Back to Journals » Cancer Management and Research » Volume 10
Tumor-suppressive function of SIRT4 in neuroblastoma through mitochondrial damage
Authors Wang Y, Guo Y, Gao J, Yuan X
Received 27 April 2018
Accepted for publication 30 June 2018
Published 9 November 2018 Volume 2018:10 Pages 5591—5603
DOI https://doi.org/10.2147/CMAR.S172509
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Harikrishna Nakshatri
Yumei Wang,1 Yinmou Guo,2 Jianzhi Gao,3 Xiangdong Yuan1
1Department of Children’s Rehabilitation, Shangqiu First People’s Hospital of Henan, Shangqiu, Henan, China; 2First Department of Oncology, Shangqiu First People’s Hospital of Henan, Shangqiu, Henan, China; 3Department of Oncology, Zhuozhou Hospital of Beijing 301 Hospital, Beijing, China
Background: SIRT4 is a member of the sirtuin family of nicotinamide adenine dinucleotide-dependent enzymes located in the mitochondria, and is involved in regulating energy metabolism, stress response, and cellular lifespan in mammalian cells. However, its function in human neuroblastoma (NB) remains unexplored.
Methods: Expression of SIRT4 in 158 pairs of human NB tumor tissues and adjacent normal tissues collected from March 2009 to October 2012 was analyzed by immunohistochemistry, Western blotting, and real-time fluorescence quantitative PCR. For in vitro study, SIRT4 was overexpressed in SH-SY5Y, SK-N-BE, and IMR-32 cells to study the effects of SIRT4 expression on proliferation, invasion, and migration of human NB cells and on mitochondrial function.
Results: SIRT4 gene expression in human NB tumor tissues was significantly lower than that in adjacent normal tissues (P<0.001). SIRT4 expression was lower in NB patients with higher International Neuroblastoma Staging System stage (P=0.018), with lymph node metastasis, than patients without lymph node metastasis (P<0.001). Survival times of NB patients with low expression of SIRT4 were significantly shorter than those of patients with high expression of SIRT4 (P=0.0036). Overexpression of SIRT4 significantly reduced the proliferation, invasion, and migration ability of NB cells as well as mitochondrial energy production, and caused SIRT1 upregulation and mitochondrial damage in NB cells.
Conclusion: SIRT4 exhibits a tumor suppressor function in human NB and inhibits mitochondrial metabolism and SIRT1 expression in tumor cells, thereby reducing the energy metabolism of tumor cells. These results suggest that SIRT4 may be a new therapeutic target for human NB.
Keywords: neuroblastoma, SIRT4, energy metabolism, SIRT1
Introduction
Neuroblastoma (NB) is the most common extracranial malignant solid tumor in children, accounting for 6%–10% of tumors.1,2 Clinical manifestations of NB are complex and diverse with reported high-grade malignancy and rapid disease progress. More than half of affected children are diagnosed as a high risk group at advanced stage (stage III-IVs) at the time of treatment. The prognosis of NB is poor and the case fatality rate is approximately 15% while the long-term survival rate is less than 40%.3,4 According to the National Cancer Center in China,1,2 the incidence of NB in China is on the rise. Approximately 3,000 new cases of NB are reported each year, of which more than half occurs in infants under 2 years of age. NB is a common malignant tumor in children, second only to leukemia and central nervous system tumors. However, the underlying cause of NB remains unclear. Since NB often occurs in infants, many studies have focused on investigating the impact of environmental risk factors5 such as exposure to harmful chemicals,6 maternal smoking, drinking, and taking drugs during pregnancy, and infections during pregnancy,7 but no clear results have been reported in these studies. Some studies have focused on genetic factors and it has been found that somatic mutation in anaplastic lymphoma kinase is associated with the development of familial NB,8,9 amplification mutation of N-myc gene is associated with diffusion of NB,10,11 and LMO1 gene, an oncogene, has been shown to be associated with malignancy grade of the tumor.12,13
SIRT4 is a member of the sirtuin protein family that is localized to the mitochondria in mammalian cells. Sirtuin protein family (SIRT1-7) is a highly conserved family of NAD+-dependent deacetylases and ADP-ribosyltransferases.14 Almost all members of the sirtuin family are thought to play important roles in occurrence and development of tumors.15,16 A study demonstrated that SIRT4 could repair DNA damage by inhibiting mitochondrial glutamine metabolism, thus inhibiting tumorigenesis.17 SIRT4 is expressed at low levels in many malignant tumor tissues such as colon cancer,18 gastric cancer,19 B-cell lymphoma,20 and breast cancer.21 However, no study has reported the expression of SIRT4 in human NB tissue and the clinical significance of its expression.
