Back to Journals » OncoTargets and Therapy » Volume 13
Transcriptome Analysis Reveals Key Genes and Pathways Associated with Metastasis in Breast Cancer
Authors Li W , Liu J, Zhang B , Bie Q, Qian H, Xu W
Received 10 August 2019
Accepted for publication 26 December 2019
Published 13 January 2020 Volume 2020:13 Pages 323—335
DOI https://doi.org/10.2147/OTT.S226770
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Jianmin Xu
Wei Li, 1,* Jianling Liu, 2,* Bin Zhang, 3 Qingli Bie, 3 Hui Qian, 1 Wenrong Xu 1
1Key Laboratory of Laboratory Medicine of Jiangsu Province, School of Medicine, Jiangsu University, Zhenjiang, Jiangsu, People’s Republic of China; 2Central Lab, The First Affiliated Hospital of Hebei North University, Zhangjiakou, Hebei, People’s Republic of China; 3Department of Laboratory Medicine, Affiliated Hospital of Jining Medical University, Jining, Shandong, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Wenrong Xu; Hui Qian
School of Medicine, Jiangsu University, 301 Xuefu Road, Zhenjiang, Jiangsu 212013, People’s Republic of China
Tel +86 511 85038215
; +86 511 85038334
Fax +86 511 85038483
Email [email protected]; [email protected]
Background: Metastasis is the major cause of death in breast cancer patients. Although the strategies targeting metastasis have promoted survival, the underlying mechanisms still remain unclear. In this study, we used microarray data of primary breast tumor, tumor derived from bone and liver, and skin metastatic tissue, to identify the key genes and pathways that are involved in metastasis in breast cancer.
Methods: We first calculated the differentially expressed genes (DEGs) between three metastatic tissues and primary tumor tissue, and then used it to perform Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis. Further, we analyzed the correlation of genes enriched in GO terms and KEGG pathways with survival of breast cancer patients. To identify the key genes and pathways associated with metastasis, we overlapped the DEGs and KEGG pathways. In our in vitro experiments, we knocked down the key gene, ERLIN2, and detected the PI3K expression in tumor cells to evaluate their effect on tumor metastasis.
Results: We identified six genes (ALOX15, COL4A6, LMB13, MTAP, PLA2G4A, TAT) that correlated with survival. Seven key genes (SNRPN, ARNT2, HDGFRP3, ERO1LB, ERLIN2, YBX2, EBF4) and seven signaling pathways (metabolic pathways, phagosome pathway, PI3K-AKT signaling pathway, focal adhesion, ECM-receptor interaction, pancreatic secretion, human papillomavirus infection) associated with metastasis were also identified. Our in vitro experiments revealed that ERLIN2 was highly expressed in MDA-MB231 cells compared to MCF-7 cells. Moreover, knockdown of ERLIN2 increased apoptosis, while inhibiting the proliferation, invasion, and migration ability of breast cancer cells. The PI3K/AKT signaling pathway was also found to be highly expressed in MDA-MB231 cells.
Conclusion: Our results reveal the key genes and signaling pathways that contribute to metastasis, and highlight that strategic targeting of ENLIN2 and PI3K/AKT signaling pathways could inhibit metastasis of breast cancer.
Keywords: breast cancer, metastasis, transcriptome microarray data, key genes, signaling pathways
Introduction
Globally, breast cancer is the leading cause of cancer-related deaths in women.1 Although surgery at an early stage has been reported to prolong the survival of the patients, the recurrence and metastasis pose critical challenges to the treatment of breast cancer.2 Common metastasis organs of breast cancer include brain, liver, lung, bone, and skin.3,4 Metastasis, a process occasioned by tumor cells detaching from the original tumor tissue and entering the metastatic site, is a complex process. The disseminated tumor cells may undergo four fates: death, cellular dormancy, dormant micrometastasis, or invasive growth.5,6 Over the past decades, strategies targeting breast cancer have focused on every stage and subpopulation. For instance, chemotherapy kills the rapidly growing tumor cells, while blocking agents of oestrogen receptor (ER+) and human epidermal growth factor receptor-2 (HER2+) benefit the patients with ER+ and HER2+ breast tumor.7,8 In recent years, immunotherapy has also been used for the treatment of advanced stages of breast cancer with metastasis.9 However, the prognosis of metastatic breast cancer still remains poor. Thus, it is important to investigate the mechanisms of metastasis in breast cancer.
Recent advancement in RNA detection technology such as RNA sequencing and microarray, has facilitated the understanding of the biological and genomic alterations involved in the formation and metastasis of breast cancer. The genomic landscape analysis revealed that TP53 and PIK3CA mutations are the most frequent genomic alterations in all subtypes of breast cancer.10 Currently, relatively few studies have comprehensively analyzed the genomic alterations leading to metastasis in breast cancer. Toy et al11 revealed that TP53, PIK3CA, and GATA3 were the most frequent mutations in metastatic breast cancer. In addition, Massard et al suggested that PTEN/PI3K/AKT and FGFR/FGF signaling pathways were dysregulated. Breast cancer metastasis is an evolving process which is strongly associated with mRNA expression changes. Kimbung et al12 found that Claudin-2 could predict early liver metastasis in breast cancer. Moreover, expressions of PHLPP1, UBC, ACACB, TGFB1, and ACTB were positively correlated with brain metastasis.13 MicroRNAs (miRNAs) have also been shown to play an important role in metastasis. Zhao et al14 demonstrated that miR-665 promoted metastasis by targeting NR4A3. Therefore, extensive studies on gene expression could greatly enhance the understanding of tumor metastasis mechanisms.
