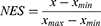Back to Journals » International Journal of General Medicine » Volume 14
Transcription Factor Signatures May Predict the Prognosis and Status of the Immune Microenvironment of Primary Lower-Grade Gliomas
Authors Liu P , Wu R, Zhang J, Zhang Y, Zhang C, Chen L, Yu S, Yang X
Received 21 August 2021
Accepted for publication 28 October 2021
Published 16 November 2021 Volume 2021:14 Pages 8173—8183
DOI https://doi.org/10.2147/IJGM.S335399
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Scott Fraser
Peidong Liu,* Ruojie Wu,* Jinhao Zhang,* Yiming Zhang, Chen Zhang, Lei Chen, Shengping Yu, Xuejun Yang
Department of Neurosurgery, Tianjin Medical University General Hospital, Tianjin, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Xuejun Yang Tel +86-22-60362255
Email [email protected]
Aim: Transcription factor (TF) in glioma, including proliferation, invasion/migration, and tumor microenvironment, has been receiving increasing attention. However, there are still no systematical analyses based on global TF. Herein, using global TF target gene sets, we comprehensively investigated their relationship with prognosis and potential biological effect in lower-grade glioma (LGG). We aimed to develop a less-biased prognostic model and provide new insight for personalized management of this disease.
Methods: TF target gene sets were collected from MSigDB and GRID database followed by ssGSEA calculating normalized enrichment score. Comprehensive survival analysis was combined with Kaplan–Meier and Cox algorithms. Consensus cluster and lasso regression were performed to develop prognostic signatures with validation of ROC and independent external cohort. Approaches of xCell/CIBERSORT/TIMER were involved in analyzing the immune microenvironment. We also correlated identified prognostic signatures with tumor mutational burden (TMB) and m6A genes.
Results: Fourteen TFs were significantly screened based on survival. Patients were classified into 2 prognosis-related clusters based on 14-TFs features. The function of differentially expressed TF target genes between Cluster1/2 was enriched mostly on glioma invasion/migration. The prognostic model was trained by 6 out of 14-TFs followed by generating risk-score as an independent prognostic indicator. We found differences between the high/low-risk group in TMB and the immune microenvironment, where the high-risk group represented “hot-tumor”. Besides, 6-TFs were correlated with m6A regulation genes.
Conclusion: Our findings suggested that the 6-TFs model could be used to predict prognosis and predict the status of the immune microenvironment in LGG.
Keywords: transcription factor target gene set, lower-grade glioma, prognostic model, tumor immune microenvironment, risk signature
Introduction
Gliomas are the most common primary cancers of the central nervous system, representing heterogeneous neuroepithelial neoplasms. Based on WHO criteria, glioma can be classified into Grade-I~IV, where Grade-II~III are defined as lower-grade glioma (LGG) by “The Cancer Genome Atlas” (TCGA). Compared with Grade-IV (glioblastoma, GBM), patients with LGG are often younger and with longer survival. However, though LGG has slower growth and locally aggressive behavior, they may be associated with greater mortality due to recurrence and malignant aggressiveness, even in the setting of optimal resection.1 Besides, some GBM patients can be recurrent and progress from LGG, especially LGG combined with IDH1 wildtype and 1p19q non-codeletion.2 Most conventional studies on glioma focused on GBM. Thus, the aim of the present study was to uncover a novel bio-signature to develop risk stratification and provide a new perspective with potential for personalized diagnosis and treatment to manage this disease.
Transcription factor (TF) is a special protein with at least one DNA-binding domain attached to a specific DNA sequence. Mounting evidence indicated that TFs have a crucial role not only in regulating biological processes but in tumorigenesis and in the micro-environment of tumors, including glioma. Our previous study found that NICD contributes to the proliferation, invasion, and maintaining the stemness of glioma cells.3–5 Other research showed that STAT3 could induce the immunosuppression microenvironment in glioma and increase the expression of PD-L1.6 NF-κB can induce CSN5 upregulation for PD-L1 stabilization, leading to eradicating anti-tumor immunity and enhanced tumor cell survival.7 Besides, targeting NF-κB could elevate ROS levels in glioma cells, and, in turn, induce cell apoptosis.8 In this study, we hypothesized that TFs could be closely associated with the progression and prognosis of gliomas. Moreover, the most important reason for less bias in our hypothesis is that TFs are the cornerstone for biological effects. However, there is still no globally comprehensive research based on TFs in LGG.
In this study, we collected TF target gene sets (TFTGS) from widely recognized and validated databases, followed by screening survival-related gene sets via normalized enrichment score (NES). A multi-TFs independent prognostic indicator was generated following unsupervised/supervised learning in both the training (TCGA) cohort and the independent external validation (CGGA) cohort. This was followed up with immune microenvironment-related enrichment analysis, somatic mutation status analysis, and epigenetic modification analysis. We also attempted to create a prognostic, predictive tool to predict survival and propensity for immune status.
Materials and Methods
Dataset
TCGA-LGG (n = 538) and GTEx RNA-seq dataset were downloaded from UCSC Xena (www.xenabrowser.net). The data was transformed into Transcripts Per Kilobase Million (TPM) followed by log2 transformed. External independent validation cohort was download from CGGA website (www.cgga.org.cn, n = 325). Since glioma in this research was supratentorial tumor, we also only selected the sample of supratentorial-brain tissue in GTEx cohort (n = 291). The batch effect among different cohorts was removed via R package “sva”. The somatic mutation dataset of TCGA-LGG was downloaded by using R package “TCGAbiolinks” and “TCGAmutations”. Recurrent glioma and missing survival data were removed in both TCGA and CGGA cohorts. This research confirms “International ethical guidelines for biomedical research involving human subjects (2002)” developed by Council For International Organizations Of Medical Sciences (CIOMS) in collaboration with World Health Organization (WHO), and research in this article are approved by Ethical Committee of Tianjin Medical University General Hospital.
