Back to Journals » Infection and Drug Resistance » Volume 13
Transmitted and Acquired HIV-1 Drug Resistance from a Family: A Case Study
Authors Yan L, Yu F, Zhang H, Zhao H, Wang L , Liang Z, Zhang X, Wu L, Liang H, Yang S, Tang Y, Zhang F
Received 16 July 2020
Accepted for publication 11 September 2020
Published 22 October 2020 Volume 2020:13 Pages 3763—3770
DOI https://doi.org/10.2147/IDR.S272232
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Suresh Antony
Liting Yan,1,2,* Fengting Yu,1,2,* Huimin Zhang,1,3 Hongxin Zhao,1,2 Linghang Wang,1,2 Zaiyan Liang,1,2 Xia Zhang,1,2 Liang Wu,1,2 Hongyuan Liang,1,2 Siyuan Yang,1,2 Yunxia Tang,1,2 Fujie Zhang1,2
1Beijing Ditan Hospital, Capital Medical University, Beijing, People’s Republic of China; 2Clinical Center for HIV/AIDS, Capital Medical University, Beijing, People’s Republic of China; 3Department of Pediatrics, Beijing Ditan Hospital, Beijing, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Fujie Zhang
Beijing Ditan Hospital, Capital Medical University, Beijing 100015, People’s Republic of China
Tel +86 10 84322581
Email [email protected]
Abstract: Antiretroviral drug resistance has become a major threat to the adequate management of human immunodeficiency virus (HIV) infection, but little attention has been paid to the spread and evolution of drug-resistant strains in the family. Here, we described a case of transmitted as well as acquired HIV drug resistance among a father, mother and infant. Epidemiological data were obtained retrospectively. Drug resistance mutations (DRMs) of three patients were tested using a validated In-house Sanger-based sequencing (SBS) method and the Vela next-generation sequencing (NGS) platform. Gene evolution analysis was also performed. According to the epidemiological history and phylogenetic data, in late pregnancy of the mother, the infant’s father transmitted HIV-1 to her, and then the mother to the baby, leading to the transmission of V106I as a common mutation of three persons. The mutant frequency was 99.57% (father), 95.38% (mother) and 99.73% (infant), respectively. Mother also acquired K101E (41.03%), K103N (27.56%) and minor mutation of V106M (4.30%) after improperly discontinuing antiretroviral regimen of lamivudine (3TC), tenofovir (TDF) and efavirenz (EFV). Such acquired mutations increased the drug resistance scores on non-nucleoside reverse transcriptase inhibitors (NNRTIs) doravirine, EFV, etravirine, nevirapine and rilpivirine from 10, 0, 10, 10 and 10 to 65, 135, 25, 150 and 55, respectively. Therefore, sexually transmitted diseases, especially DRMs of HIV-1 in families, are of concern and draw attention to the need for enhanced drug-resistance prevention efforts, and accurate surveillance by more sensitive methods in complicated cases.
Keywords: HIV-1, drug resistance, minor mutation, sexually transmitted diseases, next-generation sequencing
Introduction
The emergence of drug resistance has become a very real threat to the adequate management of human immunodeficiency virus (HIV) infection worldwide.1,2 Drug resistance mutation (DRM) is acquired due to viral replication and selective pressure in patients receiving antiretroviral therapy (ART), but can be transmitted to drug naïve persons newly infected with HIV.3,4 In the case of pregnant women, the mother-to-child transmission of DRM strains can complicate the infant management.5
All current antiretroviral drugs are at risk of emerging DRM.1 For the commonly used alternative first-line regimen based on the non-nucleoside reverse transcriptase inhibitor (NNRTI) efavirenz (EFV) in combination with two nucleoside reverse transcriptase inhibitors (NRTIs), usually tenofovir (TDF) and either lamivudine (3TC) or emtricitabine,6 DRM is more common in EFV.1,7 A major disadvantage of EFV is the low genetic barrier that certain single amino acid mutation, such as K103N, V106M, can cause a high level of drug resistance.8 Furthermore, poor adherence leads to insufficient medication exposure and selection pressure of EFV, which increases the risk of DRM.9 The plasma terminal half-life of 3TC and TDF are approximately 5 to 7 hours and 17 hours, respectively,10,11 while EFV averages 52 to 76 hours after a single dose and 40 to 55 hours after multiple doses,12 and even 136 hours in some individuals.13 Long half-life thus results in EFV monotherapy with more rapidly clearance of other drugs after treatment interruption and causes a high-risk context for the emergence of DRM.
