Back to Journals » Advances and Applications in Bioinformatics and Chemistry » Volume 14
Transcriptome Analysis of Pseudomonas aeruginosa Biofilm Following the Exposure to Malaysian Stingless Bee Honey
Authors Seder N, Abu Bakar MH, Abu Rayyan WS
Received 14 November 2020
Accepted for publication 17 December 2020
Published 14 January 2021 Volume 2021:14 Pages 1—11
DOI https://doi.org/10.2147/AABC.S292143
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Juan Fernandez-Recio
Nesrin Seder,1 Mohd Hilmi Abu Bakar,1 Walid Salem Abu Rayyan2
1Faculty of Health Sciences, University Sultan Zain Al Abidin, Kuala Nerus, Terengganu 21300, Malaysia; 2Department of Pharmacology and Biomedical Sciences, Faculty of Pharmacy, University of Petra, Amman, Jordan
Correspondence: Mohd Hilmi Abu Bakar; Walid Salem Abu Rayyan Email [email protected]; [email protected]
Introduction: Malaysian stingless bee honey (Trigona) has been aroused as a potential antimicrobial compound with antibiofilm activity. The capability of the gram-negative bacillus P. aeruginosa to sustain a fatal infection is encoded in the bacterium genome.
Methods: In the current study, a transcriptome investigation was performed to explore the mechanism underlying the biofilm dispersal of P. aeruginosa after the exposure to Trigona honey.
Results: Microarray analysis of the Pseudomonas biofilm treated by 20% Trigona honey has revealed a down-regulation of 3478 genes among the 6085 screened genes. Specifically, around 13.5% of the down-regulated genes were biofilm-associated genes. The mapping of the biofilm-associated pathways has shown an ultimate decrease in the expression levels of the D-GMP signaling pathway and diguanylate cyclases (DGCs) genes responsible for c-di-GMP formation.
Conclusion: We predominantly report the lowering of c-di-GMP through the down-regulation of DGC genes as the main mechanism of biofilm inhibition by Trigona honey.
Keywords: Pseudomonas, biofilm, stingless bee, Trigona, microarray, D-GMP
Introduction
Pseudomonas aeruginosa possesses a large genome of approximately 5–7 Mbp considerably capable of provoking life-threatening infections.1 The adaptability encoded in its genome is one of the versatile mechanisms of P. aeruginosa to sustain a fatal disease. The gram-negative bacteria is concomitantly involved with chronic infections ranged from cutaneous infections to life-threatening disorders like cystic fibrosis.2,3 Currently, the US Centers for Disease Control and Prevention (CDC) and the World Health Organization (WHO) have reported around 29,000 deaths per annum in the United States out of two million antimicrobial-resistant infections.4 Additionally, more than 33,000 deaths and 874,000 disabilities are caused by hospital-acquired and community-acquired antimicrobial-resistant infections per year in Europe.5 The notorious microorganism P. aeruginosa adopts the sessile formation to manifest an antimicrobial resistance.6 Biofilm is a collective growth of microorganisms that sustain their communities in a matrix of biopolymers comprising polysaccharides, DNA, and proteins.7 Organisms found in biofilms elicit recurring less susceptible to the host immune responses, extremely more reluctant to antibiotic treatment than planktonic bacteria, and consequently, are more difficult to be eradicated.7–9
The primitive stage for biofilm formation is mediated through the establishment of attachment proteins such as adhesions, type IV pili, and lipopolysaccharide (LPS).10,11 Environmental cues mediate direct regulation through an increase in levels of the critical player for biofilm formation bis-(3–5)-cyclic-dimeric guanosine monophosphate (c-di-GMP), an intracellular second messenger.12,13 Many types of environmental cues can cause an increase in c-di-GMP, which consequently activates the production of adhesions, Psl, Pel, alginates, and various extracellular matrix products (ECM).13,14 After the attachment of P. aeruginosa to surfaces, they start to grow, produce ECMs, synthesize water channels, and form microcolonies. Biofilm maturation requires the production of the extracellular polymeric substances (EPS) components of the matrix, which are polysaccharide synthesis locus (Psl), exopolysaccharide (Pel), and alginate (Alg). The EPS genes are up-regulated by sRNAs linked to c-di-GMP when P. aeruginosa attaches to a host surface.15,16 The detachment process is required to create new biofilms in new niches.17 The dispersal mechanism involves ECM degradation and the autolysis of a biofilm subpopulation. This process is induced by environmental cues, such as nutrients, oxygen availability, nitric oxide (NO), pH, and various chemicals.18 Flagella act as adhesion to help initial bacterial attachment to the surface, whereas, type IV pili contribute to the formation of mushroom-like biofilm cap structures.7,19
Honey is derived from two types of bee: honey bee and stingless bee.20,21 The honey bee is common all over the world, whereas, stingless bee honey is predominant in tropical regions.22 The usage of honey as Trigona honey was perceived as an antimicrobial agent for the treatment of several clinical manifestations, for instance, wound healing and cutaneous infections.23 Currently, the demands on stingless bee products are increasing because of the unique nature of honey with high osmolarity consisting of 80% (w/v) of sugars, and hydrogen peroxide has endowed a broad spectrum of antimicrobial activity. Likewise, eucalyptus honey (a sting bee honey) attenuates biofilm activity through the osmotic action combined with hydrogen peroxide and bee-derived antibacterial peptide Defensin-1.24 Recently, high concentrations of the antibacterial compound methylglyoxal (MGO) were explicitly found in Manuka honey, derived from the Manuka tree (Leptospermum scoparium).25 None the less, not all stingless bee honey have been well studied, among them Trigona honey, which hampers many clinical applications.3
Currently, in this study, we will describe the effect of Trigona honey on established biofilms of the pathogenic strain P. aeruginosa (ATCC 10145), emphasizing the molecular mechanism. Additionally, we provide shreds of evidence about the mechanism by which Trigona honey attenuates the Pseudomonas biofilm and triggers its dispersal.
