL 1% FBS DMEM medium for 1 h at 37°C, and the absorbance was measured at 450 nm at last. Colony formation assay was carried out in 6 cm dish. Briefly, cells were transfected with siULK1-1, siULK1-2, or a scramble control, and, 24 h post-transfection, 1000 cells per well were seeded and allowed to grow for 10 days; the medium was replaced every three days. Then cells were washed twice with PBS, stained with a solution of 0.5% crystal violet and 70% ethanol, washed with PBS twice, and dried. Clusters consisting of at least 50 cells were defined as colonies21 and were counted. Each assay was conducted in triplicate, and three separate assays were performed.
RNA-Sequencing and Data Analysis
Liver tissues from WT and Ulk1KO mice were obtained for RNA-seq. The process of RNA isolation and sequencing were performed by Novogene Inc. under standard procedure as we previously reported.22 In brief, RNA from liver tissue was used to construct the strand-specific library and was subjected for sequencing using Illumina 4000 platform with pair-end 150 base pair sequencing scheme, aiming for a minimum of 20 M reads per sample.
Genes showing differential expression with FDR < 0.05 and a fold change (FC) > 1.5 were defined as differentially expressed genes. The raw data of RNA-seq have been deposited in Sequence Read Archive (SRA) database, and the accession number is PRJNA907497.
Statistical Analysis
All values were expressed as mean ± SEM. A two-tailed Student’s t-test was used to compare means between two groups. Statistical analysis was performed by using GraphPad Prism 7.0. A P-value of less than 0.05 was considered statistically significant.
Results
ULK1 Was Up-Regulated in HCC Tissue and Cell Lines
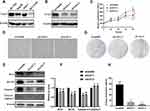
To confirm whether ULK1 was overexpressed in HCC patients as previously reported, we retrieved RNA-seq data from UCSC for further analysis, and the results showed that ULK1 was significantly up-regulated in HCC tissues (Figure 1A). However, ULK1 expression was not correlated with the stages of HCC (Figure 1B). Immunoblot was further performed to detect ULK1 protein level in human liver tissues, and it was found that ULK1 protein was also expressed at higher level in liver tumor tissues compared to non-tumor tissues (Figure 1C). In addition, the expression level of ULK1 in cancer cell lines was examined using data from the Cancer Cell Line Encyclopedia (CCLE), and liver cancer cell lines including Huh-1, Huh-7, JHH2, JHH7, and SNU761 showed relatively higher level of ULK1 expression (Figure 1D). All these data indicated a close relationship between ULK1 and cancer. To further elucidate clinicopathological relevance of ULK1 with liver cancer, Kaplan–Meier survival analysis was performed with an online tool (http://kmplot.com/analysis/). Unexpectedly, the results showed that higher ULK1 expression predicted better overall survival (OS, P = 0.013) and disease-specific survival (DSS, P = 0.02), but worse relapse-free survival (RFS, P = 0.02). ULK1 expression was also negatively correlated with progression-free survival (PFS), though this was not statistically significant (P = 0.075) (Figure S2).
Depletion of ULK1 Inhibited Cell Growth and Induced Apoptosis in Liver Cancer Cells
To further clarify the function of ULK1 in liver cancer cells, we also examined the expression of ULK1 in several HCC cell lines, among which Huh-7 showed the highest expression and was chosen for subsequent experiment (Figure 2A). ULK1 expression was then silenced in Huh-7 by siRNA, and the knockdown efficiency was confirmed for both siRNAs by Western blotting (Figure 2B). Results from CCK8 assay demonstrated that ULK1 depletion inhibited liver cancer cell growth (Figure 2C). Similar results were also obtained from microscopic observation and colony-formation assay (Figure 2D, G, and H). Furthermore, Western blotting was used to assess the expression of apoptosis-related proteins, and the results showed that ULK1 deficiency increased the expression of pro-apoptosis proteins while decreasing the expression of anti-apoptosis proteins (Figure 2E and F).