In this study, we analyzed the expression of SIRT4 in NB tissues and investigated the effect of SIRT4 overexpression on the proliferation, invasion, and migration of NB cells and its underlying mechanisms.
Materials and methods
Clinical specimens and cells
The patients (or their guardians) included in this study were informed of the study and signed informed consent, and all the experiments were approved by the Ethics Committee of Shangqiu First People’s Hospital of Henan Province.
A total of 158 pairs of human NB tumor tissues and adjacent normal tissues (>5 cm from the tumor tissues) removed during surgery were collected in Shangqiu First People’s Hospital of Henan from March 2009 to October 2012. Tumor tissues and adjacent normal tissues were fixed in formalin solution overnight at 4°C treated with gradient ethanol, and finally embedded in paraffin. Some tissue samples were directly frozen in liquid nitrogen.
The 158 NB patients enrolled in this study were not treated with radiotherapy or chemotherapy prior to surgery. The following patients were excluded from the study: incomplete information regarding age, gender, disease history, and tumor, those who were lost to postoperative follow-up or had unknown cause of death, complicated with other malignancies, or inability to identify that NB was the primary lesion, death as a result of other sudden-onset diseases (such as cardiovascular and cerebrovascular diseases), or patients with poor physical condition affecting prognosis.
Immunohistochemistry
SIRT4 protein expression in NB tissues and adjacent normal tissues was measured by immunohistochemistry using VECTASTAIN® Elite® ABC Kit (Vector Laboratories, Boston, MA, USA). Briefly, a paraffin slice was cut into 3.5 µm slices with a slicer (CUT 4062; SLEE Medical GMBH, Karlsruhe, Baden-Württemberg, Germany) and slices were placed at 60°C for 2 hours, deparaffinized, and hydrated with xylene and ethanol and then washed with PBS (135 mM NaCl, 2.7 mM KCl, 1.5 mM KH2PO4, 8 mM K2HPO4, adjusted to pH 7.2 with HCl and NaOH) and double distilled water. Slices were then stained with SIRT4 antibody (ab90485, 1:300; abcam, Cambridge, London, UK) (PBS instead of primary antibody as a negative control) and incubated overnight at 4°C. Samples were further incubated with secondary goat anti-rabbit IgG heavy and light chains of antibodies (horseradish peroxidase) (1:1,000; abcam) at 37°C for 2 hours. Finally, five fields per slice were imaged and scored as follows: SIRT4 location in mitochondria; no color was negative (−), faint yellow was weak positive (+), yellow was positive (++), claybank or tan was strongly positive (+++); negative or weak positive (+) was low expression, and positive (++) or strongly positive (+++) was high expression. Two pathology deputy chief physicians were blinded to score for SIRT4 immunohistochemical staining.
Western blotting
Frozen tissue samples stored in liquid nitrogen were placed in a mortar, an appropriate amount of liquid nitrogen was added, and the tissues were ground. Total protein was extracted with RIPA lysis buffer containing 1 mM phenylmethanesulfonyl fluoride (Beyotime, Shanghai, China). Samples were incubated on ice for 10 minutes and centrifuged at 12,000 rpm for 10 minutes to collect the supernatant which contained the total protein. SDS buffer solution was added to the supernatant at the final concentration of 1% SDS and the mixture was boiled at 100°C for 5 minutes. Protein concentration was determined with a BCA protein concentration assay kit (Beyotime). An amount of 75 μg total protein was loaded into each lane and separated by 15% SDS-PAGE (90 V, 0.5 hour; 120 V, 1 hour) and transferred (400 mA, 1.5 hour) to a polyvinylidene fluoride membrane (Amersham Biosciences, Cambridge, London, UK) previously fixed with methanol for 1 minute, washed three times (5 minutes each) with TBST (10 mM Tris-HCl, 150 mM NaCl, 0.1% tween-20, pH 7.6) and blocked with blocking buffer (5% skim milk in TBST) for 1 hour. SIRT4 antibody (ab90485, 1:1,000; abcam) or β-actin antibody (ab8227, 1:2,000; abcam) was diluted in blocking buffer at 4°C overnight. Following incubation, blots were washed with TBST three times (10 minutes each). Membranes were incubated with goat anti-rabbit secondary antibody (ab205718, 1:2,000; abcam) for 1 hour at room temperature and washed with TBST. Signal was detected using enhanced chemiluminescent substrate. Images were quantified with Image J software, and the relative expression level of target protein was normalized against β-actin.