Materials and Methods
Trigona Honey Sample
Trigona honey is natural honey produced by stingless black bees reared in a virgin rainforest in the Kelantan state on the East Coast of Peninsula, Malaysia. The honey sample was collected from Bee Haven center, Kelantan, in spring 2019. Honey samples were stored in a dark room at 25°C.
Bacterial Culture
Initially, the bacteria Pseudomonas aeruginosa (ATCC 10145) were grown aerobically in 20 mL of Tryptic Soy Broth (TSB) (Fisher Scientific, Loughborough, UK) in a shaker incubator (150 rpm) at 37°C for 24 h. After incubation, the viable bacterial count was adjusted to approximately 1.5×108 cell forming units (CFU/mL) in TSB supplemented with 1% glucose (w/v).
Biofilm Degradation Assay
Biofilm degradation assay was performed using the crystal violet method in 96-well microtiter plates (Fisher Scientific, Loughborough, UK).26
Biofilms were established for 24 h at 37°C by dispensing 200 µL of the adjusted bacterial suspension in TSB supplemented with 1% glucose (w/v) into each well of the 96-well plate, after that the liquid was aspired, discarded from each well, and displaced by 200 µL of Trigona honey which dissolved in TSB supplemented with 1% glucose (w/v) at a concentration ranging from 10% to 50% (w/v). The plates were incubated at 37°C for a further two h, bacterial suspension without honey was used as a positive control (growth control), and TSB+1% glucose was used as a negative control. Following incubation, planktonic cells were removed carefully without scratching the bottom and walls of the wells and washed with phosphate buffer saline (PBS) (Invitrogen, Leicestershire, UK). The plates were inverted upside down to be air-dried for 15 min at room temperature, and then the wells stained with 200 µL of 0.1% (w/v) crystal violet and incubated at room temperature for 15 min after which the biofilms were washed three times with PBS to remove the unabsorbed stain. The semi-quantitative assessment of biofilm formation was performed by adding 95% ethanol for bacteria to solubilize the dye in the wells.
The absorbance at 570 nm was determined using an Elisa plate reader (Tecan Infinite 200 PRO, Männedorf, Switzerland).
The percentage of biofilm mass degradation obtained for each honey concentration as calculated by the following formula:
Biofilm degradation%= [(OD growth control − OD sample)/OD growth control] x100
(The experiment was performed in triplicates, and the average value was considered).
RNA Extraction from P. aeruginosa
To study the effect of Trigona honey on established biofilms, P. aeruginosa was cultivated in 5 mL TSB supplemented with 1% glucose (w/v) for 24 h using two sterile Nunc dishes. After incubation, the liquid above biofilms was aspired manually by micropipette without scratching the bottom and edges. 5 mL of 20% of Trigona honey dissolved in TSB supplemented with 1% glucose (w/v) was added to the first dish and labeled “Treated sample.” While 5 mL of TSB supplemented with 1% glucose (w/v) was added to the other dish labeled “Untreated sample”, then all dishes were incubated for a further 2 h. Following incubation, the liquid above biofilms was removed, and the biofilms were scraped and resuspended into 1 mL sterile distilled water, then vortexed for 1 min to break up cell aggregates before treatment with 100 µL of 0.1 mg/mL lysosome (Sigma-Aldrich, Gillingham, UK) for 30 min at 37°C. RNA extraction was conducted using the SV Wizard total RNA extraction kit (Promega, Southampton, UK) according to the manufacturer’s instructions with some modifications. Total RNA concentration was measured using NanoDrop (Implen, München, Germany). RNA purity was assessed using the 260/280 absorbance ratio, samples with ratios between 1.8 and 2.1 accepted for cDNA processing. The integrity of RNA was examined using 1.5% agarose gel stained with SYBR safe green TM. Visualized under ultraviolet light gel using the Genius 3.0 gel documentation system (Genesys, Irvine, USA). The intensities of the bands were quantified densitometrically using ImageJ Software Version 1.47 (http: //www.imagej.en.softonic.com/). Later on, the RNA integrity number (RIN) was analyzed using the 2100 Agilent bioanalyzer. All samples were stored at −80°C for further analysis. (The experiment was performed in duplicate).