In the present study, we aimed to identify the key genes and pathways involved in metastasis by multiple strategies (Figure 1). We first compared the difference in gene expression level between samples from metastatic and primary tissues. The DEGs identified were subjected to GO and KEGG enrichment analysis for each group. We then evaluated the relationship between expression of the genes enriched in GO and KEGG pathways, and survival in breast cancer. To classify the key genes and pathways, we overlapped the DEGs and KEGG pathways. Subsequently, the role of the key gene and pathways, ERLIN2 and PI3K/AKT, in breast cancer cells were validated in vitro. Our results revealed the underlying mechanisms of metastasis of breast cancer, and showed that ERLIN2 and PI3K/AKT signaling pathway are potential therapeutic targets for breast cancer metastasis.
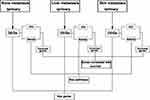 |
Figure 1 Multiple strategies used in the study. |
Materials and Methods
Microarray Data
R package (GEOquery) was used to download microarray data GSE46141 from the GEO database (https://www.ncbi.nlm.nih.gov/geo/). The data were then normalized by normalizeBetweenArrays function in limma package. A total of 88 breast tumor samples were used for this analysis, comprising 11 primary tumor tissues, 5 bone metastatic tumor tissues, 16 liver metastatic tumor tissues, 17 skin metastatic tumor tissues, and 39 lymph node metastatic tissues.
Identification and Clustering of DEGs
RVM t-test was used to filter the DEGs between the metastatic tissue and primary tumor tissue due to its ability to effectively raise the degrees of freedom in small microarray data samples. We selected the differentially expressed genes according to the p-value threshold and fold change. Differences with p value < 0.05 and fold change > 1.5 were considered significant. Hierarchical clustering was performed by EPCLUST.15
GO and KEGG Enrichment Analysis
GO enrichment analysis was used to evaluate the biological function of DEGs, while KEGG pathway analysis was used to investigate the pathways that DEGs are involved in. GO and KEGG pathway analysis was performed online on DAVID (https://david.ncifcrf.gov/). Categories with FDR < 0.05 were considered as significant GO terms and KEGG pathways.
Cell Culture
Human breast cancer cell lines MCF-7 (#SCSP-531) and MDA-MB231 (#TCHu227) were purchased from the Chinese Academy of Sciences Cell Repertoire (Shanghai, China). The MCF-7 cells were maintained in MEM medium (Invitrogen Corporation, Carlsbad, CA, USA, #11090081) with 10% fetal bovine serum (HyClone, Logan, UT, USA, #30068.03) and 0.01 mg/mL human recombinant insulin (YEASEN, Shanghai, China, #40112ES8). The MDA-MB231 cells were maintained in L-15 complete medium (GIBCO, Grand Island, NY, USA, #41300039) with 10% fetal bovine serum (HyClone, Logan, UT, USA, #30068.03). Additionally, 100 U/L of penicillin, and 100 μg/mL streptomycin (Thermo Fisher Scientific, Massachusetts, USA, #15070063) were added into the media. The cells were then cultured in an incubator with 5% CO2 at 37°C.
Real-Time RT-PCR
Total RNA was extracted from MCF-7 and MDA-MB231 cells using Trizol (Invitrogen, Carlsbad, CA, USA, #15596018), according to the manufacturer’s protocol. The RNA concentration and purity were detected using NanoDrop 2000 (Thermo Scientific, Waltham, MA, USA). One micro gram of total RNA was reverse-transcribed to cDNA using Prime Script RT reagent kit (TaKaRa, Tokyo, Japan, #RR037A). Real-time quantitative PCR was performed using Agilent Mx3005P (Santa Clara, CA, USA). All primers were purchased from Sangon Biotech (Shanghai, China) and are listed in Supplementary Table 1. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as the control, and 2−ΔΔCt method used to calculate the relative gene expression level. Each experiment was independently performed in triplicates.
Invasion and Migration Assay
To perform the cell invasion and migration assay, 2 × 105 cells were suspended in 200μL serum-free RPMI-1640 and added to the upper chamber of an 8-um pore transwell (Corning, New York, USA, #3422) with (invasion assay) or without (migration assay) Matrigel coating (Corning, New York, USA, #354234), while 600μL MEM or L-15 medium containing 10% FBS was added to the lower chamber as a chemoattractant. After incubation for 48 hrs at 37°C, the upper cells were gently wiped by a cotton swab, the other cells were fixed with 4% paraformaldehyde for 30 min and stained with 1% crystal violet for 15 min. Finally, cells attached to the filter were counted by inverted microscopy.
