Back to Journals » Journal of Inflammation Research » Volume 16
Transcriptomics Analysis Revealed Key Genes Associated with Macrophage Autophagolysosome in Male ApoE−/− Mice Aortic Atherosclerosis
Authors Zhu M , Jin T, Wu D, Zhang S , Wang A
Received 16 June 2023
Accepted for publication 14 October 2023
Published 9 November 2023 Volume 2023:16 Pages 5125—5144
DOI https://doi.org/10.2147/JIR.S426155
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Ning Quan
Meirong Zhu,1,2 Tongyu Jin,1 Ding Wu,3 Shanchao Zhang,4,* Aihua Wang1,4,*
1Department of Neurology, Shandong Provincial Qianfoshan Hospital, Shandong University, Jinan, People’s Republic of China; 2Department of Critical Medicine, Jinan Central Hospital, Jinan, People’s Republic of China; 3Vascular Surgery, Jinan Central Hospital, Jinan, People’s Republic of China; 4Department of Neurology, The First Affiliated Hospital of Shandong First Medical University & Shandong Provincial Qianfoshan Hospital, Shandong First Medical University, Jinan, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Aihua Wang, Department of Neurology, Shandong Provincial Qianfoshan Hospital, Shandong University, Jinan, People’s Republic of China, Tel +8613791120819, Email [email protected] Shanchao Zhang, Department of Neurology, The First Affiliated Hospital of Shandong First Medical University & Shandong Provincial Qianfoshan Hospital, Shandong First Medical University, Jinan, People’s Republic of China, Tel +8618678295562, Email [email protected]
Purpose: Atherosclerosis (AS) is the most common cause of cardiovascular and cerebrovascular diseases. However, the mechanisms underlying atherosclerotic plaque progression remain unclear. This study aimed to investigate the genes associated with the development of atherosclerosis in the aorta of ApoE−/− male mice, which could serve as novel biomarkers and therapeutic targets in interventions to halt plaque progression.
Methods: Eight-week-old ApoE−/− mice were fed a normal purified laboratory diet or a Western Diet (WD) for 6 or 22 weeks. High-throughput sequencing technology was used to analyze the transcriptomes of the aortas of four groups of mice that were exposed to different dietary conditions. We retrieved and downloaded the human Arteriosclerosis Disease Chip dataset GSE100927 from the Gene Expression Omnibus (GEO) database and selected 29 cases of carotid atherosclerotic lesions and 12 cases of normal carotid tissues as the experimental and control groups, respectively, to further verify our dataset. In addition, we used quantitative reverse transcription polymerase chain reaction (QT-PCR) to verify the expression levels of the core genes in an atherosclerosis mouse model.
Results: There were 265 differentially expressed genes (DEGs) between the ApoE−/− Male mice AS22W group and Sham22W group. In addition to the well-known activation of inflammation and immune response, t the autophagy-lysosome system is also an important factor that affects the development of atherosclerosis. We identified five core genes (Atp6ap2, Atp6v0b, Atp6v0d2, Atp6v1a, and Atp6v1d) in the protein-protein interaction (PPI) network that were closely related to autophagosomes. Hub genes were highly expressed in the carotid atherosclerosis group in the GSE100927 dataset (P < 0.001). QT-PCR showed that the RNA level of Atp6v0d2 increased significantly during the development of atherosclerotic plaque in ApoE−/− male mice.
Conclusion: Five core genes which affect the development of aortic atherosclerosis through the autophagy-lysosome system, especially Atp6v0d2, were screened and identified using bioinformatic techniques.
Keywords: molecular mechanism, bioinformatics, core gene, lysosome, aortic plaque development
Introduction
Atherosclerosis (AS) corresponds to the formation of fibrofatty lesions in the arterial wall and is the pathological basis of myocardial infarction, stroke, and disabling peripheral arterial diseases.1 The AS plaque formation process involves endothelial cell injury, oxygen free radical damage, lipid aggregation, foam cell formation, platelet adhesion aggregation, inflammatory cell migration and infiltration, vascular smooth muscle cell calcium overload, pathological angiogenesis, and local thrombus formation; however, its pathogenic mechanism has not been fully elucidated.2 Recent studies have shown that the autophagolysosomal system plays an important role in macrophage lipid metabolism in AS plaques.3,4 Macrophages play a central role in the progression of atherosclerosis. They develop progressive autophagic dysfunction in the developing plaques. Autophagy dysfunction triggering involves a critical factor that appears to affect the functional st atus of macrophage lysosomes.5,6 Oxidized low-density lipoprotein (LDL) and cholesterol oxidation products (ChOx) can damage the macrophage lysosomal membrane, leading to the inactivation or relocalization of some lysosomal enzymes, which in turn leads to the leakage of lysosomal contents and macrophage death. Lipid-induced lysosomal dysfunction in plaque macrophages is an essential factor in autophagy defects.7 Stimulation of autophagy-lysosomal biogenesis can reduce atherosclerosis.
High-throughput gene mapping has become an effective tool to reveal the pathogenesis of cardiovascular diseases, which provides a new direction for the study of cardiovascular diseases in the future. But at present, most of these studies have focused on comparing the differential gene expression between human atherosclerotic plaques and normal tissues. ApoE−/− mice readily form lesions similar to human atherosclerosis, even without any dietary challenge, and are currently the most commonly used mouse model for studying atherosclerosis, being successfully used to study the occurrence and progression of atherosclerosis and for drug discovery.8 In this study, we used bioinformatics methods to identify the core genes involved in the progression of atherosclerosis, hoping that our findings can provide a new therapeutic target for atherosclerosis.
Materials and Methods
All data and materials are publicly available at the NCBI Gene Expression Omnibus (GEO). The datasets can be accessed at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE232472.
Animal Experiments
The procedures involving animals and their care were conducted in accordance with institutional guidelines in compliance with national policies [Regulations on the Administration of Experimental Animals, Decree No. 676 of the State Council of the People’s Republic of China (2017); Guidelines for Ethical Review of Experimental Animal Welfare, GB/T35892-2018] and followed the principles of ethical animal research outlined in the Basel Declaration and the ethical guidelines of the International Council for Laboratory Animal Science. This study was approved by the Ethics Committee of Shandong Provincial Qianfoshan Hospital (Jinan, China) [2018; ethical approval number: S0065].