Microarray Analysis
For designing the microarray experiment, the whole genome of P. aeruginosa (ATCC 10145) was plotted on a microarray chip. The chip had six plotting wells for the P. aeruginosa genome, three were used for untreated samples while the other three were used for honey-treated samples. We used One-Color Microarray-Based Gene Expression Analysis Version 6.9.1, manufactured with Agilent SurePrint Technology. The procedure is conducted according to the vendor protocol. (https://www.agilent.com/cs/library/usermanuals/Public/G4140-90040_GeneExpression_OneColor_6.9.pdf). Data generated were analyzed using GeneSpring GX 9.0 software. Further genomic analysis conducted using Pseudomonas Genome Database (https://www.pseudomonas.com/) and KEGG: Kyoto Encyclopedia of Genes and Genomes (https://www.genome.jp/kegg/).
Gene Expression and Reverse Transcription Polymerase Chain Reaction (RT-qPCR)
Total RNA samples were converted to cDNA library using a high capacity RNA to cDNA conversion kit GoTaq® 2-Step RT-qPCR System (Promega, Southampton, UK). A mixture of 0.5 μg of total RNA, 0.5 µg oligo (dT) primer, and 0.5 µg random primers were mixed with Nuclease-Free water at a whole volume of 10 μL, then, incubated at 70°C for 5 min in the Thermal cycler (AB Applied Biosystem, USA) for denaturation and then chilled on ice for 5 min. A 4 μL of 5X GoScript RT buffer, 2 μL of MgCl2, 1 μL PCR Nucleotide Mix, 1 μL GoScript™ Reverse Transcriptase, and Nuclease-Free water was added to the total volume of 20 μL. The mixture was incubated in the Bio-Rad thermal cycle at 42°C heat block for 1 h, followed by incubation at 72°C for 15 min for enzyme inactivation.
Primers for RT-PCR analysis were retrieved, as shown in Table 1. In a final volume of 20 μL, the PCR was conducted, 10 μL of PCR Master Mix, 1 μL of 10 pM of forwarding primer, 1 μL of 10 pM of reverse primer, 2 μL of cDNA template and topped up with nuclease-free water to 20 μL. The following PCR protocol (Rotor-Gene Q, Qiagen Hilden, Germany) was used; GoTaq® Hot Start Polymerase activation at 95°C for 1 min one cycle followed by 40 cycle amplification; denaturation at 95°C for 1 min and final annealing and elongation: at 60°C for 30 s with an additional final extension at 60°C for 20 s. The expression of (RpoD) housekeeping gene was used as a reference gene. Three biological samples were used as replicates in performing the analysis.
 |
Table 1 Primer Sequence for RT-qPCR |
Statistical Analysis
Data were expressed as mean ± standard error of means. One-way analysis of variance, independent Student’s t-test was used for the study of the continuous variables and graphing were performed using the SPSS program, version 20 (SPSS, IBM, Chicago, IL, USA). Values with P < 0.05 were regarded as statistically significant.
Results
Biofilm Degradation Assay
To determine the effect of Trigona honey against established biofilms of P. aeruginosa (ATCC 10145), biofilms were grown for 24 h before honey treatment then followed by 2 h treatment with 50%, 40%, 30%, 20%, and 10% (w/v) Trigona honey. A relevant 73%, 58%, 52%, 45%, and 23% (P < 0.001) reduction in biomass was observed, respectively (Figure 1). Trigona honey has shown a significant biofilm degradation activity, and there was a sequential decomposition in relevance to the concentration. We have adopted the concentration of 20% of honey for the microarray experiment, as this concentration has shown a significant minimum inhibition concentration conducted by our research group and a significant antibiofilm activity.20
Microarray Results
Data analysis and normalizations were performed using GeneSpring software (version GX 9.0). The expression data from the two states and experiment tree were drawn using GeneSpring software (version GX 9.0) (Figure 2), clustering together the similar expression profiles.
Gene Expression in Biofilm
We used real-time PCR to compare the expression levels of nine genes potentially regulating five different pathways of biofilm formation in P. aeruginosa. Quantitative real-time PCR of (PelA, RsmA, FimW, WspA, HtpB, Alg44, PslF, BifA, and RpoD) was used to provide independent verification of the microarray results; standard and melting curves were used to verify the quality of qPCR and the fold expression was calculated using ∆∆ct method. The standard and melting curve values were within the acceptable ranges, and fold expression values were homogeneous with microarray results (not shown).