Cell transfection and construction of stable cell lines
Human NB cell lines SH-SY5Y (CRL-2266), SK-N-BE (CRL-2271), IMR-32 (CCL-127), and 293T (CRL-3216) were purchased from ATCC (Manassas, VA, USA). Cells were cultured in DMEM (D6046; Sigma-Aldrich Co., St Louis, MO, USA) containing 10% FBS (10099–141; Gibco, Thermo Fisher Scientific, Waltham, MA, USA) at 37°C and 5% CO2. No antibiotics were added to the growth medium. Cell lines overexpressing SIRT4 were established by transfection with pLenti-C-Myc-DDK-IRES-Puro Tagged Cloning Vector (PS100069; ORIGEN, Annapolis, MD, USA) and Lentiviral Packaging Kits (TR30037; ORIGEN) according to the manufacturer’s protocol. Cells transfected with empty plasmids were used as control. Puromycin at the concentration of 2 mg/mL for SH-SY5Y, 2 mg/mL for SK-N-BE and 3 mg/mL for IMR-31 to select the stable cell expression, and cell lines with SIRT4 overexpression or knockout were established.
Real-time fluorescence quantitative PCR (RT-qPCR)
Frozen tissue samples stored in liquid nitrogen were placed in a mortar, an appropriate amount of liquid nitrogen was added, and tissues were ground quickly. An amount of 1 mL of Trizol (15596026; Thermo Fisher Scientific) was added to extract RNA. RNA concentration was measured by NanoDrop 2000 Ultramicro Spectrophotometer (Thermo Scientific Scientific). The RNA was reverse transcribed into cDNA using PrimeScript™ RT Master Mix reverse transcription kit (RR036B; TaKaRa, Yokohama, Japan) according to the manufacturer’s protocol. PCR parameters were set as: 37°C/60 minutes, 85°C/5 seconds.
An amount of 20 μL of RT-qPCR reaction mix was prepared according to the SYBR Green qPCR Master Mix kit instructions (638320; TaKaRa) and qPCR was performed using ABI 7500 fluorescence qPCR instrument (Applied Biosystems, Thermo Fisher Scientific). PCR parameters were set as: 95°C/30 seconds, (90°C/5 seconds, 65°C/30 seconds) – 40 cycles. The relative expression level of the target gene was calculated by 2−∆∆ct method using β-actin as an internal control. RT-qPCR specific primer sequences were: SIRT4-F: 5′-CACAATCCAAGCACAGGA-3′; SIRT4-R: 5′-GCACACTGGGCTTTGAGC-3′; β-actin-F: 5′-TCACCCACACTGTGCCCATCTACG-3′; β-actin-R: 5′-CAGCGGAACCGCTCATTGCCAATG-3′.
Immunofluorescence
Cells grown on coverslips were washed with PBS three times (3 minutes each). Cells were fixed with 4% paraformaldehyde for 15 minutes, rinsed with PBS three times, and permeabilized with 0.5% Triton-X100. Normal goat serum was added as a blocking buffer for 30 minutes at room temperature. Cells were then incubated with the primary anti-SIRT4 antibody (ab90485, 1:1,000; abcam) overnight at 4°C. Cells were rinsed with PBST (135 mM NaCl, 2.7 mM KCl, 2.0 mM KH2PO4, 8 mM Na2HPO4, 0.05% Tween-20, pH 7.2) three times and goat anti-rabbit secondary antibody conjugated with Alexa-flour 488 (ab150077, 1:1,000; abcam) was added for 1 hour at room temperature. Cells were rinsed with PBST three times. Nuclear dye DAPI (4083; Cell Signaling Technology, Cambridge, London, UK) was added and cells were incubated in the dark for 5 minutes. After washing with PBST (5 minutes ×4 times), coverslips were mounted with anti-fluorescence quencher. Samples were observed and imaged with a fluorescence microscope.
Cell proliferation
Cells were seeded in 6-well plates at a density of 2×103 cells/mL, and 2 mL/well. The medium was changed every 3 days. Cells were routinely cultured for approximately 3 weeks until the visible clones appeared in the wells. The culture supernatant was removed and cells were washed with PBS twice, and the plates were fixed with 4% formaldehyde for 15 minutes. Subsequently, cells were stained with 0.25% crystal violet for 25 minutes and then gently rinsed with sterile water. Pictures were taken after plates were allowed to dry. The relative proliferation was measured by measuring the absorbance at 595 nm.
For counting, cells were seeded in 6-well plates at a density of 2×103 cells/mL and routinely cultured for 4 days. Trypsin-EDTA (0.25%) was used to dislodge the cells. Countstar automatic cell counter (IC1000; Shanghai Ruiyu Biological Technology Co., Ltd., Shanghai, China) was used to count the cells.
Animal experiments
The animal experiments were approved by the Ethics Committee of Shangqiu First People’s Hospital of Henan Province. Animal welfare and the relevant experiments were carried out in compliance with the guide for the care and use of laboratory animals.