Ulk1 Deficiency in Mice Liver Disturbed the Expression of Genes Involved in Immune Response
To uncover the role of Ulk1 at the molecular level, liver tissue from Ulk1KO mice and WT mice was subjected to RNA-seq. Principal component analysis showed that the two groups were distinguished well from each other, while samples within one group shared much similarity despite variance still existing in WT mice (Figure S3A). Surprisingly, only 43 genes were identified as differentially expressed genes (DEGs) in liver of Ulk1KO mice versus wild type (WT) mice (Figure 3A). Among these DEGs, 15 genes, including Slc34a2, Cry1, Leap2, and Nmi, were down-regulated, while genes such as Gucd1, Rangap1, and Fosl2 were up-regulated in Ulk1KO mice liver compared with WT mice. Gene set enrichment analysis (GSEA) was performed, and gene sets associated with up-regulated genes in Ulk1KO mice were related to interleukin 2 (IL2)-STAT5 (signal transducers and activators of transcription 5) and mitotic spindle signaling (Figure 3B, Figure S3B), while the gene sets with lower expression genes were involved in interferon (IFN)-α and IFN-γ response (Figure 3C, Figure S3C).
Ulk1 Depletion Inhibited Starvation-Induced Autophagy in Mice Liver
Considering the vital role of ULK1 in autophagy especially for the initiation stage, we evaluated the effects of ULK1 depletion on autophagy in vivo. Fasting is a known factor to induce autophagy, and results from immunoblotting showed that fasting did induce the conversion of LC3-I to LC3-II in WT mice liver tissue, indicating the occurrence of autophagy. Moreover, the autophagy inhibitor chloroquine (CQ) blocked the autophagy flux, which was demonstrated by the further accumulation of LC3-II in fasting and chloroquine double treatment group (Figure 4A). In contrast, in Ulk1KO mice, chloroquine did not lead to the further accumulation of LC3-II under fasting condition when compared with fasting treatment alone (Figure 4B). And the ratio of LC3-II/LC3-I also showed that fasting in WT mice promoted conversion of LC3-I to LC3-II, while no significant difference was found in Ulk1KO mice (Figure 4C). Taken together, these results suggested that starvation-induced autophagy was impaired in Ulk1KO mice liver.
DEN-Induced Hepatocellular Carcinoma Was Suppressed in Ulk1KO Mice
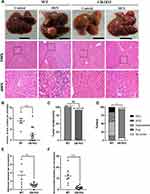
To determine the role of Ulk1 in hepatocarcinogenesis, two-week-old male Ulk1KO mice and WT mice were subjected to 25 mg/kg DEN injection as we previously reported. Thirty weeks after DEN administration, WT mice developed apparent big tumors on liver surface, while only sporadic and much smaller nodules could be found on liver surface in Ulk1KO mice based on macroscopic observation. Correspondingly, histopathological examination results using HE-staining further identified that HCC arose in WT mice, whereas most Ulk1KO mice just presented characteristics of lower-grade foci. As expected, no nodules were found in control groups for both WT and Ulk1KO mice (Figure 5A). Visible tumor nodules at liver surface were counted for each mouse, and Ulk1KO mice developed significantly fewer hepatic tumors than the WT ones (Figure 5B). Nonetheless, no statistical significance for tumor incidence was found between WT and Ulk1KO mice, although one Ulk1KO mouse was classified as no tumor formation (Figure 5C). According to the international nomenclature for classification of rodent tumors, liver histopathologic lesions were classified as foci, hyperplasia, hepatocellular adenoma (HCA), and HCC. Based on such classification, by the end of 30 weeks of DEN treatment, ten (83.3%) Ulk1KO mice developed foci, one (8.3%) developed hyperplasia, and one (8.3%) showed no tumor, whereas most WT mice progressed to hyperplasia (42.9%) and the more aggressive HCA (28.6%) and HCC (14.3%) (Figure 5D). Sizes of the tumors were also measured, and it was clear that Ulk1KO mice had much smaller liver tumors than WT mice (Figure 5E and F).
Discussion
As an essential kinase for the initiation of autophagy, the function of ULK1 has been well studied, and its role in various types of cancers was also investigated. For HCC, using 55 paired patient samples, Xu et al found that ULK1 expression was higher in HCC tissue than adjacent tissue, but not reaching a statistically significant level; on the other hand, higher ULK1 expression was associated with tumor size and worse survival time.18 Wu et al examined the expression of ULK1 in 156 HCC patients using tissue-microarray-based immunohistochemistry and found that it was highly expressed in 53.2% of the specimens; however, ULK1 expression was not related to any clinicopathological indicators. Further analysis revealed that ULK1 was not associated with 5-year OS, but was an independent prognostic biomarker for PFS.23 These contradicting results prompted us to evaluate the role of ULK1 in HCC in depth. By analyzing the TCGA database, it was found that ULK1 was overexpressed in HCC samples (Figure 1). However, by KM plot analysis, we uncovered a paradoxical role of ULK1 in predicting survival of HCC patients: high-level ULK1 expression was positively associated with better OS and DSS, but negatively correlated with RFS (Figure S2). Such results indicated that ULK1 alone may not be a good biomarker in predicting prognosis of HCC. Indeed, Wu et al demonstrated that combining autophagic biomarkers of ULK1 and LC3B was better in predicting the prognosis than used individually.