We observed 2.6-fold repression for PA3064 (PelA), in treated biofilm using RT-qPCR versus 2.4-fold repression in the microarray experiment (Figure 3C); for PA0905 (RsmA), 3.00-fold induction versus 2.1-fold induction; for PA4958 (FimW), 2.1-fold repression versus 1.4-fold repression; for PA3708 (WspA), 2.5-fold repression versus 1.93-fold repression, for PA3345 (HtpB), 3.1-fold repression versus 2.51-fold repression, for PA3542 (Alg44), 0.9-fold induction versus 1.6-fold induction; for PA2236 (PslF), 2.3-fold induction versus 1.87-fold induction; and for PA4367 (BifA), 1.3-fold repression versus 1.1-fold repression.
Pseudomonas Transcriptome Analysis
The array contained 6085 of the predicted 6232 P. aeruginosa genes. The majority of P. aeruginosa (ATCC10145) genes in the biofilm were within the analysis criterion. Regrettably, 76.5% of the 6085 genes of honey treated biofilms under the study showed low statistical significance (P> 0.05) in comparison with untreated biofilms. In contrast, only 23.5% of the genes showed a high statistical significance (P<0.05) of more than 2X fold expression.
A total of 3478 genes showed down-regulation among them 832 genes showed a fold change above 2X (Figure 3B). On the other hand, 2607 genes showed up-regulation, and only 602 genes showed above 2X fold changes (Figure 3A). Further analysis in the 3478 down-regulated genes showed 470 genes involved in biofilm formation pathways. Honey had caused down-regulation of 57% of the microorganism genes, and more specifically, 13.5% were biofilm formation associated genes.
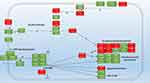
The highest up-regulation (114X) was measured in PA1319 (cyoC) Cytochrome O ubiquinol oxidase subunit III, followed by (95X) PA5481 lysosomal catalytic activity inhibitor gene then the energy correlated genes. Among the findings we obtained in this study, the impressive outcome was the total number of genes that showed biofilm-specific gene regulation is small 76 gene compared to a total of 1434 genes that showed differential expression (Figure 3B) (at least a two-fold difference; Supplementary 1). Overall, there were 36 genes up-regulated, whereas 40 were down-regulated in the biofilm population after treatment with Trigona honey. Around 18% (1104 genes) of the genes involved in the assay were code for hypothetical proteins of unknown function. This is significantly lower than the overall percentage of such genes (44%) derived from the genome-sequencing project of P. aeruginosa.27,28 To track the changes in biofilm stage genes, we draw a map for the gene expression after the treatment with Trigona honey (Figure 4). We relied on several databases among them Kegg pathway (https://www.genome.jp/kegg/) and Pseudomonas genome database (https://www.pseudomonas.com/) in drawing the map. Noticeably, the expression levels for the stage-specific genes were not showing high fold changes as the highest fold change 2.5X was demonstrated in HptB gene, but ultimately there was a comprehensive fold change in all biofilm-associated genes (Figure 4)
Discussion
Biofilm is one of the notorious forms of microorganisms commonly found in nature, industry, and medical fields.29 In this study, the reduction in the biomass was not a result of massive repression in the expression levels of biofilm genes, on the contrary, the changes were comprehensive and entailed several biofilm formation genes as expression levels ranged from 1.1 to 2.6X.
Furthermore, an in-depth analysis of the array results regarding the intimate interactions between the different genes regulating the biofilm process showed aberrantly the highest expression levels during Trigona treatment in P. aeruginosa biofilms were those of energy-related genes. This points to intracellular activity to exert a substantial modification that is closely related to escape from the current exposure to the bactericidal compound, for instance: the filamentous switching.30 These results argue strongly against the proposal that existence in biofilm results in dramatic differences in the overall make-up of bacterial cells. As we show only 5% of the significantly expressed genes were in contribution to the process of biofilm formation.