Logarithmic phase human NB cells at the density of 5×106/0.2 mL were used for administration to animals. One-week-old nude mice (male/female =1:1, quality certificate: no 0006045) that were fed (room temperature of 20°C–24°C, 12 hour light/dark cycle, air humidity of 60%) were selected. Middle axillary lateral skin of mice was selected as the cell inoculation site. Based on the inoculated cell type, mice were divided into: control SH-SY5Y (five mice), control SK-N-BE (five mice), control IMR-32 (five mice), SIRT4 SH-SY5Y (five mice), SIRT4 SK-N-BE (five mice), and SIRT4 IMR-32 (five mice), this arrangement was used for SIRT4-up group, too. After 10–14 days post-inoculation, mice were sacrificed and tumor tissues were collected and weighed.
Transwell invasion experiment
To determine the migration and invasion ability of the cells, 100 µL of Matrigel (BD354248; BD Biosciences, San Jose, CA, USA) was pipetted into ice-cold 300 µL serum-free medium with an ice-cold pipette and mixed well. An amount of 25 μL of the diluted Matrigel was added to the Transwell plate’s upper chamber (Costar; Corning Incorporated, Corning, NY, USA) and the entire polycarbonate film was coated at 37°C for 30 minutes to allow polymerization of Matrigel. Cells were digested with trypsin (Gibco, Thermo Fisher Scientific), washed with PBS, and resuspended in serum-free DMEM (D6046; Sigma Aldrich Co.). The cell density was adjusted to 0.5×106 cells/mL before plating in a 24-well Transwell upper chamber (Corning Incorporated). Medium containing 20% FBS (Gibco, Thermo Fisher Scientific) was added to Transwell lower chamber and the plate was incubated at 37°C for 24 hours. After incubation, the plate was washed twice with PBS, and methanol was for 30 minutes to fix the cells after which the plate was air dried. Transwell was then stained with crystal violet for 20 minutes and the relative migration was determined by counting the cells under the microscope.
Cell scratch test
Cells (5×105) were seeded in 6-well plate with 2 mL/well. The cell layer was scratched with a tip perpendicular to the bottom of the well. Cells were washed with PBS three times to remove the scratched cells and serum-free DMEM was added. Cells were cultured at 37°C in a 5% CO2 incubator for 24 hours and imaged using a microscope.
Glutamate dehydrogenase (GDH) activity assay
GDH activity was determined using the Glutamate Dehydrogenase Assay Kit (K729-100; Biovision, Milpitas, CA, USA) according to the manufacturer’s instructions.
Mitochondrial respiration
Oxygen consumption rate (OCR) and spare respiratory capacity (SRC) were measured using XFe-24 Extracellular Flux Analyzer (Agilent Technologies, Santa Clara, CA, USA) and XFe-24 Extracellular Flux Assay Kit (B35616; Agilent Technologies) to evaluate the mitochondrial oxidative phosphorylation reaction according to the kits’ instructions.22 SH-SY5Y, SK-N-BE, and IMR-32 cells were used at a density of 2×105/well. The final concentrations of oligomycin, carbonyl cyanide-p-trifluoromethoxyphenylhydrazone and rotenone/antimycin A added were 1.0, 0.5, and 0.5 µM, respectively. After the experiment was completed, the total protein was extracted and the concentration determined. We normalized the OCR/SRC values against the protein concentration.
Statistical analysis
Data were analyzed by SPSS 20.0 software package and expressed as mean ± SD. Student’s t-test, chi-squared test, and Fisher’s exact test were used to compare differences between the groups. Cox regression model was used to test univariate and multivariate analysis for survival times of NB patients. Survival curves of NB patients based on SIRT4 expression were drawn by Kaplan–Meier method. Log-rank test was used to compare differences of survival curves. P<0.05 indicated a significant difference.
Results
SIRT4 expression in NB tissues
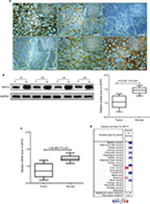
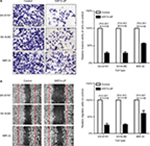
Fifty-two patients had a high expression of SIRT4 protein and 106 patients showed a low expression of SIRT4 protein in NB tissues, while all patients showed high expression of SIRT4 protein in adjacent normal tissues (Figure 1A). The relative expression level of SIRT4 protein in NB tissues was (1.03±0.23), which was significantly lower than that in normal adjacent tissues (1.45±0.13) (Figure 1B). The relative expression level of SIRT4 mRNA in NB tissues was (0.87±0.20), which was significantly lower than that in normal adjacent tissues (1.22±0.09) (Figure 1C). We also searched the ONCOMINE website (https://www.oncomine.com/resource/login.html) for the expression of SIRT4 in other malignant tumor tissues (cancer vs normal). The results showed that SIRT4 was downregulated in most tumor tissues (Figure 1D). However, no information was available on the expression information of SIRT4 in human NB tissues.