Importantly, development of DRM can reduce the virological response,14 increase the probability of transmission of resistant viruses to people newly infected with HIV,15,16 and even lead to multidrug resistance in naïve patients.17 Therefore, genotyping resistance testing is recommended as a routine laboratory monitoring before starting ART.18,19 Sanger-based sequencing (SBS), the most commonly used method to diagnose HIV drug resistance,20 was reported to have high repeatability and interpretability.21 However, only virus strains with mutation frequency more than 10% to 20% can be detected by direct sequencing,22 and the minority variants missed by standard genotyping may be lead to viral failure.23–25 Thus, more sensitive methods of drug resistance testing are necessary to detect minority variants, so as to better guide clinical decision-making.
Here, we reported a case of transmitted HIV-1 drug resistance from a family as well as acquired drug resistance after improper discontinuation of 3TC, TDF and EFV, and analyzed the application of Vela next-generation sequencing (NGS) platform in detection of minority variants in complex cases.
Case Description
Patient M (Infant’s mother), a 27-year-old woman, was tested HIV-negative at 12 and 32 weeks of gestation, respectively (Figure 1), but was suspected HIV-positive in a local hospital on July 7, 2019 (one day before delivery) and was confirmed five days after childbirth by local Centers for Disease Control. She had no sexual history before marriage and also had no history of intravenous drug use, surgical trauma or blood transfusion. She and her husband did not screen for sexually transmitted diseases (STDs) before pregnancy but had unprotected sexual contact at 32 weeks of gestation. Patient M stopped breastfeeding after the diagnosis and initiated on a first-line regimen of 3TC, TDF and EFV on September 15, 2019. However, she took only eight days of antiviral drugs and stopped them simultaneously because of a severe systemic skin allergy. Then, she presented to our clinic on November 12. The viral load (VL) was tested to be 17,500 copies/mL and CD4+T was 306 cells/ul.
 |
Figure 1 Summary of the case. |
Her contact, Patient F (Infant’s father) was a 30-year-old man and had a history of sexual contact with other female before marriage. However, he was confirmed HIV-positive for the first time after patient M was diagnosed. Baseline bloods showed VL 8920 copies/mL and CD4+T 130 cells/ul.
Their infant, patient I, was born naturally at full term on July 8, 2019. He was found to be positive for HIV antibody at 42 and 53 days after birth, respectively. Then, presented to our clinic on October 10. Baseline bloods showed VL 311,620 copies/mL and CD4+T 1469 cells/ul.
Methods
Data Source and Genotype Analysis
Clinical data and blood samples were obtained between October 10 and November 12, 2019. Genotypic resistance mutation analysis was performed using a validated In-house SBS method. In this reaction, the entire protease gene and the first 300 codons of reverse transcriptase gene, as well as the entire integrase gene were amplified, respectively. Stanford HIV-1 drug resistance database (HIVdb version 8.9–1) was used to analyze the mutations and generate the final reports including drug resistance scores, with 0–9 for susceptible, 10–14 for potential low-level resistance, 15–29 for low-level resistance, 30–59 for intermediate resistance, and ≥ 60 for high-level resistance.