Feeding and Grouping
Six-week-old (19 ± 2 g) ApoE−/− mice were purchased from Beijing Vital River Laboratory Animal Technology Co., Ltd. (Beijing, China). To prevent the possible impact of the hormonal changes of female mice on the results, only male mice were employed in the study. After two weeks of adaptive feeding, the 8-week-old male mice were divided into four groups (n = 12), two of which were fed a Western diet containing 21% anhydrous milk fat and 0.15% cholesterol (Keao Xieli Feed Co., Ltd., Beijing, China) for 6 and 22 weeks, respectively, and which were used as atherosclerosis models (AS6W and AS22W), and the other two groups were fed a normal purified diet (AIN93M, Keao Xieli Feed Co., Ltd., Beijing, China) for 6 and 22 weeks, respectively (Sham6W and Sham22W). All mice were housed in the SPF experimental animal center of Shandong Provincial Qianfoshan Hospital at a temperature of 22 ± 1°C, an ambient humidity of 50% ± 5%, a 12-hour light/dark cycle, and received food and water at will.
Ultrasound Examination
One week before the end of the dietary treatment, after an overnight fast, the mice were anesthetized using 2% isoflurane (RWD, Shenzhen, China). The left carotid artery of each ApoE−/− mouse was scanned from the proximal end to the bifurcation, and then longitudinally scanned along the common carotid artery after transverse scanning using a small animal ultrasound machine, VISUALSONICSVevo3100 (FUJIFILM Japan), a 55-MHz scanning probe, and a 4.5 mm focus, and the B-ultrasound, M-ultrasound, and PW images were preserved. The stored images were imported into the VevoLAB3.2.6 system to measure the maximum carotid systolic flow velocity (PSV), end-diastolic velocity, velocity time integral, and vascular resistance index (RI) (Figure 1A). All the measurements were performed twice at the same site.
Blood and Tissue Harvesting
At the end of the dietary treatments, after an overnight fast, the mice were euthanized under general anesthesia using 2% isoflurane (RWD, Shenzhen, China). Blood was collected from the retro-orbital plexus into tubes containing 0.1% (w/v) EDTA and centrifuged in a microcentrifuge for 20 min at 4°C at 2000 rpm. Plasma was stored at −80°C until the analyses were performed. The chest of each mouse was opened quickly and blood was removed via perfusion using 1× PBS. The aorta was rapidly detached from the aortic root to the iliac bifurcation and the periadventitial fat was removed (Figure 1C). Then, each aorta was snap-frozen in liquid nitrogen for the RNA-seq (n = 4) and qRT-PCR (n = 6), or externally fixated with 4% paraformaldehyde (n = 2) for oil red O staining. The hearts with attached aortic roots were obtained immediately, fixed overnight in 4% paraformaldehyde, embedded in optimum cutting temperature (OCT) compound, and then cut into serial 6-µm cross sections for hematoxylin and eosin, oil red O, Masson (n = 6), and immunofluorescence staining (n = 3). Aortic sinus histological analysis was performed according to the American Heart Association recommendations.9
Plasma Lipid Measurements
The plasma total cholesterol (TC), triglyceride (TG), and low-density lipoprotein cholesterol (LDL-C) levels were measured using GPO-PAP enzymatic methods (A110-1-1, A111-1-1, A113-1-1, Nanjing Jiancheng Bioengineering Institute) and a multimode detection platform (SpectraMax i3x Molecular Devices, LLC., USA).
RNA-Seq Analyses
The mRNA quality was tested before performing RNA-seq. The sequencing platform used was an Illumina Novaseq 6000 in sequencing mode PE150 (LC-Bio Technology Co., Ltd., Hangzhou, China).
RNA-Seq Data Processing and Visualization
Data Processing of the Differentially Expressed Genes (DEGs)
Network analyst10 is an online visualization platform for gene expression analysis and meta-analysis, which can be used for comparison, quantitative analysis, gene expression difference analysis and enrichment analysis, protein interaction analysis, and integration analysis of multiple datasets, among others. Here, differential expression analysis was performed using Network analyst to determine the RNA-seq counts. The DEGs were grouped based on cut-off values of adj.P < 0.05 and |logFC| > 1.0. Then, R heatmap and ggplot2 packages were used to draw the heatmap, volcano, and PCA plots to visualize the DEGs.
Protein-Protein Interaction (PPI) Network and Hub Gene Identification
STRING database was used to construct the protein-protein interaction network of the DEGs between the AS22W and Sham22W groups. The maximum neighborhood component (MNC)11 was then calculated using the CytoHubba plug-in, and the genes with the top nine MNC values were selected as the hub genes.
TRRUST12 is a human-annotated transcriptional regulatory network database that includes 8444 and 6552 transcription factor target regulatory relationships for 800 human and 828 mouse transcription factors, respectively. TRRUST was used to predict the hub gene-related transcription regulators to construct mRNA-TF interaction networks. The StarBase database,13 in particular, the high-throughput CLIP-seq and degradation group experimental data, which provide a variety of visual interfaces, was searched for miRNA targets. This database contains abundant data on miRNA-ncRNAs, miRNA-mRNAs, RBP-RNAs, and RNA-RNAs. Here, we predicted the RBP combined with hub genes using the StarBase database. The parameters used were the Clade (mammal), Genome (mouse), Assembly (mm10), CLIP-Data (≥ 3), and pan-Cancer (≥ 0).14 CTD is a public database that links the toxicological information on chemicals, genes, phenotypes, diseases, and exposures. Through this platform, we have predicted chemical drugs related to the hub genes, while retaining the chemical drugs with ≥ 10 interactions. Finally, an interaction network diagram was created using the R package iGraph.