Genes Modulate Cyclic-Di-GMP Expression
In the current study, we focused on six pathways involved in biofilm formation which are proportionally or directly regulate or under the regulation of C-di-GMP production; Gac/Rsm pathway; the D-GMP signaling pathway; the Psl polysaccharide biosynthesis pathway; Pel polysaccharide biosynthesis pathway; Alginate biosynthesis pathway and Type IV Pilli biosynthesis pathway. Interestingly, the usage of minimum inhibitory concentration (20% w/v) of Trigona honey against P. aeruginosa was significantly effective in biofilm degradation. Trigona honey has endorsed repression of about 71% of genes involved in the previously mentioned pathways, whereas, there was an up-regulation in 29% of biofilm genes (Figure 3B). This concomitantly supports the usage of Trigona honey in the treatment of Pseudomonas infections as the honey drastically decomposed the biofilm and conferred more susceptible Pseudomonas microorganisms for treatment. Morphological studies on Pseudomonas aeruginosa after the treatment with mellifera honey have demonstrated a cellular change either by cell wall thickening or cytoplasmic impurities observed by transmission electron microscopy.31
In the current study, repression in the level of wsp operon (WspA-R) was shown. The chemosensory receptor WspA drastically functions as a receptor in a chemosensory-like system leading to c-di-GMP production.32,33 Moreover, the phosphorelay signal transduction system, Wsp B, D, E, and F, tailored with the regulator WspR which augments the cytoplasmic cyclic-di-GMP levels in response to growth on surfaces.34 The sequential decrease in Wsp levels endows a reduction in intracellular c-di-GMP levels, which mediates the switch to the planktonic state. O’Connor et al, 2012 reported repression in the expression levels around 3x in WspA abrogating the phenotypic transition between planktonic and biofilm forms.33
The process of c-di-GMP synthesis in Pseudomonas aeruginosa is encoded by essential 17 diguanylate cyclase (DGC) genes.35 In this work, we focused on six genes encoding DGC genes; TpbB, SadC, WspR, SiaD, RoeA, and MucR which are subsequently involved in the c-di-GMP regulation by converting GMP to cyclic-di-GMP.36 A relevant suppression for the critical player SadC under the influence of RsmA was shown. There was a noticeable down-regulation in the expression levels for the DGCs gene, which conferred lower levels of the cyclic-di-GMP. Ueda and Wood, 2009 reported reduced cytoplasmic levels of diguanylate cyclase genes aberrantly reduce c-di-GMP production amending matrix exopolysaccharide formation and accomplishing reduced biofilm formation.37 Laventie et al, 2019, and Valentini et al, 2016 reported an essential role of c-di-GMP in the regulation of biofilm formation in P. aeruginosa and the provision of versatile growth forms in different microorganisms.38,39
Another regulator for the c-di-GMP levels in the cytoplasm is the BifA gene encoding a c-di-GMP phosphodiesterase, a negative regulator for biofilm formation and enhancer for swarming motility in P. aeruginosa.40 BifA is upstream of the Pel locus in the biofilm pathway, the overexpression of BifA endowed a down-regulation of the polysaccharide synthesis and lowered the cytoplasmic concentration of c-di-GMP by converting it to P1-(5’-Guanosyl)-P2-(5’-guanosyl)-(3’→5’)-diphosphate (pGpG). Kuchma et al, 2007 reported the direct correlation between deletion mutants of BifA and the reconstitution of the flagellar movement.40
In the current study, the level of RsmA, a negative regulator for biofilm formation, has been elevated in response to the down-regulation in GacS/GacA genes, which facilitate the swarming transformation.10,41–44 GacA and ladS regulate the biofilm formation of single species positively in Pseudomonas aeruginosa biofilms as GacA is the primary response regulator of the GacA/GacS two-component regulatory system where gacA knockout strains show a 10-fold reduction in biofilm formation in relevance to wild-type PA14.45 More precisely, the two-component system GacS/GacA extensively regulates the expression of secondary metabolism global regulator RsmA. The increase in population density is significantly combined with the interpretation of RsmZ under the control of GacA levels, whereas RsmA is under the negative power of RsmZ. This, in turn, RsmA up-regulation directly affects the Quorum sensing system through the suppression of the LasR gene, which activates the switching from sessile to planktonic form.43 In our study, the down-regulation of RsmZ/Y has conferred more free levels of the RsmA leading to a negative role on the down-regulated genes and specifically on SadC gene.
Genes Under the Regulation of Cyclic-Di-GMP
In our study, there was down-regulation in the PelA gene along with a down-regulation pattern in Pel B, C, D, E, and F genes as the polysaccharide biosynthesis is a critical player in biofilm formation. Additionally, the repression of the pel operon is under the regulation of the FleQ gene.46 FleQ a pleiotropic functional gene encoding the c-di-GMP-responsive transcriptional regulator in Pseudomonas aeruginosa that mediates regulation of bacterial-type flagellum-dependent cell motility and negative regulation of extracellular matrix assembly.47 Vasseur et al, 2005, have shown that Pel pathway is consisting of seven genes involved in the early and late biofilm formations, and Pel knockout strains were less capable in biofilm formation compared to the wild-type P. aeruginosa PAO1 strains.48
Psl cluster plays a role in cell-cell and cell-surface interaction in biofilm formation and extracellular polysaccharide biosynthetic process.49 Psl operon consists of 15 genes; Psl A, B, C, D, E, F, G, H, I, J, K, L, M, N, and O all are involved in biofilm formation.49,50 In this study, 11 Psl genes A, B, C, E, F, G, H, I, J, M, and N were up-regulated, whereas only Psl D, K, and L were down-regulated. This points to the role of genes Psl D, K, and L in other pathways besides the polysaccharide biosynthetic process. This result is rationally being aberrant since we expected down-regulation of most PSL operon genes. The genetic map and pathway interactions are still a point of concern for many scientists, and their interactions are a mystery. The pelD has shown a down-regulation in transcription levels. Wu et al, 2019 reported pelD as an exopolysaccharide transporter.51 This could explain the trial of the cell to abrogate the transformation from the biofilm form by up-regulation of the exopolysaccharide synthesis.