Then, the Vela NGS-based HIV-1 DRM test was also performed. HIV RNA was extracted from plasma sample and was used for one-step reverse transcription-polymerase chain reaction. The amplicons were enzymatically fragmented and subjected to NGS on the Sentosa® SQ301 sequencing instrument. Subsequently, Sentosa® SQ Suite software performed primary analysis on the raw sequencing data and then transferred to Sentosa® SQ Reporter Server for secondary analysis and report generation.
Gene Evolution Analysis
Homologous sequence comparison was analyzed by Basic Local Alignment Search Tool (BLAST, http://blast.ncbi.nlm.nih.gov) using HIV-1 pol gene obtained by In-house method. And these pol sequences from three patients were uploaded to the GenBank website (https://www.ncbi.nlm.nih.gov/genbank/; Accession numbers: MT939518, MT939519 and MT939520, respectively). The reference sequences of other subtypes were downloaded from Genbank. Phylogenetic tree was constructed using MEGA (Version 7.0, USA) software and the between group mean distance as well as bootstrapping test were performed.
Results
Three Patients Had a Common Transmitted Mutation of V106I
Patient I was firstly performed DRM testing using SBS method on October 14 before starting ART and was found to be subtype B infection and V106I mutation (Table 1). Then, patient M and patient F were performed DRM testing on November 12. Both of them were subtype B and had no resistance mutation to NRTIs, protease inhibitors or integrase inhibitors, but had a common V106I mutation. Patient F also had an E138A mutation. Subsequently, the DRMs were confirmed using Vela NGS method and the mutant frequency of V106I for three patients were 99.73% (Patient I), 95.38% (Patient M) and 99.57% (Patient F), respectively. The mutant frequency of E138A was 67.78% and was not transmitted to patient M.
 |
Table 1 Genotype Analysis of Drug Resistance Mutation |
To determine the evolutionary relationship of HIV strains in three patients, phylogenetic tree analysis were performed (Figure 2). These three patients were shown to be subtype B and the genetic distance among them were 0.0017 (mother and infant), 0.0075 (mother and father) and 0.0083 (father and infant), respectively. Moreover, the virus they infected came from a homologous strain and the percent identity was 97.33% (patient M), 97.52% (Patient I) and 97.94% (Patient F). Surprisingly, the homologous strain itself had V106I mutation according to the Stanford HIV-1 drug resistance database.
 |
Figure 2 Gene evolution analysis of patients. |
Patient M Acquired K101E, K103N and Minor Mutation of V106M After Improperly Discontinuing Antiretroviral Drugs
Patient I and patient F tested for DRM before the initiation of ART. However, patient M took eight days’ first-line regimen (3TC, TDF and EFV) two months before presented to our clinic. In addition to the transmitted drug resistance of V106I mutation, she acquired K101E and K103N mutations with mutant frequency of 41.03% and 27.56%, respectively. Subsequently, the sensitivity comparison of In-house SBS and Vela NGS methods to detect minority variants of HIV-1 DRM was performed. As it turned out, a minor V106M mutation with a frequency of 4.30% was found by NGS platform in patient M but not identified by SBS method.
Acquired Mutations Aggravate the Drug Resistance Scores of Patient M
These five DRMs have varying degrees of effect on NNRTIs based on the mutation scores (Table 2). V106I mutation shows susceptible to EFV (0), and potential low-level resistance to doravirine (DOR, 10), etravirine (ETR, 10), nevirapine (NVP,10) and rilpivirine (RPV, 10), which increases to low-level resistance to ETR (20) and RPV (25) when E138A is also present. However, for patient M, V106M alone is associated with a high-level reduction in NVP (60) and EFV (60) susceptibility as well as an intermediate reduction in DOR (50) susceptibility, while together with V106I, especially K101E and K103N, the mutation scores are increased to 65, 135, 25, 150 and 55 for DOR, EFV, ETR, NVP and RPV.