Enrichment Analysis
Gene Ontology (GO) analysis is a method commonly used for large-scale functional enrichment studies, including biological processes (BPs), molecular functions, and cellular components.15,16 The Kyoto Encyclopedia of Genes and Genomes (KEGG) is a widely used database that contains information on genomes, biological pathways, diseases, and drugs. The “clusterProfiler” R package17 was used to perform GO and KEGG enrichment analyses of the DEGs between the AS22W and Sham22W groups. A P value < 0.05 was considered significant. Moreover, to study the differences in BPs between different groups, gene set enrichment analysis (GSEA) against the DEGs between the AS22W and Sham22W groups was performed using GSEA Java.18 The gene set “c2.cp.v7.2. symbols.gmt [Curated]” was downloaded from MSigDB19 for GSEA, and a P value < 0.05 was considered significant. Furthermore, we used R-package gene set variation analysis (GSVA)20 to calculate the scores of the related pathways based on the gene expression matrices of the AS22W and Sham22W samples and performed differential screening for the enrichment functions using the “limma” package.
Evaluation of Immune Cell Infiltration
We used CIBERSORT to evaluate the abundance of immune cells in the samples from the AS22W and Sham22W groups. Moreover, we analyzed the correlation between the hub genes and immune cells.
Verified by the Published Human Datasets
The dataset GSE10092721 was downloaded from GEO and used for transcriptome analysis of the atherosclerotic and non-atherosclerotic tissues of the carotid, femoral, and popliteal arteries. The dataset analysis platform used was GPL17077 from the Agilent-039494 SurePrint G3 Human GE v28×60K Microarray 039381 (probe name version). Here, 29 carotid atherosclerotic tissues and 12 normal carotid tissues were selected as the experimental and control groups, respectively, to test our RNA-seq dataset.
Real-Time Quantitative PCR
SteadyPure universal RNA extraction kit (Accurate Biotechnology Co., Ltd., Hunan, China) was used to extract the total RNA from the aortic samples and Hifair III 1st Strand cDNA Synthesis SuperMix for qPCR (Yeasen Biotechnology Co., Ltd., Shanghai, China) was used to reverse transcribe the total RNA into complementary DNA (cDNA). The polymerase chain reaction (PCR) was performed using Hieff qPCR SYBR Green Master Mix (No Rox)(Yeasen Biotechnology Co., Ltd., Shanghai, China) and specific primer pairs (sequences are listed in Table 1). Amplification was performed in duplicate using LightCycler® 480 II (Roche). The experiment was repeated three times for each gene and β-actin was used as the endogenous control.
 |
Table 1 Gene-Specific Primers Used for Real-Time RT-PCR |
Immunofluorescence
Mice were sacrificed using 2% isoflurane (RWD Shenzhen, China), and the heart of each mouse was obtained immediately, fixed in 4% paraformaldehyde overnight, embedded in OCT compound, and serially cut into 6-µm-thick sections for immunofluorescence. Rabbit anti-ATP6V0D2 antibody (bs-12548R, Bioss, Beijing, China) was used for ATP6V0D2 staining. Anti-CD68 rabbit pAb (GB113109, Servicebio, Wuhan, China) was used for macrophage staining.
Statistical Analysis
Data analysis and plot drawing were performed using GraphPadPrism 8.0. The quantitative data were expressed as the mean ± standard deviation, and the normality test was carried out. The data of multiple groups were compared by single factor analysis of variance (ANOVA) (normal distribution) or KruskalWallis test (non-normal distribution). P value < 0.05 was considered significant.
Results
Verification of the Atherosclerosis ApoE−/− Male Mouse Model
Ultrasonic Evaluation of Carotid Atherosclerosis
The most commonly used ultrasonic parameter to assess arteriosclerosis is the intima-media thickness (IMT), which shows part of the blood-intima and media-adventitia interfaces in the B-mode image program. However, its disadvantage is the large variability within and among observers, particularly for blood vessels with a small inner diameter. The RI indirectly reflects the degree of atherosclerosis through hemodynamic values, which are equivalent to the IMT. The RI is easier to obtain, with fewer inter- and intra-observer variations and side-to-side differences. The RI of the ApoE−/− mouse carotid artery was significantly higher in the AS22W group than in the other three groups (P < 0.05), which is consistent with our histological results and indirectly confirmed atherosclerosis progression (Figure 1B).
Histopathological Examination of the Aorta
After 6 weeks of NLD, no atherosclerotic plaques were visible in the Sham6w group, whereas initial atherosclerosis lesions were observed in the Sham22W group; plaques size was comparable to that of plaque measured after 6 weeks of WD. As expected, 22 weeks of WD dramatically accelerated lesion progression and increased atherosclerosis in the AS22w group. Limited plaque progression was observed in ApoE−/− mice, with no dietary influence and only a time challenge. The dietary challenge (WD) accelerated plaque development, with approximately two-times larger plaques being observed in the AS22W group than in the Sham22W group (Figures 2 and 3A–E).
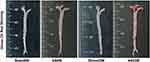 |
Figure 2 Aortic gross oil red O staining. |
Plasma Lipids
The total plasma cholesterol levels were affected by the dietary therapy. LDL-C, triglycerides, and cholesterol accumulated significantly more in the WD-fed AS group than in the ND-fed sham group (Figure 4A–C).
RNA-Seq Analysis
Identification of the DEGs
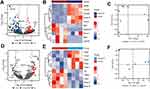
There were 220 DEGs in AS6w group compared with Sham6w group, including 123 upregulated genes and 97 downregulated genes (Figure 5A). The expression heat map of top 5 high-expressed genes including Myb, Tenm1, Serpinb11, Bex1, Galnt9, and top 5 low-expressed genes including Hoxa10, Hoxc5, Atp4b, Hoxc9, Slc2a5 is shown in Figure 5B. As shown in Figure 5D, the volcanic map of difference analysis between the AS22w group and the Sham22W group showed 45 differentially expressed genes, 32 with high expression and 13 with low expression. The expression heat map of top 5 high-expressed genes including Fabp3, Mmp13, Insig1, Tg, Mmp12, and top 5 low-expressed genes including Ppbp, Nipal4, Tubb1, Fmo3, Saa3 is shown in Figure 5E. PCA figures are shown in Figure 5C and F respectively.