Relationship between SIRT4 expression and clinical pathology of NB
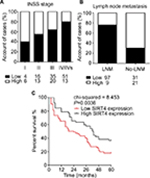
We analyzed the relationship between SIRT4 protein expression and clinicopathological data in NB patients. The results showed that gender, age at onset, pathological type, and pathogenesis of NB patients were not related to SIRT4 expression (P>0.05). International Neuroblastoma Staging System (INSS) stage (P=0.018) and lymph node metastasis of NB patients (P<0.001) were inversely correlated with SIRT4 protein expression (Table 1). With the increase of INSS stage in NB patients, the proportion of patients with low expression of SIRT4 protein was gradually increased (Figure 2A). The proportion of patients with low expression of SIRT4 protein with lymph node metastasis was higher than that in patients without lymph node metastasis (Figure 2B).
  | Table 1 Relationship between SIRT4 expression and clinical pathology of neuroblastoma Abbreviations: INSS, International Neuroblastoma Staging System; NB, neuroblastoma. |
Effect of SIRT4 expression on the prognosis of patients
A total of 158 NB patients were followed-up at least once a month. The factors influencing the survival times of NB patients were analyzed by Cox regression model. The results showed that age (OR =2.739, 95% CI =1.127–12.414), INSS stage (OR =2.612, 95% CI =1.004–10628), and SIRT4 expression level (OR =6.028, 95% CI =3.042–15.917) were independent risk factors influencing the prognosis of NB patients (Table 2). Postoperative survival in NB patients with low expression of SIRT4 was significantly lower than that in patients with high expression of SIRT4 (P=0.0036) (Figure 2C).
Establishment of human NB cell line with upregulated expression of SIRT4
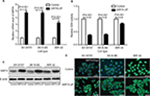
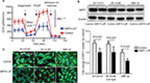
SIRT4 gene expression in SIRT4-expressing cell lines was stably upregulated compared to human NB cell lines transfected with the empty vector (SH-SY5Y, SK-N-BE, and IMR-32), as shown by RT-qPCR, Western blotting, and immunofluorescence, and the gene expression of SIRT4 in SIRT4-over-express cell lines was stably upregulated (Figure 3A, C, and D). It is well-known that upregulation of SIRT4 inhibits GDH activity.23,24 In our study, we also found that SIRT4 significantly inhibited GDH activity in NB cell lines SH-SY5Y, SK-N-BE, and IMR-32 (P<0.001) (Figure 3B).
Effect of SIRT4 expression on the proliferation of human NB cells
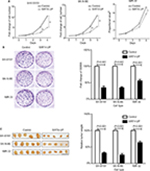
We first investigated the effects of SIRT4 expression on the proliferation of cell lines in vitro and in vivo. The results showed that upregulation of SIRT4 significantly inhibited the cell growth rate (Figure 4A), colony forming ability (Figure 4B), and in vivo tumor growth (Figure 4C) of human NB cell lines (SH-SY5Y, SK-N-BE, and IMR-32).
Effect of SIRT4 expression on the invasion and migration ability of human NB cells
Effect of SIRT4 expression on in vitro migration ability of the cells was detected by cell scratch test. Upregulation of SIRT4 significantly reduced the number of migrating cells in human NB cell lines (SH-SY5Y, SK-N-BE, and IMR-32) (Figure 5A). The effect of SIRT4 expression on the invasiveness of cells was detected by Transwell assay. Upregulation of SIRT4 expression significantly reduced the number of cells in human NB cell lines (SH-SY5Y, SK-N-BE, and IMR-32) that invaded the lower chamber of the Transwell (Figure 5B).
Effect of SIRT4 expression on mitochondrial metabolism in human NB cells
In SH-SY5Y cells, we investigated the effects of SIRT4 expression on mitochondrial OCR and SRC. We found that upregulation of SIRT4 significantly reduced the basic OCR value of SH-SY5Y cells (P<0.001), suggesting that upregulation of SIRT4 inhibited mitochondrial respiration and reduced mitochondrial energy production.22 Simultaneously, upregulation of SIRT4 significantly reduced the SRC value of SH-SY5Y cells (P<0.001), suggesting that upregulation of SIRT4 may reduce the maximum capacity of SH-SY5Y mitochondria energy production and increase mitochondrial damage (Figure 6A).25
Next, we analyzed the effect of SIRT4 on SIRT1 protein expression in human NB cell lines. We found that there was a reduced SIRT1 protein expression in SH-SY5Y, SK-N-BE, and IMR-32 cells overexpressing SIRT4 (Figure 6B and C).