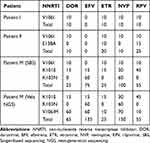 |
Table 2 Mutation Scores of Non-Nucleoside Reverse Transcriptase Inhibitors |
Discussion
Antiretroviral drug resistance becomes a major threat for the adequate management of HIV infection.2 Resistant mutation in the HIV genome results in hard-to-treat infections, increases health-care costs,1 and can be transmitted from person to person,26 as described in this case. Uniquely, for the first time, we proposed the necessity of screening for STDs in both couples at the same time during pregnancy testing and also emphasized the importance of monitoring for minor mutations in complex cases by NGS method.
Currently, the guidelines recommend that pregnant women should screen for STDs at the first prenatal visit, including HIV, a serologic test for syphilis, hepatitis B surface antigen, hepatitis C antibodies and Neisseria gonorrhoeae,27,28 while no recommendation for simultaneous screening for sexual partners. However, STDs can also occur during pregnancy as well as lead to intrauterine or perinatal transmission. In this case, patient F was infected with the V106I mutant of HIV-1 according to phylogenetic tree analysis and developed to chronic infection status without knowing it. He transmitted the drug-resistant mutant viruses to patient M because he did not screen for STDs before or after his wife became pregnant. To make matters worse, the HIV programs to prevent mother-to-child transmission have failed, probably mainly due to the limitations of local medical conditions. This leads to the spread of HIV-1 mutant strains in this family, and causes a serious public health problem, which has far-reaching consequences for the long-term health and health care costs.29 But worse, the overall incidence of STDs has been increasing in more recent years.30,31 In China, an average of more than 1 baby per hour was reported to be born with congenital syphilis in 2008, for a total of 9480 cases.32 Therefore, the case we described maybe just the epitome of many families with STDs.
The common V106I mutant of three patients, transmitted through sexual transmission and mother-to-child transmission, respectively, is also a strong evidence of how DRM spreads in HIV-1 naïve individuals. In contrast to transmitted drug resistance, acquired drug resistance develops because of viral replication and selective pressure in patients receiving ART.1,4 Patient M took eight days of 3TC, TDF and EFV and stopped three drugs simultaneously because of a severe systemic skin allergy, which may result in EFV monotherapy due to its longer half-life.10–12 As expected, she rapidly acquired K101E, K103N and V106M mutations.
The proportion of wild type virus, and the resistant viruses evolved under the sustained selective pressure of drugs is different in the presence and absence of ART.33 Persistent viremia in the presence of therapy leads to further accumulation of mutations and reduces replication of the wild type virus, which increases resistance.4 However, the discontinuation of ART when viral suppression was not achieved causes a relatively fast decay of mutants, and the rapid re-emergence of wild type virus for their higher fitness or replication capacity without drug selective pressure.34,35 Patient M performed genotypic resistance mutation testing two months after the cessation of ART, when the circulating viruses may have changed, so the mutants frequency of K101E and K103N were 41.03% and 27.56%, respectively. Long detection time interval after drug withdrawal may even cause resistant strains to decline to levels undetectable by conventional In-house SBS approach. It must be noted, however, such minor mutant can also contribute to the failure of salvage regimens.24,36,37 Consequently, we compared the sensitivity of In-house SBS and NGS methods to detect minority variants of HIV-1 DRM, and a minor V106M mutation with a frequency of 4.30% was found by NGS method in patient M but not identified by SBS, which is associated with high-level reductions in NVP and EFV susceptibility.
Our study also supports the recommendation of WHO guidelines that dolutegravir together with two NRTIs should be the preferred first-line regimen for HIV-infected adults initiating ART,38 especially in resource limited settings. For the reason that resistance to NRTIs and integrase inhibitors was relatively stable, while resistance to NNRTIs increased significantly in the past few years.1 However, in combination with our recommendations, their own economic conditions and local health insurance policies, patient M chose the national free antiviral drugs 3TC/TDF/lopinavir and ritonavir, patient F chose elvitegravir/cobicistat/emtricitabine/tenofovir alafenamide regimen together with compound sulfamethoxazole for CD4+T was 130 cells/ul, patient I was given 3TC/abacavir/lopinavir and ritonavir regimen to return to the local home for treatment.