PPI Network and Modular Analysis
Using the STRING database, we constructed a PPI network of the DEGs between the AS22W and Sham22W groups (Figure 6A). Screening of the hub genes using the CytoHubba plug-in in Cytoscape revealed that the top nine hub genes were Atp6v1d, Atp6v1a, Atp6v1h, Atp6v0c, Atp6v0d2, Atp6v0b, Atp6ap1, Atp6ap2, and Slc38a9 (Figure 6B). The expression of the hub genes matched between the AS6W and Sham6W groups (Figure 7A); Atp6ap2 was significantly overexpressed in the AS6w group (P < 0.05). The expression of hub genes matched between the AS22W and Sham22W groups (Figure 7B); Atp6Ap2, Atp6v0b, Atp6v0d2, Atp6v1a, and Atp6v1d were significantly overexpressed in the AS22w group (P < 0.05). In the GSE100927 dataset (patients with carotid atherosclerosis and normal carotid arteries), the expression of the hub genes was significantly higher in the carotid atherosclerosis group (P<0.001) (Figure 7C).
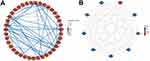 |
Figure 6 PPI network construction. (A) PPI networks of DEGs in AS22w and Sham22W groups. (B) Top 9 hub genes based on PPI network. |
The interaction network based on the TRRUST database for predicting transcription factors binding to DEGs of the AS22w group and Sham22W group is shown in Figure 8A. The interaction network based on the TRRUST database for predicting transcription factors binding to AS22w group and Sham22W group DEGs is shown in Figure 8B; the interaction network based on the StarBase database for predicting RPB combined with AS22w group and Sham22W group DEGs is shown in Figure 8B; and the network based on the CTD database for predicting chemical drugs related to AS22w group and Sham22W group DEGs is shown in Figure 8C.
 |
Figure 8 Construction of related regulatory network. (A) mRNA-TF regulatory network. (B) mRNA-RBP regulatory network. (C) mRNA-Chemical network. |
Enrichment Analysis
We conducted GO annotation and KEGG signaling pathway enrichment analyses based on the DEGs between the AS22W and Sham22W groups (Figure 9, Tables 2–3). The GO annotation results showed that the DEGs were mainly related to BPs, such as regulation of the body fluid levels, regulation of small molecule metabolic processes, regulation of cytokine-mediated signaling pathways, regulation of ion transmembrane transport, and regulation of the response to cytokine stimulus (Figure 9A). At the same time, it is mainly enriched in Cellular Components (CC) such as extracellular matrix, transporter complex, transmembrane transporter complex, ion channel complex, cation channel complex (Figure 9B). In terms of Molecular Function (MF), it is mainly enriched in receptor ligand activity, endopeptidase activity, organic acid binding, proton transmembrane transporter activity, scaffold protein binding, collagen binding (Figure 9C). The KEGG pathway enrichment analysis showed that the DEGs were mainly enriched in the prion disease, cardiac muscle contraction, Alzheimer’s disease, neurodegeneration-multiple diseases, oxidative phosphorylation, amyotrophic lateral sclerosis, and Parkinson’s disease pathways (Figure 9D). Moreover, GSEA enrichment analysis showed that the atherosclerotic samples were significantly enriched in the KEGG fatty acid metabolism, KEGG peroxisome, WP amino acid metabolism, KEGG PPAR signaling pathway, WP ferroptosis, KEGG tryptophan metabolism, KEGG proteasome, and PID integrin two pathways (Figure 10, Table 4).
 |
Table 2 GO Enrichment Analysis |
 |
Table 3 GSEA Enrichment Analysis |
 |
Table 4 KEGG Signaling Pathway |
Furthermore, the GSVA enrichment analysis showed that the atherosclerotic samples were significantly enriched in hallmark oxidative phosphorylation, PI3K-AKT-mTOR signaling, glycolysis, peroxisome, adipogenesis, mTORC1 signaling, fatty acid metabolism, reactive oxygen species pathway, KRAS signaling, and xenobiotic metabolism (Figure 11, Table 5).
 |
Table 5 GSVA Enrichment Analysis |
 |
Figure 11 GSVA Enrichment analysis. (A and B) Enrichment pathway in AS22w group and Sham22W difference analysis box map and heat map (ns p≥0.05, *p< 0.05, **p<0.01, *** p<0.001). |
Immunoassay
Finally, we used CIBERSORT to evaluate the abundance of immune cells in the AS22W and Sham22W samples (Figure 12A). Significant negative correlations between Atp6v1d and follicular helper T cells, Atp6v1a and T cells, CD8 and regulatory T cells (Tregs), Atp6v0b and macrophages M1, and Atp6ap2 and resting mast cells were observed (Figure 12B)
 |
Figure 12 Immunoassay. (A) Abundance of immune cells in AS22w and Sham22W samples. (B) Heat map of correlation between Hub genes and levels of immune cell infiltration (*p< 0.05). |
Corroboration of the Hub Genes Using QT-PCR
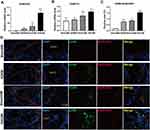
PPI network analysis revealed that Atp6ap2, Atp6v0b, Atp6v0d2, Atp6v1a, and Atp6v1d were significantly expressed in the AS22w group. QT-PCR performed on the ApoE−/− mouse aortas showed that the expression of two core genes (Atp6v0d2, Atp6v1a), especially Atp6v0d2 (P < 0.0001), increased during atherosclerotic plaque progression (Figure 13A and B)
Co-Localization of Atp6v0d2 and Macrophages in Atherosclerotic Lesions
Macrophages, endothelial cells, and vascular smooth muscle cells are the main cell types present in atherosclerotic plaques.22 The colocalization of Atp6v0d2 and macrophages in the atherosclerotic lesions in mice was verified via double immunofluorescence of Atp6v0d2 and CD68 (a macrophage marker) (Figure 13C and D).
Discussion
Atherosclerosis is the root cause of cardiovascular and cerebrovascular diseases and is associated with high morbidity and mortality.23,24 The mechanisms underlying the formation and development of atherosclerotic plaques are complex. In recent decades, the rapid development of high-throughput detection technology and the progress made in bioinformatics have resulted in the development of new methods for studying the pathophysiological mechanisms of atherosclerosis. However, owing to the complexity of its pathogenesis, effective diagnostic and treatment strategies have not been employed in routine clinical practice.