Wound Healing Assay
To perform the cell wound healing assay, 2 × 105 MCF-7 or MDA-MB231 cells were seeded on 12-well plates and grown to full confluence in a complete medium. The monolayer of cells was scratched by a 10μL pipette tip and then washed thrice with PBS to remove the detached cells, 1 mL serum-free MEM or L-15 medium was added. After incubation for 24 hrs at 37°C, the wounded areas were imaged by inverted microscopy. All experiments were independently repeated thrice.
RNA Interference
To perform RNA interference, the siARNT2 and negative control constructs were designed and purchased from Gene Pharma Company (Shanghai, China). The siRNAs sequences are listed in the Supplementary Table 2. All siRNAs were diluted in diethyl pyrocarbonate water to a final concentration of 20μM according to the manufacturer’s protocol. MCF-7 and MDA-MB231 cells were incubated on six-well plates at a density of 6 × 105 cells/well. The cells were then transfected with siRNAs or negative control once they reached approximately 60% confluence, using Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA, #L3000001). After 48 hrs, the cells were collected for further analysis. All experiments were independently repeated thrice.
Flow Cytometry for Apoptosis Analysis
After transfection with siRNAs or negative control, the cells were harvested and washed with PBS. Cell apoptosis was detected by TACS Annexin V-FITC Apoptosis Detection Kit (R&D Systems, Minneapolis, USA, #4830-01-k) according to the manufacturer’s protocol. Briefly, the cells were re-suspended in Annexin-V-binding buffer at a concentration of 106 cells/mL and then 100μL of cell suspension was transferred to a 1.5 mL centrifuge tube and incubated with 5μL Annexin-V (FITC) for 15 min in the dark. After incubation, the samples were treated with propidium iodide (5μL) and immediately analyzed with flow cytometry (BD, San Diego, CA, USA, FACSCanto II).
Western Blotting
After washing the cells with cold PBS, 100μL RIPA (Solarbio, China, #R0010) lysate containing protease and phosphatase inhibitors, was added into the plates, and lysed on ice for 10 min. The mixture was transferred to a new 1.5 mL EP tube, denatured at 100°C, 300 rpm metal bathed for 10 min and then used for Western blot assay. A total of 20 μg (20 μL volume) protein was added to the gel, and electrophoresis performed at step 1: 80 v, 30 min, step 2: 100 v, 60 min. The proteins were then transferred to polyvinylidene fluoride membrane (PVDF) for 90 min. Ponceau S staining was carried out to confirm the success of electrotransfer. The PVDF membrane was blocked with 5% skimmed milk in TBST for 1 hr. The membrane was cut according to the molecular weight of the protein and placed into the primary antibody, dissolved in 5% bovine serum albumin (BSA). The primary antibodies were anti-β-actin (1:1000 dilution; Cell Signaling Technology, MA, USA, #3700S), anti-total PI3K (1:1000 dilution; Cell Signaling Technology, MA, USA, #4249), and anti p-PI3K (1:1000 dilution; Cell Signaling Technology, MA, USA, #4228). After they were incubated overnight at 4°C in a shaker, the membrane was washed 3 times with TBST for 5 min and then treated with secondary antibody in BSA for 1 hr at room temperature. The membrane was exposed to highly sensitive ECL luminescent liquid and imaged using the Bio-Rad ChemiDoc™ XRS+ System.
Statistical Analysis
Data were analyzed with GraphPad Prism7.00 (version 5.0; San Diego, CA, USA) and the results were presented as mean ± SEM. The two-tailed unpaired t-test was used to compare differences in key gene expression, proliferation rates and apoptosis ratios between experimental and control groups. The results were presented as histograms with error bars. Each experiment was performed with at least three biological replicates and p < 0.05 was considered statistically significant.
Results
Identification of DEGs Between Metastatic Tumor Tissue and Primary Tumor Tissue
The microarray data GSE46141 was downloaded from GEO database. The dataset consisted of 88 samples, of which 11 were primary tumor tissues, five were bone metastatic tissues, 16 were liver metastatic tissues, 17 were skin metastatic tissues and 39 were lymph node metastatic tissues. First, we identified the DEGs between the metastatic and primary tumor tissues. The detail information of the top 10 DEGs are listed in Supplementary Tables 3–5 for each group. Subsequently, we performed cluster analysis of DEGs using EPCLUST, and the findings showed differences in gene expression between metastatic tumor tissue and primary tissue, except for the lymph node metastasis group (Figure 2A–C, Supplementary Figure 1). We therefore excluded this group in the subsequent analysis to enhance accuracy of the analysis.
 |
Figure 2 Clustering analysis of DEGs in three groups. (A) Bone metastatic group. (B) Liver metastatic group. (C) Skin metastatic group. |
GO Enrichment Analysis for the DEGs
To explore the function of DEGs in tumor metastasis, we performed GO enrichment analysis for each group. The results revealed that 486 GO terms were significantly enriched in bone metastasis group, of which 224 were upregulated and 262 downregulated. A total of 1068 GO terms were significantly enriched in liver metastasis group, of which 559 were unregulated and 509 downregulated. A total of 670 GO terms were significantly enriched in skin metastasis group, of which 357 were upregulated and 313 downregulated. The top 20 GO terms are shown in Figure 3A–F. We observed that GO terms related to transcription, lipid transport, and cell adhesion were mainly enriched in bone, skin, and liver. However, most GO terms differed among three groups, indicating that the biological function of tumor cells is altered according to the specific microenvironment.