Conclusion
In summary, simultaneous screening of STDs for both husband and wife during pregnancy testing is necessary. Besides, in some complicated cases like intermittent ART, the Vela NGS platform has an advantage in identifying minority variants of HIV-1 drug resistance than the In-house SBS method.
Abbreviations
HIV, human immunodeficiency virus; DRM, drug resistance mutation; SBS, Sanger-based sequencing; NGS, next-generation sequencing; 3TC, lamivudine; TDF, tenofovir; EFV, efavirenz; NNRTIs, non-nucleoside reverse transcriptase inhibitors; NRTIs, nucleoside reverse transcriptase inhibitors; ART, antiretroviral therapy; STDs, sexually transmitted diseases; VL, viral load; DOR, doravirine; ETR, etravirine; NVP, nevirapine; RPV, rilpivirine; PR, protease; PI, protease inhibitor; IN, integrase; RT, reverse transcriptase.
Ethics and Consent
This study was part of a research project that had been approved by the ethics committee of Beijing Ditan Hospital of Capital Medical University (Approval number: 2019-037-002). Two adult patients provided written informed consent and signed consent for their infant, and they agreed to publish details of the three cases.
Acknowledgment
The authors would like to acknowledge Beijing Ditan Hospital, Capital Medical University and the Beijing Key Laboratory of Emerging Infectious Diseases to support the study. We also thank all the implementing partners of Vela company for providing technical guidance on second-generation sequencing. L.Y. and F.Y. are co-first authors for this study.
Funding
This study was supported by the National Science and Technology Major Project, Ministry of Science and Technology of China (grant number 2018ZX10302-102).
Disclosure
The authors report no conflicts of interest in this work.
References
1. WHO. HIV drug resistance report 2019. Geneva: World Health Organization; 2019. Available from: https://www.who.int/hiv/pub/drugresistance/hivdr-report-2019/en/.
2. Beyrer C, Pozniak A. HIV drug resistance - an emerging threat to epidemic control. N Engl J Med. 2017;377(17):1605–1607. doi:10.1056/NEJMp1710608
3. Clavel F, Hance AJ. HIV drug resistance. N Engl J Med. 2004;350(10):1023–1035. doi:10.1056/NEJMra025195
4. Paredes R, Clotet B. Clinical management of HIV-1 resistance. Antiviral Res. 2010;85(1):245–265. doi:10.1016/j.antiviral.2009.09.015
5. Yeganeh N, Kerin T, Ank B, et al. Human immunodeficiency virus antiretroviral resistance and transmission in mother-infant pairs enrolled in a large perinatal study. Clin Infect Dis. 2018;66(11):1770–1777. doi:10.1093/cid/cix1104
6. WHO. Consolidated guidelines on the use of antiretroviral drugs for treating and preventing HIV infection: recommendations for a public health approach. Geneva: World Health Organization; 2016. Available from: https://www.who.int/hiv/pub/arv/arv-2016/en/.