We used high-throughput sequencing to analyze four groups of aortic transcripts of atherosclerosis ApoE−/− mouse models under different experimental conditions. A total of 265 DEGs were found. GO annotation showed that the DEGs were mainly involved in BPs such as the regulation of body fluid levels, regulation of small molecule metabolic processes, regulation of cytokine-mediated signaling pathways, regulation of ion transmembrane transport, and regulation of the response to cytokine stimuli. GSEA and GSVA showed that the atherosclerosis samples were significantly enriched in fatty acid metabolism, peroxisome, amino acid metabolism, oxidative phosphorylation, PI3K-AKT-mTOR signaling, glycolysis, among other pathways. These signaling pathways are related to the pathogenesis of atherosclerosis.25 Glycolysis is upregulated in atherosclerosis. Atherosclerotic plaques are characterized by local hypoxia. Under hypoxic conditions, HIF-1α activates the glycolytic pathway, thus reducing cell dependence on oxidative phosphorylation (OXPHOS). Oxidized lipids generated by LPO play an important role in inflammatory responses in atherosclerosis and CVD.26–28 It is reported that the signal pathways of typical cytokines such as interleukin, interferon, tumor necrosis factor, chemokine, and TGF- β are related to atherosclerosis.29 Amino acid metabolism has been identified as a key regulator of vascular homeostasis and immune cell function. In the process of L-arginine (Arg) and L-hyperarginine (HArg), there are circulatory effects among endothelial cells, innate immune cells, and acquired immune cells, and among L-tryptophan (Trp) metabolism, which may have different effects on the development of atherosclerosis.30 It is well known that inhibition of the PI3K-Akt signaling pathway may reduce the vulnerability of atherosclerotic plaques.31 PPARs regulate multiple genes related to lipid metabolism, inflammation, and oxidative stress in the cardiovascular system.32,33 The involvement of fatty acids and their lipid metabolites in cross-linking with inflammation in atherosclerosis is of interest. Fatty acids are involved in atherogenesis in different ways, demonstrating both pro- and anti-atherogenic functions. They may contribute to inflammation and endothelial dysfunction by participating in many signaling pathways.34 Ferroptosis is a non-apoptotic form of cell death, different from cell necrosis and autophagy. It is characterized by high levels of iron-catalyzed free radicals and the accumulation of lipid reactive oxygen species induced by iron, resulting in oxidative stress and subsequent DNA, protein, and lipid damage. It has been proved that it is involved in the occurrence and progression of atherosclerosis through various signal pathways.35 Busnelli et al used high-throughput sequencing technology to analyze the mouse aortic transcriptome. They showed that in addition to the well-known activation of inflammation and immune response, the damage of the phagosome-lysosome system and osteoclast differentiation was also related to the development of atherosclerosis.
To identify the core genes involved in the progression of aortic atherosclerotic plaque in male ApoE−/− mice, We constructed DEG PPI networks and screened the hub genes involved in atheromatous plaque progression using the STRING database and the CytoHubba plug-in in Cytoscape. Atp6ap2, Atp6v0b, Atp6v0d2, Atp6v1a, and Atp6v1d were significantly overexpressed in the AS22w group. The hub genes were also significantly overexpressed in human carotid atherosclerotic lesions based on GSE100927 data. Finally, qRT-PCR showed that the mRNA levels of two core genes (Atp6v0d2 and Atp6v1a) were upregulated during plaque progression in atherosclerosis ApoE−/− mouse models; in particular, the RNA levels of Atp6v0d2 were significantly increased.
Atp6v0d2 (V-ATPaseD2 subunit), as a specific macrophage subunit of vacuolar ATPases (V-ATPases), has been shown to promote the formation of autophagy lysosomes in vitro.36 The V-ATPases are large protein complexes with a peripheral V1 domain that hydrolyzes ATP and an integral V0 domain that transports hydrogen ions, acidifying intracellular compartments, including lysosomes, which are necessary to complete autophagy.37 After severe burns, the activity of myocardial lysosomal V-ATPase decreases in rats, which inhibits myocardial autophagy flux and causes myocardial injury.38 ATPases also work through a pathway that does not depend on lysosomal acidification, such as the fusion of lysosomes with other organelles to form larger compartments. This suggests that V-ATPases may play a role in the fusion of autophagosomes and lysosomes.39,40 V-ATPases subunits in V1 and V0 domains, including D subunits, play a coordinating role in controlling the coupling of proton transport and ATP hydrolysis.41 Previous studies have shown that in vitro, the D1 subunit collaboratively promotes the acidification of macrophage lysosomes, and the D2 subunit promotes macrophage autophagosome-lysosome fusion by promoting STX17 binding to VAMP8.39 The expression of Atp6v0d2 in liver macrophages was upregulated after liver ischemia-reperfusion, and by gradually promoting the formation of autophagolysosomes to increase autophagy flux to limit the activation of liver inflammation.42
