Back to Journals » Infection and Drug Resistance » Volume 11
Two residues in Staphylococcus aureus α-hemolysin related to hemolysis and self-assembly
Authors Du Y, Liu L, Zhang C, Zhang Y
Received 9 March 2018
Accepted for publication 12 May 2018
Published 21 August 2018 Volume 2018:11 Pages 1271—1274
DOI https://doi.org/10.2147/IDR.S167779
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Joachim Wink
Yufeng Du,1 Li Liu,2 Chunping Zhang,3 Yani Zhang1
1College of Life Sciences, Northwest University, Xi’an 710069, China; 2Ultrasonic Diagnosis Department, Shaanxi Provincial People’s Hospital, Xi’an 710068, China; 3College of Chemistry and Material Science, Northwest University, Xi’an 710069, China
Abstract: Staphylococcus aureus is becoming increasingly intractable because of its ability to acquire antimicrobial resistance and secrete numerous virulence factors that can exacerbate inflammation. Alpha-hemolysin (Hla) is a pore-forming virulence factor produced by S. aureus that can self-assemble into heptameric mushroom-structured pores in target cell membranes, leading to cell lysis and death. In the present study, we sought to better understand the mechanism underlying hemolysis and the oligomerization of Hla by creating nine mutants with single amino acid changes in different positions of the Hla protein: N17C, T18C, P103C, N105C, M113C, T117C, N121C, D128C, and T129C. The results showed that the P103C and N105C mutations, which are located in the triangle region, significantly diminished hemolysis and heptamer formation when compared with the wild-type Hla protein. This suggests that the P103 and N105 residues play key roles in the assembly of the Hla pore. These results improve our understanding of the mechanism underlying the pore-forming ability of Hla.
Keywords: α-hemolysin, hemolysis, heptamer oligomers, cysteine mutants, assembly
Introduction
Staphylococcus aureus is a major bacterial pathogen that invades and damages host tissue by expressing harmful toxins. α-hemolysin (Hla), a pore-forming toxin secreted by S. aureus, is very important for pathogenesis. Hla can form a mushroom-shaped pore in eukaryotic cell membranes, causing cell damage and subsequent cell death.1,2 Previous studies have reported that strains lacking Hla exhibited obviously attenuated pathogenicity in a mouse model of S. aureus infection.3 Hla is a water-soluble monomer of 33.2 kDa, and seven copies of H1a self-assemble to form a heptameric pore of 232.4 kDa that is composed of three structural regions, the cap, rim, and stem, as observed based on X-ray diffraction.4 However, the mechanism involved in Hla-induced hemolysis and oligomer pore assembly are not well understood. In this study, we generated nine mutants, each with a single amino acid change in different regions of the Hla protein, to investigate their role in Hla pore formation and cell hemolysis.
Characterization of the assembly and hemolysis activities of nine different mutant Hla proteins
In the present study, nine different amino acids in wild-type (WT) H1a were individually changed to a cysteine residue to determine their role in pore formation. These nine sites, N17, T18, P103, N105, M113, T117, N121, D128, and T129, are located on the cap, rim, and stem regions of the heptameric mushroom-shaped pore structure (Figure 1). Site-directed mutagenesis was used to change these nine different sites to cysteine codons by using the Muta-direct™ Kit (catalog number: SDM-15; SBS Genetech, Beijing, China) and a reconstructed WT Hla gene in a T7 plasmid (pT7-Hla) as the template. Then, the desired mutation was confirmed by sequencing. The primers used in this study are shown in Table 1. The pT7-Hla and mutant plasmids were individually transformed into Escherichia coli BL21 (DE3) pLysS cells for protein expression. Ultrafiltration centrifuge tubes with three different pore sizes. 100-, 50-, and 30-kDa molecular weight cut off (MWCO) (catalog numbers: UFC910096, UFC905096, and UFC503096, respectively; Millipore, Burlington, MA, USA) were used for protein isolation and concentration, and all steps were conducted at 4°C. Initially, the 100-kDa MWCO centrifuge filter tube was used to eliminate macromolecular proteins from the cell lysate. Then, this filtrate was subsequently centrifuged in a 50-kDa MWCO tube, and then a 30-kDa MWCO tube. Finally, the retentate remaining on the top of the 30-kDa MWCO filter was used as the Hla protein solution. The protein solution was subsequently analyzed by SDS-PAGE.
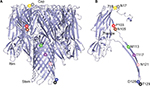  | Figure 1 Ribbon representation of seven protomers from Hla heptamer (A) and a protomer from heptamer (B) in a PyMoL model. Notes: The cap, rim, and stem domains are labeled. In (A) and (B), mutated positions in Hla nanopore structure were labeled with different colors. The figure was drawn using PyMoL software (https://pymol.org). Abbreviation: Hla, alpha-hemolysin. |
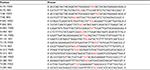  | Table 1 Primers used in the study Notes: Red indicates the mutated base. Abbreviations: FWD, forward; REV, reverse. |
To compare the hemolytic potencies of the WT and mutant Hla proteins, their pore-forming activities were measured by determining the rate of lysis of rabbit red blood cells (rRBCs) in a 96 well plate.5,6 New Zealand white rabbit was used to prepare rRBCs as previously described.7 All animal experiments were performed in strict accordance with the Regulations for the Administration of Affairs Concerning Experimental Animals approved by the State Council of China (November 14, 1988). All animal procedures were approved by the Institutional Animal Care and Use Committee of the college of life sciences of Northwest University with a permit number: NW-02-2014. The protein solutions were incubated with the rRBCs at 37°C for 1 h. The results are shown in Figure 2A. Compared to WT Hla, the P103C and N105C mutants showed far less hemolytic activity toward the rRBCs, the N121C mutant showed decreased hemolytic activity, and the other six mutants showed activity similar to the WT protein. In all experiments, the concentration of the Hla protein was adjusted to 0.20 mg/mL.