Discussion
In this study, we demonstrated a profound impact of SIRT4 on mitochondrial metabolism and biological functions in NB cells in vitro and in vivo. SIRT4 was expressed at low levels in the human NB tissues, and increased expression of SIRT4 in human NB cell line inhibited the proliferation, invasion, and migration of human NB cells by affecting the mitochondrial function of the cells.
Our findings are consistent with the previous studies showing that SIRT4 acts as a tumor suppressor by modulating mitochondrial glutamine metabolism. SIRT4 is a NAD+-dependent ADP-ribose transferase in mitochondria that catalyzes ADP-ribosylation of a target protein such as GDH. SIRT4 can also regulate fatty acid oxidation and insulin secretion.26 Numerous studies have found that SIRT4 expression is associated with a significant downregulation of malignant tumors such as bladder cancer, breast cancer, colon cancer, gastric cancer, ovarian cancer, thyroid cancer, and leukemia.27,28
This study found that SIRT4 gene expression in human NB tissue was significantly downregulated and decreased with an increase in INSS staging and lymph node metastasis in patients with NB. Moreover, survival time of NB patients with a lower expression of SIRT4 was significantly lower than the patients with high expression of SIRT4. Therefore, the lack of expression of SIRT4 in human NB tissue may provide a more favorable environment for tumor cell proliferation and migration. We confirmed this in our in vitro study using human NB cells. Interestingly, differential expression of SIRT4 was found in cell proliferation and migration ability. In this study, the proliferation, invasion, and migration ability of transfected IMR-32 cells were significantly different from that of SH-SY5Y and SK-N-BE transfected with SIRT4 plasmid. This may be related to the expression of SIRT4, which is consistent with the finding that SIRT4 expression was decreased with the increased INSS stage and lymph node metastasis in NB patients.
It has been reported that SIRT4 expression was low in cancer cells, and its overexpression inhibited cancer cell proliferation and tumor progression.29 Huang et al observed SIRT4 expression by immunofluorescence and compared the histopathological features of the tumors and found that SIRT4 was significantly decreased in gastric cancer, and the degree of expression was inversely related to the extent of tumor differentiation, depth of invasion, and the number of involved lymph nodes.30 Jeong et al reported that SIRT4 could inhibit Myc-induced B-cell lymphoma by inhibiting the metabolism of mitochondrial glutamine.20 Studies have found that after DNA damage, a large number of genotoxic agents induce SIRT4 production, inhibit glutamine uptake into the tricarboxylic acid cycle (TCA) cycle metabolism, inhibit cell cycle, and eventually inhibit tumorigenesis. SIRT4 catalyzes glutamate to form α-ketoglutarate. Studies have shown that SIRT4 plays an important role in α-ketoglutarate-dependent enzymes involved in tumorigenesis, such as TET enzyme and jmjC histone demethylase.31 It was first demonstrated that abnormal epigenetic regulation caused by metabolic disorders eventually caused tumorigenesis due to mutations in isocitrate dehydrogenase.32
The main function of SIRT is to regulate mitochondrial glutamine metabolism, a raw material for α-ketoglutarate in tricarboxylic acid cycle.33 It is the “engine” that provides energy support for cancer cell proliferation, invasion, and migration.34,35 This study found that overexpression of SIRT4 in human NB cells significantly reduced the proliferation, invasion, and migration ability. SIRT4 overexpression has been found to inhibit mitochondrial respiration of NB cells, reduce mitochondrial production, reduce maximal capacity of mitochondria in NB, and increase cell mitochondrial damage. These findings suggest that SIRT4 may be a potential therapeutic target for treating human NB tumors. A study found that loss of SIRT4 resulted in the dysregulation of mTORC1, which caused a loss in the stability of CREB2, a transcriptional mediator of SIRT4, thereby increasing GDH activity and glutamine metabolism.36,37 Glutamine is a metabolite necessary for cell proliferation and facilitating the transition from G1 phase to S phase, and rapid proliferation is one of the characteristics of tumor cell metabolism.38 Therefore, the low expression of SIRT4 in tumor cells promotes glutamine into the TCA cycle, increases cellular energy supply, accelerates cell cycle, and enhances cell proliferation, invasion, and migration.