The activation of the phagocytic-lysosomal pathway in macrophages is considered a critical event in atherosclerotic plaque progression.4 Lysosomes are tiny acidic organelles that have the activity of up to 60 different hydrolases, including proteases, lipases, and nucleases.43 The dynamic process of autophagy usually begins with the formation of two-membrane autophagosomes, engulfs cytoplasmic substances, and ends with the fusion and degradation of autophagosomes and lysosomes.44,45 Therefore, it is essential to enhance the formation/maturation of autophagosomes and promote the fusion of autophagosome/lysosome for the function of autophagy.46,47 Rajat Singh first reported in 2009 that autophagy regulates intracellular lipid storage.48 Subsequent studies have shown that lysosomal acid lipases mediate the hydrolysis of cholesterol esters, which are transferred from lipid droplets to lysosomes via autophagy.49,50 Within macrophages, lysosomes degrade extracellular substances, including lipids, through heterophagy and intracellular substances, such as lipid droplets, through autophagy.51 Some studies have shown that lysosome / autophagy pathway is the key regulator of cell metabolism, and the genetic and environmental factors that affect lysosome homeostasis can affect systemic metabolism, especially lipid metabolism.52 Under oxidative stress, low-density lipoprotein, and cholesterol esters are easily oxidized to complex oxidation products through free radical-induced lipid peroxidation (LPO).53 OxLDL is usually transported to cells through receptor-mediated endocytosis, which not only partially inactivates lysosomal enzymes but also destroys the stability of acid vacuolar chambers and destroys lysosomal membranes, resulting in lysosomal enzymes and cathepsin leakage into the cytoplasm, thus inducing cell apoptosis.54 Cholesterol oxidation products (ChOx) have been reported to be the significant cytotoxic components of oxidized LDL/LDL-, 7b-hydroxycholesterol (7bOH) and 7-ketocholesterol (7keto) are the main toxic components of oxLDL, which have toxic effects on arterial wall cells, including monocytes, macrophages, and smooth muscle cells. The mechanism may be related to early lysosomal lipid accumulation, activation of the lysosome, cellular oxidative stress, and mitochondrial pathway.6,55 In addition, the expression of lysosomal cathepsin L was significantly increased in atherosclerotic plaques, which was mainly related to cd68 positive macrophages, apoptosis, and stress protein ferritin. Macrophage apoptosis was significantly correlated with the expression of cathepsin L in the nucleus and cell membrane.56 Hyperlipidemia and atherosclerosis affect the autophagy lysosomal system. Oxidative stress and lysosomal iron induced by increased permeability and rupture of lysosomal membrane lead to ferritin induction, accompanied by mitochondrial pathway activation and cell apoptosis. Impaired autophagy and apoptosis of macrophages lead to a vicious cycle that aggravates atherosclerosis.57 The negative association between autophagy activity and human atherosclerotic progression suggests that autophagy activity in early human atherosclerotic lesions may be a transient self-defense that then declines with prolonged lipid oxidation and oxidative stress and that the flux of autophagy through lysosomes decreases with plaque progression in advanced human atherosclerotic lesions. Autophagy activity may play a key role in maintaining the stability of atherosclerotic plaques by regulating lipid accumulation and necrotic core formation.58 Therefore, improving autophagy damage and promoting autophagy-lysosome fusion is one of the treatment ideas to improve atherosclerosis. Our study shows that Atp6v0d2 is significantly up-regulated in aortic atherosclerosis in male ApoE−/− mice, which is considered as the compensatory protective response of pressure overload, and its effect on autophagy-lysosome can be used as a target for the study of atherosclerosis mechanism.
Conclusion
In summary, Atp6v0d2 was significantly upregulated during the progression of atherosclerotic plaques in ApoE−/− mice and the double immunofluorescence confirmed its colocalization with the macrophages. Atp6v0d2 may affect atherosclerosis progression by regulating atherosclerotic macrophage autophagy, and the upregulation of Atp6v0d2 promotes autophagy as a compensatory protective response to pressure overload. This study is only a preliminary screening of the hub genes involved in the atherosclerosis process, and the specific pathophysiological mechanisms in which these hub genes are involved require further research. In a future study, we will clarify the mechanism by which Atp6v0d2 affects autophagy in atherosclerotic macrophages in vitro and in vivo and provide new potential targets for atherosclerosis treatment.
Abbreviations
BP, Biological processes; DEG, Differentially expressed genes; GEO, Gene Expression Omnibus; GO, Gene Ontology; GSEA Gene set enrichment analysis; GSVA, Gene set variation analysis; IMT, Intima-media thickness; KEGG, Kyoto Encyclopedia of Genes and Genomes; LDL, Low-density lipoprotein; MNC, Maximum neighborhood component; OCT, Optimum cutting temperature; PCR, Polymerase chain reaction; PPI, Protein interaction; RI, Resistance index.
Ethics Approval and Informed Consent
The procedures involving animals and their care were conducted in accordance with institutional guidelines in compliance with national policies [Regulations on the Administration of Experimental Animals, Decree No. 676 of the State Council of the People’s Republic of China (2017); Guidelines for Ethical Review of Experimental Animal Welfare, GB/T35892-2018] and followed the principles of ethical animal research outlined in the Basel Declaration and the ethical guidelines of the International Council for Laboratory Animal Science. All procedures performed in the studies involving animals were approved by the Ethics Committee of Shandong Provincial Qianfoshan Hospital [2018; ethical approval number: S0065].