In contrast, there is no transportation for the exopolysaccharides outside the cell to buildup and maintain the biofilm. Moreover, PeLG has shown to have a catalytic activity as a degrader of ePsl in the extracellular matrix, so consistently, it will cause an exopolysaccharide degradation.51 Byrd et al, 2009 showed overlapping of the enzymatic roles in the biosynthesis of Psl, LPS production, and the part of Psl genes in surface conditioning and attachment. These results aroused an amending trial of Pseudomonas to transform into a planktonic state to render the inhibitory effect of honey.49
Remarkably, in our analysis, Alg44 showed overexpression of 1.61 X despite the direct regulation of the reduced levels of c-di-GMP. Rational thinking points toward a summative decrease in Alg44 expression since the cells will bear to switch to motile form rather than keeping in biofilm form. By studying the biofilm pathway, a cross-correlation with other paths was found; Alg44 is a match point for three pathways; Fructose and mannose metabolism pathway, metabolic pathway, and biofilm formation pathway, so it will not be governed only by a single upstream gene.52–54 This explains the overall switching of the P. aeruginosa gene profile towards the planktonic motile form.
Finally, the type IV pili system has shown a down-regulation in the response of low c-di-GMP levels and down-regulation of the intracellular c-di-GMP receptor (FimW), which regulates Type IV pili elongation.38 Choi et al, 2011 reported repression in the expression levels around 2x in FimW abrogating the phenotypic transition between planktonic and biofilm forms.32
Conclusion
Trigona honey attenuates established Pseudomonas biofilms through an overall down-regulation in biofilm-specific genes. We strongly propose the antibiofilm activity for Trigona honey is exerted through repression in the expression level of diguanylate cyclase genes in the D-GMP signaling pathway and overexpression of BifA (cyclic-guanylate-specific phosphodiesterase). Which induces lowering of c-di-GMP levels in the bacterial cytoplasm conferring planktonic growth and biofilm dispersal.
Acknowledgment
We are much thankful for the medical campus of UniSZA (University Sultan Zain Al Abidin), this work was funded by the award of Dana Penyelidikat No. UniSZA /2018/DPU/13 R0034-R013. Also, we thank the Deanship of Scientific Research at the University of Petra and the University of Petra Pharmaceutical Center (UPPC) for their cooperation in providing the infrastructure to conduct the research.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Stover CK, Pham XQ, Erwin AL, et al. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature. 2000;406(6799):959–964. doi:10.1038/35023079
2. Bustami MR, Abu-Qatouseh LF, Hussein NA, et al. The Impact of the Complexity of Cystic Fibrosis in Jordanian Patients on the Spectrum of Cystic Fibrosis Transmembrane Conductance Regulator Mutations. Jordan J Biol Sci. 2018;11:5.
3. Althunibat OY, Qaralleh H, Sya A-D, et al. Effect of thymol and carvacrol, the major components of Thymus capitatus on the growth of Pseudomonas aeruginosa. J Pure Appl Microbiol. 2016;10(1):367–374.
4. CDC. Antibiotic Resistance Threats In The United States. https://www.cdc.gov/drugresistance/pdf/threats-report/2019-ar-threats-report-508.pdf.
5. Cassini A, Högberg LD, Plachouras D, et al. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: a population-level modelling analysis. Lancet Infect Dis. 2019;19(1):56–66.
6. Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284(5418):1318–1322.
7. Wei Q, Ma LZ. Biofilm matrix and its regulation in Pseudomonas aeruginosa. Int J Mol Sci. 2013;14(10):20983–21005.
8. Bardoel BW, van der Ent S, Pel MJ, et al. Pseudomonas evades immune recognition of flagellin in both mammals and plants. PLoS Pathog. 2011;7(8):e1002206.
9. Rayyan WA, Singh A, Al-Jaafreh A, et al. The Role of Glutamine-Rich Region of Candida Albicans Tec1p in Mediating Morphological Transition and Invasive Growth. Int J Med Health Sci. 2019;13(4):150–158.