In addition, our study also found that SIRT4 overexpression significantly reduced SIRT1 protein expression. SIRT1 regulation of tumors through energy metabolism has been confirmed by numerous studies.39,40 Studies have shown that SIRT1, as a new tumor biomarker, regulates the occurrence and development of tumors through various pathways involved in energy metabolism. For example, SIRT1 and PGC-1α promote the development of tumors by interfering with the biosynthesis of mitochondria.41,42 SIRT1 also interferes with the glycolysis of tumor cells by activating HIF-2α, thereby regulating tumor energy metabolism.43
Conclusion
In this study, a lower expression of SIRT4 was found in human NB tissues and tumor cells. Conversely, overexpression of SIRT4 in human NB cells effectively inhibited proliferation, invasion, and migration, which may be related to the role of SIRT4 in regulating mitochondrial glutamine metabolism to exert its tumor-suppressive effects.
Disclosure
The authors report no conflicts of interest in this work
References
Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. | ||
Chen W, Zheng R, Zhang S, et al. Cancer incidence and mortality in China in 2013: an analysis based on urbanization level. Chin J Cancer Res. 2017;29(1):1–10. | ||
Sharp SE, Gelfand MJ, Shulkin BL. Pediatrics: diagnosis of neuroblastoma. Semin Nucl Med. 2011;41(5):345–353. | ||
Marimpietri D, Petretto A, Raffaghello L, et al. Proteome profiling of neuroblastoma-derived exosomes reveal the expression of proteins potentially involved in tumor progression. PLoS One. 2013;8(9):e75054. | ||
Rios P, Bailey HD, Lacour B, et al. Maternal use of household pesticides during pregnancy and risk of neuroblastoma in offspring. A pooled analysis of the ESTELLE and ESCALE French studies (SFCE). Cancer Causes Control. 2017;28(10):1125–1132. | ||
Heck JE, Park AS, Qiu J, Cockburn M, Ritz B. An exploratory study of ambient air toxics exposure in pregnancy and the risk of neuroblastoma in offspring. Environ Res. 2013;127(11):1–6. | ||
Mazul AL, Siega-Riz AM, Weinberg CR, et al. A family-based study of gene variants and maternal folate and choline in neuroblastoma: a report from the Children’s Oncology Group. Cancer Causes Control. 2016;27(10):1209–1218. | ||
Siaw JT, Wan H, Pfeifer K, et al. Brigatinib, an anaplastic lymphoma kinase inhibitor, abrogates activity and growth in ALK-positive neuroblastoma cells, Drosophila and mice. Oncotarget. 2016;7(20):29011–29022. | ||
Umapathy G, Guan J, Gustafsson DE, et al. MEK inhibitor trametinib does not prevent the growth of anaplastic lymphoma kinase (ALK)-addicted neuroblastomas. Sci Signal. 2017;10(507):eaam7550. | ||
Tsuda T, Obara M, Hirano H, et al. Analysis of N-myc amplification in relation to disease stage and histologic types in human neuroblastomas. Cancer. 1987;60(4):820–826. | ||
Tavana O, Li D, Dai C, et al. HAUSP deubiquitinates and stabilizes N-Myc in neuroblastoma. Nat Med. 2016;22(10):1180–1186. | ||
Wang K, Diskin SJ, Zhang H, et al. Integrative genomics identifies LMO1 as a neuroblastoma oncogene. Nature. 2011;469(7329):216–220. | ||
Oldridge DA, Wood AC, Weichert-Leahey N, et al. Genetic predisposition to neuroblastoma mediated by a LMO1 super-enhancer polymorphism. Nature. 2015;528(7582):418–421. | ||
Mcglynn L, Reilly J, Edwards J, Shiels P, et al. The Role of SIRT2, SIRT3 and SIRT4 in Breast Cancer. Cancer Res. 2009;69(24 Supplement):3032–3032. | ||
Kim EJ, Juhnn YS. Cyclic AMP signaling reduces sirtuin 6 expression in non-small cell lung cancer cells by promoting ubiquitin-proteasomal degradation via inhibition of the Raf-MEK-ERK (Raf/mitogen-activated extracellular signal-regulated kinase/extracellular signal-regulated kinase) pathway. J Biol Chem. 2015;290(15):9604–9613. | ||
Igci M, Kalender ME, Borazan E, et al. High-throughput screening of Sirtuin family of genes in breast cancer. Gene. 2016;586(1):123–128. | ||
Jeong SM, Xiao C, Finley LW, et al. SIRT4 has tumor-suppressive activity and regulates the cellular metabolic response to DNA damage by inhibiting mitochondrial glutamine metabolism. Cancer Cell. 2013;23(4):450–463. | ||