Acknowledgments
We thank all participants who participated in this study and the Central Laboratory of Ji Nan Central Hospital that provided support.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This study was supported by grants from the Natural Science Foundation of Shandong Province (No. ZR2019MH062, ZR2016HP04), the National Natural Science Foundation of China (No. 81601020), the China Postdoctoral Science Foundation (No. 2021M691227), Shandong Provincial Traditional Chinese Medicine Science and Technology Development Plan Project (No. 2019-0369), Key Research and Development Program of Shandong Province (public welfare)(No. 2019GSF108008, 2019GSF108033), the National Natural Science Foundation of China Incubation Fund (No. QYPY2019NSFC0616, QYPY2021NSFC0618), and the Shandong Provincial Medical and Health Science and Technology Development Program (Healthcare Project)(No. 2021BJ000005).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Libby P, Buring JE, Badimon L, et al. Atherosclerosis. Nat Rev Dis Primers. 2019;5(1):56. doi:10.1038/s41572-019-0106-z
2. Libby P. The changing landscape of atherosclerosis. Nature. 2021;592(7855):524–533. doi:10.1038/s41586-021-03392-8
3. Liao X, Sluimer JC, Wang Y, et al. Macrophage autophagy plays a protective role in advanced atherosclerosis. Cell Metab. 2012;15(4):545–553. doi:10.1016/j.cmet.2012.01.022
4. Sergin I, Evans TD, Razani B. Degradation and beyond: the macrophage lysosome as a nexus for nutrient sensing and processing in atherosclerosis. Curr Opin Lipidol. 2015;26(5):394–404. doi:10.1097/MOL.0000000000000213
5. Yuan XM, Li W, Olsson AG, Brunk UT. The toxicity to macrophages of oxidized low-density lipoprotein is mediated through lysosomal damage. Atherosclerosis. 1997;133(2):153–161. doi:10.1016/S0021-9150(97)00094-4
6. Yuan XM, Li W, Brunk UT, Dalen H, Chang YH, Sevanian A. Lysosomal destabilization during macrophage damage induced by cholesterol oxidation products Free. Radic Biol Med. 2000;28(2):208–218. doi:10.1016/S0891-5849(99)00220-8
7. Sergin I, Evans TD, Zhang X, et al. Exploiting macrophage autophagy-lysosomal biogenesis as a therapy for atherosclerosis. Nat Commun. 2017;8(1):15750. doi:10.1038/ncomms15750
8. Emini Veseli B, Perrotta P, De Meyer GRA, et al. Animal models of atherosclerosis. Eur J Pharmacol. 2017;816:3–13. doi:10.1016/j.ejphar.2017.05.010
9. Daugherty A, Tall AR, Daemen M, et al. Recommendation on design, execution, and reporting of animal atherosclerosis studies: a scientific statement from the American Heart Association. Arterioscler Thromb Vasc Biol. 2017;37(9):e131–e157. doi:10.1161/ATV.0000000000000062
10. Zhou G, Soufan O, Ewald J, Hancock REW, Basu N, Xia J. NetworkAnalyst 3.0: a visual analytics platform for comprehensive gene expression profiling and meta-analysis. Nucleic Acids Res. 2019;47(W1):W234–W41. doi:10.1093/nar/gkz240
11. Chin CH, Chen SH, Wu HH, Ho CW, Ko MT, Lin CY. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol. 2014;8(Suppl 4):S11. doi:10.1186/1752-0509-8-S4-S11
12. Han H, Cho JW, Lee S, et al. TRRUST v2: an expanded reference database of human and mouse transcriptional regulatory interactions. Nucleic Acids Res. 2018;46(D1):D380–D386. doi:10.1093/nar/gkx1013
13. Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42(Database issue):D92–D97. doi:10.1093/nar/gkt1248
14. Davis AP, Grondin CJ, Johnson RJ, et al. Comparative Toxicogenomics Database (CTD): update 2021. Nucleic Acids Res. 2021;49(D1):D1138–D1143. doi:10.1093/nar/gkaa891
15. Gene Ontology Consortium. Gene Ontology Consortium: going forward. Nucleic Acids Res. 2015;43(Database issue):D1049–D1056. doi:10.1093/nar/gku1179
16. Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. doi:10.1093/nar/28.1.27
17. Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. doi:10.1089/omi.2011.0118
18. Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi:10.1073/pnas.0506580102
19. Liberzon A, Birger C, Thorvaldsdottir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1(6):417–425. doi:10.1016/j.cels.2015.12.004
20. Hanzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013;14:7. doi:10.1186/1471-2105-14-7
21. Steenman M, Espitia O, Maurel B, et al. Identification of genomic differences among peripheral arterial beds in atherosclerotic and healthy arteries. Sci Rep. 2018;8(1):3940. doi:10.1038/s41598-018-22292-y
22. Moore KJ, Sheedy FJ, Fisher EA. Macrophages in atherosclerosis: a dynamic balance. Nat Rev Immunol. 2013;13(10):709–721. doi:10.1038/nri3520
23. Verhaar BJH, Prodan A, Nieuwdorp M, Muller M. Gut microbiota in hypertension and atherosclerosis: a review. Nutrients. 2020;12(10):2982. doi:10.3390/nu12102982
24. Mensah GA, Roth GA, Fuster V. The global burden of cardiovascular diseases and risk factors: 2020 and beyond. J Am Coll Cardiol. 2019;74(20):2529–2532. doi:10.1016/j.jacc.2019.10.009
25. van Tuijl J, Joosten LAB, Netea MG, Bekkering S, Riksen NP. Immunometabolism orchestrates training of innate immunity in atherosclerosis. Cardiovasc Res. 2019;115(9):1416–1424. doi:10.1093/cvr/cvz107
26. Mantovani A, Garlanda C, Locati M. Macrophage diversity and polarization in atherosclerosis. Arterioscler Thromb Vasc Biol. 2009;29(10):1419–1423. doi:10.1161/ATVBAHA.108.180497
27. Hume DA. The many alternative faces of macrophage activation. Front Immunol. 2015;6:370. doi:10.3389/fimmu.2015.00370
28. Mantovani A, Sica A, Sozzani S, Allavena P, Vecchi A, Locati M. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol. 2004;25(12):677–686. doi:10.1016/j.it.2004.09.015
29. Nitz K, Lacy M, Atzler D. Amino acids and their metabolism in atherosclerosis. Arterioscler Thromb Vasc Biol. 2019;39(3):319–330. doi:10.1161/ATVBAHA.118.311572