To further explore the properties of these polypeptides, the ability of the mutant monomers to form heptameric oligomers in rRBC membranes (rRBCMs) was evaluated. The mutant monomers were reacted with 0.5% rRBCMs for 3 h at 37°C to allow oligomer assembly.8 Then, oligomer formation was evaluated by SDS-PAGE. The oligomers were separated by electrophoresis using 7% SDS-PAGE gels with a prestained protein marker at 120 V for 6 h at 4°C. As expected, SDS-PAGE revealed that compared to WT Hla, the extent of oligomer formation by the P103C and N105C mutants was very low but detectable, which was consistent with the results showing low hemolytic activity. In contrast, the mutations at positions 17, 18, 113, 117, 121, 128, and 129 did not appear to affect binding and oligomer formation, and these seven mutants had phenotypes similar to that of WT Hla (Figure 2B).
Conclusion
Hla plays a critical role in the virulence of S. aureus. Here, we used single amino acid Hla mutants to examine the mechanisms involved in hemolysin and oligomer assembly, which are important for understanding the actions of cytolytic toxins and immune proteins. In previous studies, using cysteine mutants and generating targeted chemical modifications at various positions, it was shown that some residues and regions of Hla are critical for heptamer assembly.9 Specifically, conversion of three residues in the triangle region, R104, K110, and D152, to cysteine residues diminished membrane binding, cell lysis, and heptamer-forming activities.4 Two other residues, D108 and K154, were shown to be critical for assembly of Hla but not for membrane binding.9 Here, our experiments showed that the P103C and N105C mutations diminished the hemolytic and heptamer-forming activities of the H1a protein. These results suggest that P103 and N105 play important roles in hemolysis and heptamer formation. The triangle region of the Hla heptameric oligomer is the linker between the protomer core and the stem-forming strands, and it participates in crucial protomer–protomer interactions.4,10 The cysteine mutants at positions 103 and 105 in the triangle region failed to form oligomers, which may explain their low hemolytic activity. These results confirmed that the triangle region plays an essential role in the conversion of monomers to heptamers. The glycine-rich stem region is translocated across the membrane. The five cysteine mutants we generated in this region, at residues 113, 117, 121, 128, and 129, did not alter monomer assembly in cell membranes. These findings provide a foundation for new studies into the mechanism underlying Hla assembly and function.
Disclosure
The authors report no conflicts of interest in this work.
References
Rokitskaya TI, Nazarov PA, Golovin AV, Antonenko YN. Blocking of Single α-Hemolysin Pore by Rhodamine Derivatives. Biophys J. 2017;112(11):2327–2335. | ||
DuMont AL, Torres VJ. Cell targeting by the Staphylococcus aureus pore-forming toxins: it’s not just about lipids. Trends Microbiol. 2014;22(1):21–27. | ||
Zhang B, Teng Z, Li X, et al. Chalcone Attenuates Staphylococcus aureus Virulence by Targeting Sortase A and Alpha-Hemolysin. Front Microbiol. 2017;8:1715. | ||
Gouaux E. alpha-Hemolysin from Staphylococcus aureus: an archetype of beta-barrel, channel-forming toxins. J Struct Biol. 1998;121(2):110–122. | ||
Raychaudhuri P, Li Q, Mason A, Mikhailova E, Heron AJ, Bayley H. Fluorinated amphiphiles control the insertion of α-Hemolysin pores into lipid bilayers. Biochemistry. 2011;50(10):1599–1606. | ||
Cheley S, Malghani MS, Song L. Spontaneous oligomerization of a staphylococcal α-hemolysin conformationally constrained by removal of residues that form the transmembrane β-barrel. Protein Eng. 1997;10:1433–1443. | ||
Mendez AJ, He JL, Huang HS, Wen SR, Hsia SL. Interaction of rabbit lipoproteins and red blood cells with liposomes of egg yolk phospholipids. Lipids. 1988;23(10):961–967. | ||
Hammerstein AF, Jayasinghe L, Bayley H. Subunit dimers of alpha-hemolysin expand the engineering toolbox for protein nanopores. J Biol Chem. 2011;286(16):14324–14334. | ||
Kawate T, Gouaux E. Arresting and releasing Staphylococcal alpha-hemolysin at intermediate stages of pore formation by engineered disulfide bonds. Protein Sci. 2003;12(5):997–1006. | ||
Song L, Hobaugh MR, Shustak C, Cheley S, Bayley H, Gouaux JE. Structure of staphylococcal alpha-hemolysin, a heptameric transmembrane pore. Science. 1996;274(5294):1859–1866. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.