Although the above results cast doubt on the prognostic value of ULK1, its function in liver cancer growth is relatively clear. For example, it was shown that silencing ULK1 inhibited liver cancer cell growth and proliferation, and deletion of ULK1 abrogated tumor growth in a xenograft mouse model.17 In this study, our results also confirmed that silencing ULK1 led to the inhibition of liver cancer cell growth and colony formation; in addition, it was shown that the expression of anti-apoptosis proteins was down-regulated, indicating apoptosis might be responsible for the decreased cell growth (Figure 2). Compared with the corresponding normal cells, tumor cells always exhibit aberrant proliferation.24 To fulfill the rapid biosynthetic demands associated with proliferation, cancer cells usually promote autophagy as a self-catabolic process for recycling engulfed cargos to survive and proliferate in metabolically unfavorable conditions.25 Therefore, the loss of ULK1 could cause decreased autophagy, which might lead to decreased proliferation of cancer cells.
Autophagy is regarded as a double-edged sword in cancer since it can suppress tumor formation by maintaining homeostasis of normal cells or promote tumor progression by enabling cancer cells to survive under stress.26 For instance, on one hand, monoallelic deletion of Becn1 in mice resulted in diminished but intact autophagy, and the mice developed spontaneous hepatocellular carcinomas.27 On the other hand, Atg7 deficiency dramatically changed the nature of lung tumors driven by BrafV600E from adenomas to benign oncocytomas.28 These results indicated that autophagy was important for the suppression of spontaneous tumorigenesis but was required for tumor progression to malignancy. In our study, first it was found that knockout of Ulk1 impaired starvation-induced autophagy but not basal autophagy (Figure 4). Then in the hepatocarcinogenesis experiment, no spontaneous tumor formation was found in Ulk1KO mice, but DEN-induced primary liver tumor formation was inhibited (Figure 5). Such results indicated that Ulk1 deficiency did not promote tumor formation but suppressed tumor progression in mice, which suggested the role of ULK1 in tumor was different from other autophagy-related genes such as beclin1, ATG5, and ATG7.
Several studies have indicated the close relationship between ULK1 and immunity.29,30 In our study, GSEA results revealed that IFN-α and IFN-γ signaling were down-regulated. This is inconsistent with the conclusion of previous studies in which ULK1 was a key mediator of type I IFN signals and was required in IFN-γ-inducible antiviral responses.31,32 It was reported that IFN-α could either stimulate immune cells to eliminate cancer cells or enable cancer cells to escape immune clearance with immune exhaustion because of prolonged stimulation.33 IFN-γ could either promote melanoma development or inhibit breast cancer by reducing cancer stem cells.34,35 However, what exact role the IFN-α and IFN-γ signaling played in DEN-induced hepatocarcinogenesis remained to be clarified. Conversely, GSEA results showed that IL-2-STAT5 and mitotic spindle pathway were up-regulated in Ulk1KO mice. STAT5 is a critical downstream mediator of IL-2 signaling and affects immune function in many aspects.36 STAT5 acts both as a tumor suppressor by affecting STAT3 signaling and as a tumor promoter under other circumstances.37 Interestingly, it was reported that IFN-α could inhibit IL-2 signal transduction in T-lymphocytes,38 thus the activation of IL-2-STAT5 pathway might be a consequence of the down-regulation of IFN-α. Still, more clear evidence is required to verify the relationship between the different immune signaling pathways, as well as their impacts on cancer development.
In summary, by using DEN-induced HCC model, we expanded our understanding of ULK1 function in cancer by uncovering that ULK1 deficiency suppressed primary HCC. Clues from RNA-sequencing of Ulk1KO mice pointed to the direction of altered immune status in cancer development. Taken together, we believe that ULK1 could be a potential target for HCC prevention and treatment.
Funding
This work was supported in parts by grants from the National Natural Science Foundation of China (Nos. 31971138 and 32270186 to J.Y.), Zhejiang Provincial Natural Science Foundation of China (No. LQ20H160013 to T.D.).
Disclosure
The authors declare that they have no conflicts of interest.