30. Wang N, Zhang X, Ma Z, et al. Combination of tanshinone IIA and astragaloside IV attenuate atherosclerotic plaque vulnerability in ApoE(-/-) mice by activating PI3K/AKT signaling and suppressing TRL4/NF-κB signaling. Biomed Pharmacother. 2020;123:109729. doi:10.1016/j.biopha.2019.109729
31. X-H Y, Zheng X-L, Tang C-K. Peroxisome proliferator-activated receptor α in lipid metabolism and atherosclerosis. Adv Clin Chem. 2015;71:171–203. doi:10.1016/bs.acc.2015.06.005
32. Li H, Li H, Bao Y, Zhang X, Yu Y. Free fatty acids induce endothelial dysfunction and activate protein kinase C and nuclear factor-κB pathway in rat aorta. Int J Cardiol. 2011;152(2):218–224. doi:10.1016/j.ijcard.2010.07.019
33. Ghosh A, Gao L, Thakur A, Siu PM, Lai CWK. Role of free fatty acids in endothelial dysfunction. J Biomed Sci. 2017;24(1):50. doi:10.1186/s12929-017-0357-5
34. Li J, Xu L, Zuo YX, Chang XQ, Chi HT. Potential intervention target of atherosclerosis: ferroptosis (Review). Mol Med Rep. 2022;26(5):343. doi:10.3892/mmr.2022.12859
35. Busnelli M, Manzini S, Chiara M, et al. Aortic gene expression profiles show how ApoA-I levels modulate inflammation, lysosomal activity, and sphingolipid metabolism in murine atherosclerosis. Arterioscler Thromb Vasc Biol. 2021;41(2):651–667. doi:10.1161/ATVBAHA.120.315669
36. Xia Y, Liu N, Xie X, et al. The macrophage-specific V-ATPase subunit ATP6V0D2 restricts inflammasome activation and bacterial infection by facilitating autophagosome-lysosome fusion. Autophagy. 2019;15(6):960–975. doi:10.1080/15548627.2019.1569916
37. Cotter K, Stransky L, McGuire C, Forgac M. Recent insights into the structure, regulation, and function of the V-ATPases. Trends Biochem Sci. 2015;40(10):611–622. doi:10.1016/j.tibs.2015.08.005
38. Yan XP, Zhang DX, Yan TT, Zhang Q, Jia JZ, Huang YS. Effects of change in the activity of vacuolar adenosine triphosphatase of myocardial lysosome on myocardial damage in rats after severe burn and its mechanism. Zhonghua Shao Shang Za Zhi. 2017;33(5):295–300. doi:10.3760/cma.j.issn.1009-2587.2017.05.008
39. Bayer MJ, Reese C, Buhler S, Peters C, Mayer A. Vacuole membrane fusion: V0 functions after trans-SNARE pairing and is coupled to the Ca2+-releasing channel. J Cell Biol. 2003;162(2):211–222. doi:10.1083/jcb.200212004
40. Hiesinger PR, Fayyazuddin A, Mehta SQ, et al. The v-ATPase V0 subunit a1 is required for a late step in synaptic vesicle exocytosis in Drosophila. Cell. 2005;121(4):607–620. doi:10.1016/j.cell.2005.03.012
41. Nishi T, Kawasaki-Nishi S, Forgac M. Expression and function of the mouse V-ATPase d subunit isoforms. J Biol Chem. 2003;278(47):46396–46402. doi:10.1074/jbc.M303924200
42. Wang Z, Wang H, Chen X, et al. Inhibiting ATP6V0D2 aggravates liver ischemia-reperfusion injury by promoting NLRP3 activation via impairing autophagic flux independent of Notch1/Hes1. J Immunol Res. 2021;2021:6670495. doi:10.1155/2021/6670495
43. Settembre C, Ballabio A. Lysosome: regulator of lipid degradation pathways. Trends Cell Biol. 2014;24(12):743–750. doi:10.1016/j.tcb.2014.06.006
44. Mijaljica D, Prescott M, Devenish RJ. Autophagy in disease. Methods Mol Biol. 2010;648:79–92. doi:10.1007/978-1-60761-756-3_5
45. Zhang H, Ge S, He K, et al. FoxO1 inhibits autophagosome-lysosome fusion leading to endothelial autophagic-apoptosis in diabetes. Cardiovasc Res. 2019;115(14):2008–2020. doi:10.1093/cvr/cvz014
46. Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147(4):728–741. doi:10.1016/j.cell.2011.10.026
47. Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451(7182):1069–1075. doi:10.1038/nature06639
48. Singh R, Kaushik S, Wang Y, et al. Autophagy regulates lipid metabolism. Nature. 2009;458(7242):1131–1135. doi:10.1038/nature07976
49. Razani B, Feng C, Coleman T, et al. Autophagy links inflammasomes to atherosclerotic progression. Cell Metab. 2012;15(4):534–544. doi:10.1016/j.cmet.2012.02.011
50. Ouimet M, Franklin V, Mak E, Liao X, Tabas I, Marcel YL. Autophagy regulates cholesterol efflux from macrophage foam cells via lysosomal acid lipase. Cell Metab. 2011;13(6):655–667. doi:10.1016/j.cmet.2011.03.023
51. Zhang H, Ge S, Ni B, et al. Augmenting ATG14 alleviates atherosclerosis and inhibits inflammation via promotion of autophagosome-lysosome fusion in macrophages. Autophagy. 2021;17(12):4218–4230. doi:10.1080/15548627.2021.1909833
52. Martinez-Lopez N, Athonvarangkul D, Mishall P, Sahu S, Singh R. Autophagy proteins regulate ERK phosphorylation. Nat Commun. 2013;4:2799. doi:10.1038/ncomms3799
53. Zhong S, Li L, Shen X, et al. An update on lipid oxidation and inflammation in cardiovascular diseases. Free Radic Biol Med. 2019;144:266–278. doi:10.1016/j.freeradbiomed.2019.03.036
54. Li W, Yuan XM, Olsson AG, Brunk UT. Uptake of oxidized LDL by macrophages results in partial lysosomal enzyme inactivation and relocation. Arterioscler Thromb Vasc Biol. 1998;18(2):177–184. doi:10.1161/01.atv.18.2.177
55. Li W, Ghosh M, Eftekhari S, Yuan XM. Lipid accumulation and lysosomal pathways contribute to dysfunction and apoptosis of human endothelial cells caused by 7-oxysterols. Biochem Biophys Res Commun. 2011;409(4):711–716. doi:10.1016/j.bbrc.2011.05.071
56. Li W, Kornmark L, Jonasson L, Forssell C, Yuan XM. Cathepsin L is significantly associated with apoptosis and plaque destabilization in human atherosclerosis. Atherosclerosis. 2009;202(1):92–102. doi:10.1016/j.atherosclerosis.2008.03.027
57. Ghosh M, Carlsson F, Laskar A, Yuan XM, Li W. Lysosomal membrane permeabilization causes oxidative stress and ferritin induction in macrophages. FEBS Lett. 2011;585(4):623–629. doi:10.1016/j.febslet.2010.12.043
58. Li W, Sultana N, Siraj N, et al. Autophagy dysfunction and regulatory cystatin C in macrophage death of atherosclerosis. J Cell Mol Med. 2016;20(9):1664–1672. doi:10.1111/jcmm.12859
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.