KEGG Enrichment Analysis of DEGs
To identify the signaling pathways altered during metastasis, we performed KEGG pathway enrichment analysis of DEGs. We found that 56 signaling pathways were abnormally activated in the bone metastatic group, of which 40 were upregulated and 16 downregulated. A total of 139 KEGG pathways were enriched in the liver metastatic group, of which 88 were upregulated and 51 downregulated. A total of 105 KEGG pathways were enriched in skin metastatic group, of which 56 were upregulated and 49 downregulated. The top 20 signaling pathways for each group are shown in Figure 4A–F. There was significant differential activation of signaling pathways among the three groups, suggesting the organ specificity in cancer metastasis.
Screening for Genes with Prognostic Value in Breast Cancer
To screen for genes correlated with the survival of breast cancer patients, we selected genes that were enriched in GO and KEGG pathways. A total of 172 genes were subjected to Kaplan-Meier analysis. We identified six genes to be correlated with the survival, of which high expression of collagen type IV alpha 6 chain (COL4A6), laminin subunit beta 3(LMB3), phospholipase A2 group IVA (PLA2G4A), and tyrosine aminotransferase (TAT) negatively correlated with survival. However, expression of arachidonate 15-lipoxygenase (ALOX15) and methylthioadenosine phosphorylase (MTAP) were positively correlated with survival (Figure 5A–F). We also found that the role of these genes in tumor metastasis are not well understood, and should be explored further.
Identification of the Key Genes and Pathways Associated with Metastasis
To identify the key genes and pathways involved in metastasis, we overlapped the DEGs and KEGG pathways of the three groups. We observed that seven genes, including small nuclear ribonucleoprotein polypeptide N(SNRPN), aryl hydrocarbon receptor nuclear translocator 2 (ARNT2), hepatoma-derived growth factor, related protein 3 (HDGFRP3), endoplasmic reticulum oxidoreductase 1 beta (ERO1LB), ER lipid raft associated 2 (ERLIN2), Y-Box binding protein 2(YBX2) and EBF family member 4 (EBF4), had significant change of expression in different metastatic sites compared to primary tissue. These genes were then subjected to further analysis (Supplementary Figure 2). The details of these genes are presented in Table 1. In addition, seven signaling pathways were aberrantly activated or inhibited in the metastatic tumor tissue, including metabolic pathways, phagosome pathway, phosphotylinosital 3 kinase/serine threonine protein kinase (PI3K-AKT) signaling pathway, focal adhesion, ECM-receptor interaction, pancreatic secretion, and human papillomavirus infection. The details are presented in Table 2. However, we did not identify any signaling pathway that was consistent in activation or inhibition within the three groups.
 |
Table 1 The Detail Information of Key Genes Overlapped in the Three Groups |
 |
Table 2 The Detail Information of Key Pathways Overlapped in the Three Groups |
Validation of the Key Genes and Pathways in Breast Cancer Cells
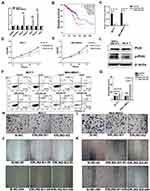
To validate the function of the key genes and pathways in breast cancer, we analyzed the expression of the seven genes by RT-PCR in MCF-7 and MDA-MB231 cells. We found that ERLIN2 was highly expressed in MDA-MB231 tumor cells than MCF-7 cells (Figure 6A). By analyzing the RNA seq data in the TCGA database, we observed that ERLIN2 was negatively correlated with survival (Figure 6B), but they have different expression in subtypes of primary breast cancer (Supplementary Figure 3). To further explore the role of ERLIN2 in the tumor cells, we knocked down ERLIN2 in the two cell lines, and the knockdown efficiency was confirmed by RT-PCR (Figure 6C). ERLIN2 knockdown inhibited the proliferation and increased the apoptosis of tumor cells (Figure 6D–G). The invasion, transwell assay, and wound healing assay revealed that ERLIN2 knockdown decreased the ability of cancer cells to invasion (Figure 6H–K, Supplementary Figure 4). These results suggest that ERLIN2 regulates the malignancy of tumors. In addition, the above results revealed that the PI3K/AKT signaling pathway plays an important role in metastasis. Therefore, we performed further tests to determine the expression of PI3K and phosphorylated PI3K (p-PI3K) in MCF-7 and MDA-MB231 cells. We observed that expression level of p-PI3K was higher in MDA-MB231 cells (Figure 6L), an indication that PI3K signaling could promote the metastasis of tumor cells. These results suggest that targeting ERLIN2 or inhibiting PI3K/AKT signaling could prevent breast cancer from metastasizing.