7. Phillips AN, Cambiano V, Nakagawa F, et al. Cost-effectiveness of public-health policy options in the presence of pretreatment NNRTI drug resistance in sub-Saharan Africa: a modelling study. Lancet HIV. 2018;5(3):e146–e154. doi:10.1016/s2352-3018(17)30190-x
8. Young SD, Britcher SF, Tran LO, et al. L-743, 726 (DMP-266): a novel, highly potent nonnucleoside inhibitor of the human immunodeficiency virus type 1 reverse transcriptase. Antimicrob Agents Chemother. 1995;39(12):2602–2605. doi:10.1128/aac.39.12.2602
9. Wallis CL, Godfrey C, Fitzgibbon JE, Mellors JW. Key factors influencing the emergence of human immunodeficiency virus drug resistance in low- and middle-income countries. J Infect Dis. 2017;216(suppl_9):S851–s856. doi:10.1093/infdis/jix409
10. Johnson MA, Moore KH, Yuen GJ, Bye A, Pakes GE. Clinical pharmacokinetics of lamivudine. Clin Pharmacokinet. 1999;36(1):41–66. doi:10.2165/00003088-199936010-00004
11. Kearney BP, Flaherty JF, Shah J. Tenofovir disoproxil fumarate: clinical pharmacology and pharmacokinetics. Clin Pharmacokinet. 2004;43(9):595–612. doi:10.2165/00003088-200443090-00003
12. Vrouenraets SM, Wit FW, van Tongeren J, Lange JM. Efavirenz: a review. Expert Opin Pharmacother. 2007;8(6):851–871. doi:10.1517/14656566.8.6.851
13. Delobel P, Saliou A, Nicot F, et al. Minor HIV-1 variants with the K103N resistance mutation during intermittent efavirenz-containing antiretroviral therapy and virological failure. PLoS One. 2011;6(6):e21655. doi:10.1371/journal.pone.0021655
14. Wittkop L, Gunthard HF, de Wolf F, et al. Effect of transmitted drug resistance on virological and immunological response to initial combination antiretroviral therapy for HIV (EuroCoord-CHAIN joint project): a European multicohort study. Lancet Infect Dis. 2011;11(5):363–371. doi:10.1016/s1473-3099(11)70032-9
15. Vercauteren J, Wensing AM, van de Vijver DA, et al. Transmission of drug-resistant HIV-1 is stabilizing in Europe. J Infect Dis. 2009;200(10):1503–1508. doi:10.1086/644505
16. Wheeler WH, Ziebell RA, Zabina H, et al. Prevalence of transmitted drug resistance associated mutations and HIV-1 subtypes in new HIV-1 diagnoses, U.S.-2006. AIDS. 2010;24(8):1203–1212. doi:10.1097/QAD.0b013e3283388742
17. Markowitz M, Mohri H, Mehandru S, et al. Infection with multidrug resistant, dual-tropic HIV-1 and rapid progression to AIDS: a case report. Lancet. 2005;365(9464):1031–1038. doi:10.1016/s0140-6736(05)71139-9
18. Hirsch MS, Gunthard HF, Schapiro JM, et al. Antiretroviral drug resistance testing in adult HIV-1 infection: 2008 recommendations of an International AIDS Society-USA panel. Clin Infect Dis. 2008;47(2):266–285. doi:10.1086/589297
19. Saag MS, Benson CA, Gandhi RT, et al. Antiretroviral drugs for treatment and prevention of HIV infection in adults: 2018 recommendations of the International Antiviral Society-USA panel. JAMA. 2018;320(4):379–396. doi:10.1001/jama.2018.8431
20. Moscona R, Ram D, Wax M, et al. Comparison between next-generation and Sanger-based sequencing for the detection of transmitted drug-resistance mutations among recently infected HIV-1 patients in Israel, 2000–2014. J Int AIDS Soc. 2017;20(1):21846. doi:10.7448/ias.20.1.21846
21. Tzou PL, Ariyaratne P, Varghese V, et al. Comparison of an in vitro diagnostic next-generation sequencing assay with sanger sequencing for HIV-1 genotypic resistance testing. J Clin Microbiol. 2018;56(6):e00105–00118. doi:10.1128/jcm.00105-18