10. Mulcahy LR, Burns JL, Lory S, Lewis K. Emergence of Pseudomonas aeruginosa strains producing high levels of persister cells in patients with cystic fibrosis. J Bacteriol. 2010;192(23):6191–6199. doi:10.1128/JB.01651-09
11. Ryder C, Byrd M, Wozniak DJ. Role of polysaccharides in Pseudomonas aeruginosa biofilm development. Curr Opin Microbiol. 2007;10(6):644–648. doi:10.1016/j.mib.2007.09.010
12. Tolker-Nielsen T. Biofilm Development. Microbiol Spectr. 2015;3(2):
13. Gjermansen M, Nilsson M, Yang L, Tolker-Nielsen T. Characterization of starvation-induced dispersion in Pseudomonas putida biofilms: genetic elements and molecular mechanisms. Mol Microbiol. 2010;75(4):815–826. doi:10.1111/j.1365-2958.2009.06793.x
14. Chambers JR, Sauer K. Small RNAs and their role in biofilm formation. Trends Microbiol. 2013;21(1):39–49. doi:10.1016/j.tim.2012.10.008
15. Petrova OE, Sauer K. A novel signaling network essential for regulating Pseudomonas aeruginosa biofilm development. PLoS Pathog. 2009;5(11):e1000668. doi:10.1371/journal.ppat.1000668
16. Donlan RM. Biofilms: microbial life on surfaces. Emerg Infect Dis. 2002;8(9):881–890. doi:10.3201/eid0809.020063
17. Kim SK, Lee JH. Biofilm dispersion in Pseudomonas aeruginosa. J Microbiol. 2016;54(2):71–85. doi:10.1007/s12275-016-5528-7
18. Yang L, Hu Y, Liu Y, Zhang J, Ulstrup J, Molin S. Distinct roles of extracellular polymeric substances in Pseudomonas aeruginosa biofilm development. Environ Microbiol. 2011;13(7):1705–1717. doi:10.1111/j.1462-2920.2011.02503.x
19. O’Toole GA, Kolter R. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol. 1998;30(2):295–304. doi:10.1046/j.1365-2958.1998.01062.x
20. Al-kafaween MA, Hilmi ABM, Jaffar N, Al-Jamal HAN, Zahri MK, Jibril FI. Antibacterial and Antibiofilm activities of Malaysian Trigona honey against Pseudomonas aeruginosa ATCC 10145 and Streptococcus pyogenes ATCC 19615. Jordan J Biol Sci. 2020;13:1.
21. Al-Nahari AA, Almasaudi SB, El Sayed M, Barbour E, Al Jaouni SK, Harakeh S. Antimicrobial activities of Saudi honey against Pseudomonas aeruginosa. Saudi j Biol Sci. 2015;22(5):521–525. doi:10.1016/j.sjbs.2015.04.006
22. Al-Nadaf AH, Seder NJ, Rayyan WA. Wound healing; antimicrobial and anti-oxidant activity for Jordanian Juglans Regia L. unripe fruits. J Innovations Pharm Biol Sci. 2018;5:26–34.
23. Garedew A, Schmolz E, Lamprecht I. The antimicrobial activity of honey of the stingless bee Trigona spp. J Apicultural Sci. 2003;47(1):37–49.
24. Proaño A, Coello D, Villacrés-Granda I, et al. The osmotic action of sugar combined with hydrogen peroxide and bee-derived antibacterial peptide Defensin-1 is crucial for the antibiofilm activity of eucalyptus honey. LWT. 2014;136:110379. doi:10.1016/j.lwt.2020.110379
25. Johnston M, McBride M, Dahiya D, Owusu-Apenten R, Nigam PS. Antibacterial activity of Manuka honey and its components: an overview. AIMS Microbiology. 2018;4(4):655. doi:10.3934/microbiol.2018.4.655
26. Shukla SK, Rao TS. An improved crystal violet assay for biofilm quantification in 96-well microtitre plate. Biorxiv. 2017;100214.
27. Klockgether J, Cramer N, Wiehlmann L, Davenport CF, Tümmler B. Pseudomonas aeruginosa genomic structure and diversity. Front Microbiol. 2011;2:150. doi:10.3389/fmicb.2011.00150
28. Vasquez-Rifo A, Veksler-Lublinsky I, Cheng Z, Ausubel FM, Ambros V. The Pseudomonas aeruginosa accessory genome elements influence virulence towards Caenorhabditis elegans. Genome Biol. 2019;20(1):1–22. doi:10.1186/s13059-019-1890-1
29. O’Toole G, Kaplan HB, Kolter R. Biofilm formation as microbial development. Annu Rev Microbiol. 2000;54(1):49–79. doi:10.1146/annurev.micro.54.1.49
30. Mikkelsen H, Sivaneson M, Filloux A. Key two-component regulatory systems that control biofilm formation in Pseudomonas aeruginosa. Environ Microbiol. 2010;75(4):1666–1681. doi:10.1111/j.1462-2920.2011.02495.x
31. Morroni G, Alvarez-Suarez JM, Brenciani A, et al. Comparison of the antimicrobial activities of four honeys from three countries (New Zealand, Cuba, and Kenya). Front Microbiol. 2009;5:1378. doi:10.3389/fmicb.2018.01378
32. Choi D-S, Kim D-K, Choi SJ, et al. Proteomic analysis of outer membrane vesicles derived from Pseudomonas aeruginosa. Proteomics. 2011;11(16):3424–3429. doi:10.1002/pmic.201000212
33. O’Connor JR, Kuwada NJ, Huangyutitham V, Wiggins PA, Harwood CS. Surface sensing and lateral subcellular localization of WspA, the receptor in a chemosensory-like system leading to c-di-GMP production. Mol Microbiol. 2012;86(3):720–729. doi:10.1111/mmi.12013
34. D’Argenio DA, Calfee MW, Rainey PB, Pesci EC. Autolysis and autoaggregation in Pseudomonas aeruginosa colony morphology mutants. J Bacteriol. 2002;184(23):6481–6489. doi:10.1128/JB.184.23.6481-6489.2002
35. Chen G, Gan J, Yang C, et al. The SiaA/B/C/D signaling network regulates biofilm formation in Pseudomonas aeruginosa. EMBO J. 2020;39(6):e103412.