Discussion
Breast cancer is the leading cause of cancer-related deaths, and metastasis poses a major obstacle to its treatment. There are many aspects that affect the process of metastasis from the dissemination of primary tumor to the colonization in the secondary organ. The host cells that have been re-educated by the tumor cells, including vascular and lymphatic endothelial cells, fibroblasts, neuronal cells, and leukocytes, gain the function of promoting angiogenesis, therefore creating an immune barrier to meet the requirement of nutrition and survival of breast cancer cells in the process of metastasis.16–19 The complex metastatic microenvironment composed of the tumor cells, immune cells and stromal cells, which determine the outcome of these opposing effects and the pattern of growth of the metastases, are not yet well understood. The roles of cells and signaling pathways are different in each metastatic organ, thus, targeting specific cells or signaling pathways in the tumor environment is extremely challenging.20 It has however been reported that the balance between the activity of the mitogen-activated protein kinases (MAPK) ERK1/2 and p38, influences the proliferation status of cancer cells. Activation of ERK1/2 promotes the proliferation of cancer cells, while activation of p38 leads to quiescence of cells involved in metastasis.21,22 Harper et al proved that Her2 expression and p38 downregulation led to early dissemination of breast cancer cells to the lungs and bones, and also promoted the formation of metastatic lesions. Besides, the high expression of PI3K/AKT signaling pathway in MDA-MB231 cells is essential for the outgrowth of bone metastasis.23 A well-studied metastatic factor is the breast cancer metastasis suppressor 1 (BRMS-1) and CXCL12 (SDF-1)/CXCR4 chemo-axis. Overexpression of BRMS-1inhibits the MDA-MB231 metastasis and blocks the outgrowth of cancer cells by inducing apoptosis,24,25 while overexpression of CXCR4 in cancer cells is essential for the metastasis of bone marrow.26 Although the current research provides a better understanding of the metastasis of breast cancer, further research is still needed to explore the key genes and mechanisms involved in metastasis.
In this study, we analyzed the gene expression data from GEO database to identify the key genes and pathways that regulate metastasis of breast cancer. We used 49 samples in this study and observed a significant difference in the gene expression between the metastatic tissues and primary tissues from different metastatic organs. The difference was highest between liver metastatic tumor tissue and the primary tumor, which had 1028 DEGs. Liver is a common site of metastasis, mainly because of its abundant blood supply, as well as the abundant stromal matrix, which promotes tumor colonization and growth.20 Therefore, the interaction between tumor cells and stroma in the liver could alter the biological behavior of the tumor cells. We also observed different enriched GO and KEGG pathways in the three groups. We found that TGF-β signaling was activated in bone metastatic tissue. However, PPAR signaling was inactivated, while other genes involved in Wnt signaling pathway were downregulated, in bone metastatic tissue. These results were consistent with the findings of Sadanandam et al Our results were also consistent with the findings of Vesuna et.al, who reported that PPAR signaling and Wnt signaling were inactive in lung metastatic tissue.27,28 We also observed that the signaling patterns differed in different metastatic tissues. For instance, Phagosome and PI3K/AKT signaling were downregulated in bone and liver metastatic tissue, but upregulated in skin metastatic tissue, an indication that the alteration of biological function and pathways in tumor cells depends on the microenvironment of the metastatic organ. These results, apart from providing a potential target for tumor metastasis treatment, suggest that the strategies of targeting metastasis should consider organ specificity. However, these results should be further validated in in vitro or in vivo experiments.
To identify the key genes and pathways that regulate metastasis, we overlapped the DEGs and KEGG pathways of the three groups. Seven genes were identified as the key genes, of which ERLIN2 recorded the highest difference between the two cell lines. ERLIN2 is a member of the 8p11-12 region and encodes for SPFH domain-containing proteins. The encoded protein is localized to lipid rafts in the endoplasmic reticulum (ER) and plays a critical role in inositol 1,4,5-trisphosphate (IP3) signaling, by mediating ER-associated degradation of activated IP3 receptors, and is considered an oncogene in breast cancer and pancreatic ductal adenocarcinoma.29,30 However, the function of ERLIN2 in tumor metastasis remains unclear. A recent study reported that overexpression of ERLIN2 promoted the survival of breast cancer cells by modulating endoplasmic reticulum stress pathways, and could be targeted by miR-410.31,32 In agreement with the study, our results revealed that the knockdown of ERLIN2 increased the apoptosis of MCF-7 and MDA-MB231 cells. ERLIN2 had a high expression in highly metastatic cells, however, its knockdown inhibited the ability of tumor cells to migrate. In the TCGA database, we discovered that the expression of ERLIN2 negatively associated with cancer survival. While, we also found that RRLIN2 downregulated in tumor tissue than normal and had lowest expression in triple-negative breast cancer. These results indicate that the high expression of ERLIN2 is to response to the new survival pressure at the metastatic site and overexpression of ERLIN2 may break the dormancy of tumor cells and could be responsible for the outgrowth of breast cancer in the metastatic site, indicating that ERLIN2 could serve as a metastatic gene in breast cancer. It is still unclear why he expresses the lowest in the more malignant subtypes. Since ERLIN2 is also expressed in normal tissue sites, unintended toxicity to normal organs should be considered when targeting ERLIN2. Moreover, the functions of other key genes, such as SNRPN, EBF4, and genes correlated with survival in metastasis of tumor are poorly understood and should be investigated further.