References
1. Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. doi:10.3322/caac.21660
2. McGlynn KA, Petrick JL, El-Serag HB. Epidemiology of hepatocellular carcinoma. Hepatology. 2021;73(Suppl 1):4–13. doi:10.1002/hep.31288
3. Raffetti E, Portolani N, Molfino S, et al. Is survival for hepatocellular carcinoma increasing? A population-based study on survival of hepatocellular carcinoma patients in the 1990s and 2000s. Clin Res Hepatol Gastroenterol. 2021;45(1):101433. doi:10.1016/j.clinre.2020.04.004
4. Chidambaranathan-Reghupaty S, Fisher PB, Sarkar D. Chapter One - Hepatocellular carcinoma (HCC): epidemiology, etiology and molecular classification. In: Sarkar D, Fisher PB editors. Advances in Cancer Research. Vol 149. Mechanisms and Therapy of Liver Cancer. Academic Press; 2021:1–61. doi:10.1016/bs.acr.2020.10.001
5. Llovet JM, Pinyol R, Kelley RK, et al. Molecular pathogenesis and systemic therapies for hepatocellular carcinoma. Nat Cancer. 2022;3(4):386–401. doi:10.1038/s43018-022-00357-2
6. Klionsky DJ, Abdel-Aziz AK, Abdelfatah S, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2006;17(1):1–382. doi:10.1080/15548627.2020.1797280
7. Mathea S, Salah E, Tallant C, et al. Conformational plasticity of the ULK3 kinase domain. Biolchem J. 2021;478(14):2811–2823. doi:10.1042/BCJ20210257
8. Zachari M, Ganley IG. The mammalian ULK1 complex and autophagy initiation. Essays Biochem. 2017;61(6):585–596. doi:10.1042/EBC20170021
9. Demeter A, Romero-Mulero MC, Csabai L, et al. ULK1 and ULK2 are less redundant than previously thought: computational analysis uncovers distinct regulation and functions of these autophagy induction proteins. Sci Rep. 2020;10(1):10940. doi:10.1038/s41598-020-67780-2
10. Li TY, Sun Y, Liang Y, et al. ULK1/2 constitute a bifurcate node controlling glucose metabolic fluxes in addition to autophagy. Mol Cell. 2016;62(3):359–370. doi:10.1016/j.molcel.2016.04.009
11. Joshi A, Iyengar R, Joo JH, et al. Nuclear ULK1 promotes cell death in response to oxidative stress through PARP1. Cell Death Differ. 2016;23(2):216–230. doi:10.1038/cdd.2015.88
12. Zhang L, Fu L, Zhang S, et al. Discovery of a small molecule targeting ULK1-modulated cell death of triple negative breast cancer in vitro and in vivo. Chem Sci. 2017;8(4):2687. doi:10.1039/c6sc05368h
13. Mao L, Zhan Y, Wu B, et al. ULK1 phosphorylates Exo70 to suppress breast cancer metastasis. Nat Commun. 2020;11(1):117. doi:10.1038/s41467-019-13923-7
14. Tang F, Hu P, Yang Z, et al. SBI0206965, a novel inhibitor of Ulk1, suppresses non-small cell lung cancer cell growth by modulating both autophagy and apoptosis pathways. Oncol Rep. 2017;37(6):3449–3458. doi:10.3892/or.2017.5635
15. Liu J, Long S, Wang H, et al. Blocking AMPK/ULK1-dependent autophagy promoted apoptosis and suppressed colon cancer growth. Cancer Cell Int. 2019;19:336. doi:10.1186/s12935-019-1054-0
16. Singha B, Laski J, Ramos Valdés Y, et al. Inhibiting ULK1 kinase decreases autophagy and cell viability in high-grade serous ovarian cancer spheroids. Am J Cancer Res. 2020;10(5):1384–1399.
17. Xue S-T, Li K, Gao Y, et al. The role of the key autophagy kinase ULK1 in hepatocellular carcinoma and its validation as a treatment target. Autophagy. 2020;16:1823–1837. doi:10.1080/15548627.2019.1709762
18. Xu H, Yu H, Zhang X, et al. UNC51-like kinase 1 as a potential prognostic biomarker for hepatocellular carcinoma. Int J Clin Exp Pathol. 2013;6(4):711–717.