36. Merritt JH, Ha D-G, Cowles KN, et al. Specific control of Pseudomonas aeruginosa surface-associated behaviors by two c-di-GMP diguanylate cyclases. MBio. 2010;1:4.
37. Ueda A, Wood TK. Connecting quorum sensing, c-di-GMP, pel polysaccharide, and biofilm formation in Pseudomonas aeruginosa through tyrosine phosphatase TpbA (PA3885). PLoS Pathog. 2009;5(6):e1000483.
38. Laventie B-J, Sangermani M, Estermann F, et al. A surface-induced asymmetric program promotes tissue colonization by Pseudomonas aeruginosa. Cell Host Microbe. 2019;25(1):140–152.
39. Valentini M, Laventie B-J, Moscoso J, Jenal U, Filloux A. The diguanylate cyclase HsbD intersects with the HptB regulatory cascade to control Pseudomonas aeruginosa biofilm and motility. PLoS Genet. 2016;12(10):e1006354.
40. Kuchma SL, Brothers KM, Merritt JH, Liberati NT, Ausubel FM, O’Toole GA. BifA, a cyclic-Di-GMP phosphodiesterase, inversely regulates biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J Bacteriol. 2007;189(22):8165–8178.
41. Heurlier K, Williams F, Heeb S, et al. Positive control of swarming, rhamnolipid synthesis, and lipase production by the posttranscriptional RsmA/RsmZ system in Pseudomonas aeruginosa PAO1. J Bacteriol. 2004;186(10):2936–2945.
42. Brencic A, Lory S. Determination of the regulon and identification of novel mRNA targets of Pseudomonas aeruginosa RsmA. Mol Microbiol. 2009;72(3):612–632.
43. Pessi G, Williams F, Hindle Z, et al. The global posttranscriptional regulator RsmA modulates production of virulence determinants and N-acylhomoserine lactones in Pseudomonas aeruginosa. J Bacteriol. 2001;183(22):6676–6683.
44. Winsor GL, Griffiths EJ, Lo R, Dhillon BK, Shay JA, Brinkman FS. Enhanced annotations and features for comparing thousands of Pseudomonas genomes in the Pseudomonas genome database. Nucleic Acids Res. 2016;44(D1):D646–D653.
45. Parkins MD, Ceri H, Storey DG. Pseudomonas aeruginosa GacA, a factor in multihost virulence, is also essential for biofilm formation. Mol Microbiol. 2001;40(5):1215–1226.
46. Hickman JW, Harwood CS. Identification of FleQ from Pseudomonas aeruginosa as ac‐di‐GMP‐responsive transcription factor. Mol Microbiol. 2008;69(2):376–389.
47. Dasgupta N, Wolfgang MC, Goodman AL, et al. A four‐tiered transcriptional regulatory circuit controls flagellar biogenesis in Pseudomonas aeruginosa. Mol Microbiol. 2003;50(3):809–824.
48. Vasseur P, Vallet-Gely I, Soscia C, Genin S, Filloux A. The pel genes of the Pseudomonas aeruginosa PAK strain are involved at early and late stages of biofilm formation. Microbiology. 2005;151(3):985–997.
49. Byrd MS, Sadovskaya I, Vinogradov E, et al. Genetic and biochemical analyses of the Pseudomonas aeruginosa Psl exopolysaccharide reveal overlapping roles for polysaccharide synthesis enzymes in Psl and LPS production. Mol Microbiol. 2009;73(4):622–638.
50. Franklin MJ, Nivens DE, Weadge JT, Howell PL. Biosynthesis of the Pseudomonas aeruginosa extracellular polysaccharides, alginate, Pel, and Psl. Front Microbiol. 2011;2:167.
51. Wu X, Fan G, He S, Gu X, Yang Y. Comparison of Clinical Outcomes of Two-Level PELD and Foraminoplasty PELD for Highly Migrated Disc Herniations: A Comparative Study. Biomed Res Int. 2019;2019.
52. Remminghorst U, Rehm BH. Alg44, a unique protein required for alginate biosynthesis in Pseudomonas aeruginosa. FEBS Lett. 2006;580(16):3883–3888.
53. Oglesby LL, Jain S, Ohman DE. Membrane topology and roles of Pseudomonas aeruginosa Alg8 and Alg44 in alginate polymerization. Microbiology. 2008;154(Pt 6):1605.
54. Tatnell PJ, Russell NJ, Gacesa P. Chemical analysis of alginates from mucoid strains of Pseudomonas aeruginosa. Biochem Soc Trans. 1994;22(3):S310–S310.
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.