Among the key pathways, the metabolic pathways were the most altered in the three groups. Metabolism is the process of converting nutrients into small metabolites. In tumor environment, however, immune cells experience metabolic stress caused by tumor cells, leading to impaired antitumor immune response.33 Accumulating evidence has shown that metabolic reprogramming is essential for the invasion of tumor cells.34 The hexokinase (HK) 2 and pyruvate kinase (PK) M2 were found to be correlated with the invasive capacity of various tumor types.35,36 Lactate promotes breast cancer progression by promoting the chemo-attraction.37 It also has also been shown that the metabolism in secondary tumor sites differs from the primary tumor. Christen et al revealed that tumor tissue from lung metastatic site thrives in pyruvate carboxylase-dependent refilling of the TCA cycle due to the enrichment of pyruvate in the lung. In this study, we identified various metabolic alterations in primary and metastatic tumors, this could be attributed to the nutrient composition in the secondary tumor sites. The glutamate metabolism gene (GLS2) was upregulated in liver metastasis. Previous studies have demonstrated that blocking glutamate metabolism can inhibit tumor growth, though most patients cannot tolerate the effects of this treatment.38,39 Recently, a modified glutamate antagonist that can blockade glutamate in tumor microenvironment, and enhance immune response has been reported.40 These results indicate that blocking glutamate can inhibit tumor growth and may also reduce liver metastasis in breast cancer. Moreover, we also identified several genes involved in fatty acid metabolism that were dysregulated in tumor metastatic tissue, e.g., ACACA, ACADXB, ACAOX1. Tumor cells often have increased fatty acid synthesis for the generation of plasma membrane and soluble molecules to form tumor microenvironment. However, fatty acid is also important for immune cells. Most pharmacological inhibitors of fatty acid metabolism have been developed to treat autoimmunity in the context of autoimmune diseases including systemic lupus erythematosus, and graft-versus-host disease; however, the anti-tumor effect of blocking fatty acid metabolism needs further evaluation.41,42 Metabolic changes that support the invasion of tumor cells usually activate the signaling pathways. It has been shown that increased levels of lactate activates the nuclear factor kappaB (NF-kB) signaling to secrete matrix metallopeptidase 9 (MMP9), for the degradation of extracellular matrix and thus promoting the invasion of tumor cells.43 This also suggests that the alteration of signaling pathways observed in our study may be partly due to metabolic changes. The PI3K/AKT signaling pathway is the most hyperactive intracellular signaling in most cancers. Extensive studies have shown that PI3K/AKT regulates the survival, angiogenesis, chemoresistance, and metastasis in tumor cells.44 The known PI3K–Akt activation mechanisms in cancers are those triggered by receptor tyrosine kinases (RTKs) and somatic mutations. Besides, the activation of PI3K signaling is linked to failure of cancer treatment. We also found the PI3K/AKT signaling was aberrantly activated in the three metastatic tissues. It was downregulated in the bone and liver metastatic tissue, while in the skin metastatic tissue it was upregulated, an indication that the PI3K/AKT inhibitor could benefit patients with skin metastasis. Although previous studies have shown that decreased stimulation of PTEN signaling causes PI3K/AKT activation, we did not observe any alteration of PTEN signaling in tumor metastatic tissues. Therefore, this could be attributed to the response of tumor cells to the microenvironment of secondary organs and metabolic changes. In the in vitro experiments, we also found that the PI3K/AKT signaling was highly expressed in MDA-MB231 cells, which had a high migration capacity, and partially confirmed results reported by Nobre et al that PI3K/AKT are responsible for the survival of metastatic tumor cells. A recent study also showed that PI3K/AKT signaling promotes metastasis of ovarian cancer and hepatocellular carcinoma, suggesting that inhibition of PI3K/AKT signaling could reduce tumor metastasis.45,46 This study had the following limitations. First, we did not consider the lung and brain metastasis, and this could have led to loss of important information. Second, we did not conduct in vivo experiments to validate these results.
Conclusions
In conclusion, this study provides deeper insights into the metastasis of breast cancer to different secondary organs and reveals the key genes and pathways that contribute to metastasis. From the in vitro experiments, we present that knockdown of ERLIN2 increases apoptosis and inhibit the proliferation and the migration ability of tumor cells. Moreover, the PI3K/AKT signaling is highly expressed in metastatic tumor cells, indicating that ERLIN2 and PI3K/AKT signaling pathway could serve as therapeutic targets for the metastasis of breast cancer.
Data Sharing Statement
The data sets used and/or analyzed during the current study are available from the corresponding authors on reasonable request. The data that support the findings of this study are openly available in the NCBI GEO database (GSE46141, URL: https://www.ncbi.nlm.nih.gov/gds/?term=gse46141).