19. Kundu M, Lindsten T, Yang CY, et al. Ulk1 plays a critical role in the autophagic clearance of mitochondria and ribosomes during reticulocyte maturation. Blood. 2008;112(4):1493–1502. doi:10.1182/blood-2008-02-137398
20. Mohr U, ed. International Classification of Rodent Tumors.
21. Franken NAP, Rodermond HM, Stap J, Haveman J, van Bree C. Clonogenic assay of cells in vitro. Nat Protoc. 2006;1(5):2315–2319. doi:10.1038/nprot.2006.339
22. Duan T, Sun W, Zhang M, et al. Dietary restriction protects against diethylnitrosamine-induced hepatocellular tumorigenesis by restoring the disturbed gene expression profile. Sci Rep. 2017;7:43745. doi:10.1038/srep43745
23. Wu D, Wang TT, Ruan DY, et al. Combination of ULK1 and LC3B improve prognosis assessment of hepatocellular carcinoma. Biomed Pharmacother. 2018;97:195–202. doi:10.1016/j.biopha.2017.10.025
24. Hanahan D. Hallmarks of cancer: new dimensions. Cancer Discov. 2022;12(1):31–46. doi:10.1158/2159-8290.CD-21-1059
25. Cozzo AJ, Coleman MF, Pearce JB, et al. Dietary energy modulation and autophagy: exploiting metabolic vulnerabilities to starve cancer. Front Cell Dev Biol. 2020;8:590192. doi:10.3389/fcell.2020.590192
26. Chavez-Dominguez R, Perez-Medina M, Lopez-Gonzalez JS, et al. The double-edge sword of autophagy in cancer: from tumor suppression to pro-tumor activity. Front Oncol. 2020;10:578418. doi:10.3389/fonc.2020.578418
27. Qu X, Yu J, Bhagat G, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112(12):1809–1820. doi:10.1172/JCI20039
28. Strohecker AM, Guo JY, Karsli-Uzunbas G, et al. Autophagy sustains mitochondrial glutamine metabolism and growth of braf V600E–driven lung tumors. Cancer Discov. 2013;3(11):1272–1285. doi:10.1158/2159-8290.CD-13-0397
29. Deng J, Thennavan A, Dolgalev I, et al. ULK1 inhibition overcomes compromised antigen presentation and restores antitumor immunity in LKB1-mutant lung cancer. Nat Cancer. 2021;2(5):503–514. doi:10.1038/s43018-021-00208-6
30. Qiu X, Zheng L, Liu X, et al. ULK1 inhibition as a targeted therapeutic strategy for psoriasis by regulating keratinocytes and their crosstalk with neutrophils. Front Immunol. 2021;12. doi:10.3389/fimmu.2021.714274
31. Saleiro D, Mehrotra S, Kroczynska B, et al. Central Role of ULK1 in Type I Interferon Signaling. Cell Rep. 2015;11(4):605–617. doi:10.1016/j.celrep.2015.03.056
32. Saleiro D, Blyth GT, Kosciuczuk EM, et al. IFNγ-inducible antiviral responses require ULK1-mediated activation of MLK3 and ERK5. Sci Signal. 2018;11:eaap9921. doi:10.1126/scisignal.aap9921
33. Vidal P . Interferon α in cancer immunoediting: From elimination to escape. Scandinavian Journal of Immunology. 2020;91(5):e12863. doi:10.1111/sji.12863
34. Tong S, Cinelli MA, El-Sayed NS, et al. Inhibition of interferon-gamma-stimulated melanoma progression by targeting neuronal nitric oxide synthase (nNOS). Sci Rep. 2022;12. doi:10.1038/s41598-022-05394-6
35. Zhuang X, Shi G, Hu X, et al. Interferon-gamma inhibits aldehyde dehydrogenase bright cancer stem cells in the 4T1 mouse model of breast cancer. Chin Med J. 2022;135:194–204. doi:10.1097/CM9.0000000000001558
36. Jones DM, Read KA, Oestreich KJ. Dynamic roles for IL-2-STAT5 signaling in effector and regulatory CD4+ T cell populations. J Immunol. 2020;205(7):1721–1730. doi:10.4049/jimmunol.2000612
37. Halim CE, Deng S, Ong MS, et al. Involvement of STAT5 in Oncogenesis. Biomedicines. 2020;8:316. doi:10.3390/biomedicines8090316
38. Erickson S, Matikainen S, Thyrell L, et al. Interferon-alpha inhibits Stat5 DNA-binding in IL-2 stimulated primary T-lymphocytes. European J Biochem. 2002;269(1):29–37. doi:10.1046/j.0014-2956.2002.02626.x
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.