Huang G, Cheng J, Yu F, et al. Clinical and therapeutic significance of sirtuin-4 expression in colorectal cancer. Oncol Rep. 2016;35(5):2801–2810. | ||
Shen X, Li P, Xu Y, et al. Association of sirtuins with clinicopathological parameters and overall survival in gastric cancer. Oncotarget. 2017;8(43):74359. | ||
Jeong SM, Lee A, Lee J, Haigis MC. SIRT4 protein suppresses tumor formation in genetic models of Myc-induced B cell lymphoma. J Biol Chem. 2014;289(7):4135–4144. | ||
Shi Q, Liu T, Zhang X, et al. Decreased sirtuin 4 expression is associated with poor prognosis in patients with invasive breast cancer. Oncol Lett. 2016;12(4):2606–2612. | ||
Liu TF, Vachharajani V, Millet P, Tie Fu L, Vidula V, Patrick M, et al. Sequential actions of SIRT1-RELB-SIRT3 coordinate nuclear-mitochondrial communication during immunometabolic adaptation to acute inflammation and sepsis. J Biol Chem. 2015;290(1):396–408. | ||
Haigis MC, Mostoslavsky R, Haigis KM, et al. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic beta cells. Cell. 2006;126(5):941–954. | ||
Komlos D, Mann KD, Zhuo Y, et al. Glutamate dehydrogenase 1 and SIRT4 regulate glial development. Glia. 2013;61(3):394–408. | ||
Desler C, Hansen TL, Frederiksen JB, et al. Is There a Link between Mitochondrial Reserve Respiratory Capacity and Aging? J Aging Res. 2012;2012(4):192503. | ||
Nasrin N, Wu X, Fortier E, et al. SIRT4 regulates fatty acid oxidation and mitochondrial gene expression in liver and muscle cells. J Biol Chem. 2010;285(42):31995–32002. | ||
Miyo M, Yamamoto H, Konno M, et al. Tumour-suppressive function of SIRT4 in human colorectal cancer. Br J Cancer. 2015;113(3):492–499. | ||
Wang Q, Wen YG, Li DP, Dp L, et al. Upregulated INHBA expression is associated with poor survival in gastric cancer. Med Oncol. 2012;29(1):77–83. | ||
Fernandez-Marcos PJ, Serrano M. Sirt4: the glutamine gatekeeper. Cancer Cell. 2013;23(4):427–428. | ||
Huang G, Cui F, Yu F, et al. Sirtuin-4 (SIRT4) is downregulated and associated with some clinicopathological features in gastric adenocarcinoma. Biomed Pharmacother. 2015;72:135–139. | ||
Xiao M, Yang H, Xu W, et al. Inhibition of α-KG-dependent histone and DNA demethylases by fumarate and succinate that are accumulated in mutations of FH and SDH tumor suppressors. Genes Dev. 2012;26(12):1326–1338. | ||
Lu C, Ward PS, Kapoor GS, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483(7390):474–478. | ||
Le A, Lane AN, Hamaker M, et al. Glucose-independent glutamine metabolism via TCA cycling for proliferation and survival in B cells. Cell Metab. 2012;15(1):110–121. | ||
Wise DR, Thompson CB. Glutamine addiction: a new therapeutic target in cancer. Trends Biochem Sci. 2010;35(8):427–433. | ||
Son J, Lyssiotis CA, Ying H, et al. Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature. 2013;496(7443):101–105. | ||
Csibi A, Fendt SM, Li C, et al. The mTORC1 pathway stimulates glutamine metabolism and cell proliferation by repressing SIRT4. Cell. 2013;153(4):840–854. | ||
Menon S, Manning BD. Common corruption of the mTOR signaling network in human tumors. Oncogene. 2008;27(2):S43–S51. | ||
Colombo SL, Palacios-Callender M, Frakich N, et al. Molecular basis for the differential use of glucose and glutamine in cell proliferation as revealed by synchronized HeLa cells. Proc Natl Acad Sci U S A. 2011;108(52):21069–21074. | ||
Anastasiou D, Krek W. SIRT1: linking adaptive cellular responses to aging-associated changes in organismal physiology. Physiology. 2006;21(6):404–410. | ||
Kim EJ, Um SJ. SIRT1: roles in aging and cancer. BMB Rep. 2008;41(11):751–756. | ||
Aquilano K, Vigilanza P, Baldelli S, et al. Peroxisome proliferator-activated receptor gamma co-activator 1alpha (PGC-1alpha) and sirtuin 1 (SIRT1) reside in mitochondria: possible direct function in mitochondrial biogenesis. J Biol Chem. 2010;285(28):21590–21599. | ||
Rodgers JT, Lerin C, Haas W, et al. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434(7029):113–118. | ||
Dioum EM, Chen R, Alexander MS, et al. Regulation of hypoxia-inducible factor 2alpha signaling by the stress-responsive deacetylase sirtuin 1. Science. 2009;324(5932):1289–1293. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.