Acknowledgment
This work was supported by the National Natural Science Foundation of China (81702439 and 81572075).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
2. Kodack DP, Askoxylakis V, Ferraro GB, et al. Emerging strategies for treating brain metastases from breast cancer. Cancer Cell. 2015;27(2):163–175. doi:10.1016/j.ccell.2015.01.001
3. Kennecke H, Yerushalmi R, Woods R, et al. Metastatic behavior of breast cancer subtypes. J Clin Oncol. 2010;28(20):3271–3277. doi:10.1200/JCO.2009.25.9820
4. Kang Y, Siegel PM, Shu W, et al. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell. 2003;3(6):537–549. doi:10.1016/S1535-6108(03)00132-6
5. Dittmer J. Mechanisms governing metastatic dormancy in breast cancer. Semin Cancer Biol. 2017;44:72–82. doi:10.1016/j.semcancer.2017.03.006
6. Pradhan S, Slater JH. Tunable hydrogels for controlling phenotypic cancer cell states to model breast cancer dormancy and reactivation. Biomaterials. 2019;215:119177. doi:10.1016/j.biomaterials.2019.04.022
7. Untch M, von Minckwitz G, Gerber B, et al. Survival analysis after neoadjuvant chemotherapy with trastuzumab or lapatinib in patients with human epidermal growth factor receptor 2-positive breast cancer in the GeparQuinto (G5) study (GBG 44). J Clin Oncol. 2018;36(13):1308–1316. doi:10.1200/JCO.2017.75.9175
8. Earl HM, Hiller L, Vallier AL, et al. 6 versus 12 months of adjuvant trastuzumab for HER2-positive early breast cancer (PERSEPHONE): 4-year disease-free survival results of a randomised Phase 3 non-inferiority trial. Lancet. 2019;393(10191):2599–2612. doi:10.1016/S0140-6736(19)30650-6
9. Esteva FJ, Hubbard-Lucey VM, Tang J, et al. Immunotherapy and targeted therapy combinations in metastatic breast cancer. Lancet Oncol. 2019;20(3):e175–e186. doi:10.1016/S1470-2045(19)30026-9
10. Stephens PJ, Tarpey PS, Davies H, et al. The landscape of cancer genes and mutational processes in breast cancer. Nature. 2012;486(7403):400–404. doi:10.1038/nature11017
11. Toy W, Shen Y, Won H, et al. ESR1 ligand-binding domain mutations in hormone-resistant breast cancer. Nat Genet. 2013;45(12):1439–1445. doi:10.1038/ng.2822
12. Kimbung S, Kovacs A, Bendahl PO, et al. Claudin-2 is an independent negative prognostic factor in breast cancer and specifically predicts early liver recurrences. Mol Oncol. 2014;8(1):119–128. doi:10.1016/j.molonc.2013.10.002
13. Lu X, Gao C, Liu C. et al. Identification of the key pathways and genes involved in HER2-positive breast cancer with brain metastasis. Pathol Res Pract;2019. 152475. doi:10.1016/j.prp.2019.152475
14. Zhao XG, Hu JY, Tang J, et al. miR-665 expression predicts poor survival and promotes tumor metastasis by targeting NR4A3 in breast cancer. Cell Death Dis. 2019;10(7):479. doi:10.1038/s41419-019-1705-z
15. Wright GW, Simon RM. A random variance model for detection of differential gene expression in small microarray experiments. Bioinformatics. 2003;19(18):2448–2455. doi:10.1093/bioinformatics/btg345
16. Carmeliet P, Jain RK. Molecular mechanisms and clinical applications of angiogenesis. Nature. 2011;473(7347):298–307. doi:10.1038/nature10144
17. Bhowmick NA, Neilson EG, Moses HL. Stromal fibroblasts in cancer initiation and progression. Nature. 2004;432(7015):332–337. doi:10.1038/nature03096
18. Karnoub AE, Dash AB, Vo AP, et al. Mesenchymal stem cells within tumour stroma promote breast cancer metastasis. Nature. 2007;449(7162):557–563. doi:10.1038/nature06188
19. Erez N, Coussens LM. Leukocytes as paracrine regulators of metastasis and determinants of organ-specific colonization. Int J Cancer. 2011;128(11):2536–2544. doi:10.1002/ijc.v128.11
20. Brodt P. Role of the microenvironment in liver metastasis: from pre- to prometastatic niches. Clin Cancer Res. 2016;22(24):5971–5982. doi:10.1158/1078-0432.CCR-16-0460
21. Aguirre-Ghiso JA, Liu D, Mignatti A, et al. Urokinase receptor and fibronectin regulate the ERK(MAPK) to p38(MAPK) activity ratios that determine carcinoma cell proliferation or dormancy in vivo. Mol Biol Cell. 2001;12(4):863–879. doi:10.1091/mbc.12.4.863
22. Aguirre Ghiso JA, Kovalski K, Ossowski L. Tumor dormancy induced by downregulation of urokinase receptor in human carcinoma involves integrin and MAPK signaling. J Cell Biol. 1999;147(1):89–104. doi:10.1083/jcb.147.1.89
23. Harper KL, Sosa MS, Entenberg D, et al. Corrigendum: mechanism of early dissemination and metastasis in Her2(+) mammary cancer. Nature. 2018;553(7688):366. doi:10.1038/nature24666
24. Seraj MJ, Samant RS, Verderame MF, et al. Functional evidence for a novel human breast carcinoma metastasis suppressor, BRMS1, encoded at chromosome 11q13. Cancer Res. 2000;60(11):2764–2769.