22. Halvas EK, Aldrovandi GM, Balfe P, et al. Blinded, multicenter comparison of methods to detect a drug-resistant mutant of human immunodeficiency virus type 1 at low frequency. J Clin Microbiol. 2006;44(7):2612–2614. doi:10.1128/jcm.00449-06
23. Dykes C, Najjar J, Bosch RJ, et al. Detection of drug-resistant minority variants of HIV-1 during virologic failure of indinavir, lamivudine, and zidovudine. J Infect Dis. 2004;189(6):1091–1096. doi:10.1086/382033
24. Lecossier D, Shulman NS, Morand-Joubert L, et al. Detection of minority populations of HIV-1 expressing the K103N resistance mutation in patients failing nevirapine. J Acquir Immune Defic Syndr. 2005;38(1):37–42. doi:10.1097/00126334-200501010-00007
25. Halvas EK, Wiegand A, Boltz VF, et al. Low frequency nonnucleoside reverse-transcriptase inhibitor-resistant variants contribute to failure of efavirenz-containing regimens in treatment- experienced patients. J Infect Dis. 2010;201(5):672–680. doi:10.1086/650542
26. Stekler JD, Milne R, Payant R, et al. Transmission of HIV-1 drug resistance mutations within partner-pairs: A cross-sectional study of a primary HIV infection cohort. PLoS Med. 2018;15(3):e1002537. doi:10.1371/journal.pmed.1002537
27. Workowski K, Bolan G; Prevention. CfDCa. Sexually transmitted diseases treatment guidelines, 2015. MMWR Recomm Rep. 2015;64(RR–03):1–137.
28. Owens DK, Davidson KW, Krist AH, et al. Screening for hepatitis B virus infection in pregnant women: US preventive services task force reaffirmation recommendation statement. JAMA. 2019;322(4):349–354. doi:10.1001/jama.2019.9365
29. Workowski KA, Berman SM. Centers for disease control and prevention sexually transmitted diseases treatment guidelines. Clin Infect Dis. 2007;44(Suppl 3):S73–S76. doi:10.1086/511430
30. Cingolani A, Zona S, Girardi E, et al. Increased incidence of sexually transmitted diseases in the recent years: data from the ICONA cohort. J Int AIDS Soc. 2014;17(4 Suppl 3):19653. doi:10.7448/ias.17.4.19653
31. Cingolani A, Zona S, Girardi E, et al. Incidence and factors associated with the risk of sexually transmitted diseases in HIV-infected people seen for care in Italy: data from the Icona Foundation cohort. HIV Med. 2015;16(7):412–420. doi:10.1111/hiv.12226
32. Tucker JD, Chen XS, Peeling RW. Syphilis and social upheaval in China. N Engl J Med. 2010;362(18):1658–1661. doi:10.1056/NEJMp0911149
33. Tang JW, Pillay D. Transmission of HIV-1 drug resistance. J Clin Virol. 2004;30(1):1–10. doi:10.1016/j.jcv.2003.12.002
34. Devereux HL, Youle M, Johnson MA, Loveday C. Rapid decline in detectability of HIV-1 drug resistance mutations after stopping therapy. AIDS. 1999;13(18):F123–F127. doi:10.1097/00002030-199912240-00001
35. Izopet J, Massip P, Souyris C, et al. Shift in HIV resistance genotype after treatment interruption and short-term antiviral effect following a new salvage regimen. AIDS. 2000;14(15):2247–2255. doi:10.1097/00002030-200010200-00005
36. Charpentier C, Dwyer DE, Mammano F, et al. Role of minority populations of human immunodeficiency virus type 1 in the evolution of viral resistance to protease inhibitors. J Virol. 2004;78(8):4234–4247. doi:10.1128/jvi.78.8.4234-4247.2004
37. Johnson JA, Li JF, Wei X, et al. Minority HIV-1 drug resistance mutations are present in antiretroviral treatment-naive populations and associate with reduced treatment efficacy. PLoS Med. 2008;5(7):e158. doi:10.1371/journal.pmed.0050158
38. WHO. Update of recommendations on first-and second-line antiretroviral regimens. Geneva: World Health Organization; 2019. Available from: https://www.who.int/hiv/pub/guidelines/en/.
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