25. Hedley BD, Vaidya KS, Phadke P, et al. BRMS1 suppresses breast cancer metastasis in multiple experimental models of metastasis by reducing solitary cell survival and inhibiting growth initiation. Clin Exp Metastasis. 2008;25(7):727–740. doi:10.1007/s10585-008-9184-0
26. Xu C, Zhao H, Chen H, et al. CXCR4 in breast cancer: oncogenic role and therapeutic targeting. Drug Des Devel Ther. 2015;9:4953–4964. doi:10.2147/DDDT.S84932
27. Vesuna F, Bergman Y, Raman V. Genomic pathways modulated by twist in breast cancer. BMC Cancer. 2017;17(1):52. doi:10.1186/s12885-016-3033-3
28. Sadanandam A, Futakuchi M, Lyssiotis CA, et al. A cross-species analysis of a mouse model of breast cancer-specific osteolysis and human bone metastases using gene expression profiling. BMC Cancer. 2011;11:304. doi:10.1186/1471-2407-11-304
29. Giulietti M, Occhipinti G, Principato G, et al. Weighted gene co-expression network analysis reveals key genes involved in pancreatic ductal adenocarcinoma development. Cell Oncol (Dordr). 2016;39(4):379–388. doi:10.1007/s13402-016-0283-7
30. Zhang X, Cai J, Zheng Z, et al. A novel ER-microtubule-binding protein, ERLIN2, stabilizes cyclin B1 and regulates cell cycle progression. Cell Discov. 2015;1:15024. doi:10.1038/celldisc.2015.24
31. Wang G, Liu G, Wang X, et al. ERLIN2 promotes breast cancer cell survival by modulating endoplasmic reticulum stress pathways. BMC Cancer. 2012;12:225. doi:10.1186/1471-2407-12-225
32. Wu H, Li J, Guo E, et al. MiR-410 acts as a tumor suppressor in estrogen receptor-positive breast cancer cells by directly targeting ERLIN2 via the ERS pathway. Cell Physiol Biochem. 2018;48(2):461–474. doi:10.1159/000491777
33. Li X, Wenes M, Romero P, et al. Navigating metabolic pathways to enhance antitumour immunity and immunotherapy. Nat Rev Clin Oncol. 2019;16(7):425–441. doi:10.1038/s41571-019-0203-7
34. Elia I, Doglioni G, Fendt SM. Metabolic Hallmarks of metastasis formation. Trends Cell Biol. 2018;28(8):673–684. doi:10.1016/j.tcb.2018.04.002
35. Botzer LE, Maman S, Sagi-Assif O, et al. Hexokinase 2 is a determinant of neuroblastoma metastasis. Br J Cancer. 2016;114(7):759–766. doi:10.1038/bjc.2016.26
36. Sun H, Zhu A, Zhang L, et al. Knockdown of PKM2 suppresses tumor growth and invasion in lung adenocarcinoma. Int J Mol Sci. 2015;16(10):24574–24587. doi:10.3390/ijms161024574
37. Bonuccelli G, Tsirigos A, Whitaker-Menezes D, et al. Ketones and lactate “fuel” tumor growth and metastasis: evidence that epithelial cancer cells use oxidative mitochondrial metabolism. Cell Cycle. 2010;9(17):3506–3514. doi:10.4161/cc.9.17.12731
38. Lemberg KM, Vornov JJ, Rais R, et al. We’re not “DON” yet: optimal dosing and prodrug delivery of 6-Diazo-5-oxo-L-norleucine. Mol Cancer Ther. 2018;17(9):1824–1832. doi:10.1158/1535-7163.MCT-17-1148
39. Lynch G, Kemeny N, Casper E. Phase II evaluation of DON (6-diazo-5-oxo-L-norleucine) in patients with advanced colorectal carcinoma. Am J Clin Oncol. 1982;5(5):541–543. doi:10.1097/00000421-198210000-00014
40. Leone RD, Zhao L, Englert JM, et al. Glutamine blockade induces divergent metabolic programs to overcome tumor immune evasion. Science. 2019;366:1013–1021. doi:10.1126/science.aav2588
41. McDonald G, Deepak S, Miguel L, et al. Normalizing glycosphingolipids restores function in CD4+ T cells from lupus patients. J Clin Invest. 2014;124(2):712–724. doi:10.1172/JCI69571
42. Bettencourt IA, Powell JD. Targeting metabolism as a novel therapeutic approach to autoimmunity, inflammation, and transplantation. J Immunol. 2017;198(3):999–1005. doi:10.4049/jimmunol.1601318
43. Payen VL, Porporato PE, Baselet B, et al. Metabolic changes associated with tumor metastasis, part 1: tumor pH, glycolysis and the pentose phosphate pathway. Cell Mol Life Sci. 2016;73(7):1333–1348. doi:10.1007/s00018-015-2098-5
44. Noorolyai S, Shajari N, Baghbani E, et al. The relation between PI3K/AKT signalling pathway and cancer. Gene. 2019;698:120–128. doi:10.1016/j.gene.2019.02.076
45. Sun C, Li X, Guo E, et al. MCP-1/CCR-2 axis in adipocytes and cancer cell respectively facilitates ovarian cancer peritoneal metastasis. Oncogene. 2019. doi:10.1038/s41388-019-1090-1
46. Sun F, Wang J, Sun Q, et al. Interleukin-8 promotes integrin beta3 upregulation and cell invasion through PI3K/Akt pathway in hepatocellular carcinoma. J Exp Clin Cancer Res. 2019;38(1):449. doi:10.1186/s13046-019-1455-x
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.