TFs Functional Annotation
TF functional annotations were performed via DAVID v6.8 (www.david.ncifcrf.gov) and Cistrome (www.cistrome.org).
Unsupervised Learning
LGG patients were classified into several groups based on significantly changed TFTGSs between longer/shorter-OS groups via R package “ConsensusClusterPlus”. We set 1000 iterations, a resample rate of 80%, a clustering method of “k-means”, and distance of “euclidean” to conduct consensus clustering. The optimal clustering number was validated by both results and external independent validation by R package “fpc”. Principal component analysis (PCA) and t-distributed stochastic neighbor embedding (tSNE) were conducted via R package “PCAtools” and “Rtsne”. Single sample gene set enrichment analysis (ssGSEA) was conducted by R packages “GSVA” and GMT annotation file was downloaded from MsigDb (www.gsea-msigdb.org). TFTGSs were validated by Gene Transcription Regulation Database9 (GTRD, www.gtrd.biouml.org). The NES of TFTGSs were selected for survival analysis. Enrichment scores were normalized by the formula:
Survival-Related Analysis
Kaplan–Meier (KM) method was performed based on NES of TFTGSs for overall survival (OS) analysis via R package “survival”. The Log rank test was performed to compare samples with different OS; p-value <0.001 was considered extremely significant. The median value of NES was regarded as the cut-off threshold. Cox regression algorithm (R package “survival”) was also applied to screen significant TF target gene sets contributing to OS. Schoenfeld Individual test both on the global model and separate covariate was performed to test the proportional hazards assumption for Cox regression model fit. The Cox regression model was considered to fit the proportional hazards assumption when the linear relationship between residual error and time was non-significant (p > 0.05). Marking TF target gene set was extremely significant to OS (p < 0.001, wald-test).
Differential Expression TF Target Genes (DETFTGs)
Differential expression TF target genes between Cluster1/2 were screened in primary-LGG patients using the R package “DESeq2”; p-values <0.05, as well as log2 fold change >1 were regarded as statistically significant.
Enrichment Analysis
DETFGs’ enrichment was done via R package “clusterProfiler” and “ReactomePA”, including Gene Ontology (GO) analysis, Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis, Reactome pathway analysis, and Gene Set Enrichment Analysis (GSEA).
Supervised Learning
Lasso regression algorithm was adopted with R package “glmnet” to develop a survival prediction model with a potential risk signature. To evaluate prognosis with key-TFs (kTFs) and prognostic indicators, cox algorithm was performed. Univariate Cox (uni-Cox) analyses were operated to screen the independent prognostic indicators (p < 0.05). Nomogram was built by R package “regplot” and a calibration curve was created with R package “Hmisc”.
Immune-Related Analysis and N6-Methyladenosine (m6A) Correlation
Immunocyte infiltrating analysis was performed by TIMER (www.timer.comp-genomics.org), CIBERSORT (www.cibersortx.stanford.edu), and xCell (www.xcell.ucsf.edu) algorithm. The m6A genes were acquired from previous research.10 The correlation between m6A genes and kTFs was performed with R package “ggcor”.
Somatic Mutation Analysis
Somatic mutation analysis and tumor mutational burden (TMB) were performed and visualized by R package “maftools”. T-test was adopted to compare the distribution between high/low-risk group, Cluster1/2, and grade.
Results
Screening OS-Related TFTGSs and Function Annotation in Primary-LGG
ssGSEA algorithm was conducted with TFTGSs to generate a score of each primary-LGG sample in the TCGA cohort (Figure 1A). The normalized enrichment score (NES) was used for the following analyses. There were 14-TFTGSs that extremely contributed to OS via KM survival analysis and Cox regression algorithm (Figure 1B). The 14-TFTGSs’ survival curve is shown in Figure 1B-I~XIV.
To investigate the NES consensus patterns of those 14-TFs, we performed an unsupervised cluster algorithm. Interestingly, primary-LGG samples in the TCGA cohort were clustered into 2 clusters based on the clustering result (Figure 1C-I~III). The lowest proportion of ambiguous clustering was “k=2” which seems to be the optimal choice. Moreover, we also conducted “Prediction Strength” method, an external independent clustering algorithm, to validate the k value. When the prediction value was more than the cut-off, the samples were clustering into 2 groups (Figure 1C-IV).
We also performed PCA and tSNE to compare the distribution of 14-TFs’ NES. Similarly, results indicated that samples were gathered into 2 sub-cohorts, both in PCA and tSNE. Cluster1/2 were well-grouping in the dimension of PC1 (Figure 1D-I) and tSNE2 (Figure 1D-II).
The above-reported findings suggested that cluster algorithm on 14-TFs’ NES could divide primary-LGG samples in a novel pattern. To further investigate the functions of 14-TFs, we firstly performed differentially expression genes (DEGs). Then, 285-TF target genes were matched out of 2679 DEGs and were marked as DETFTGs followed by enrichment analysis. As a result, twenty of the terms in GO and Reactome databases were significantly enriched (Figure S1A and B, Supplementary Table) and top 12 out of 25 terms in GSEA (Figure S1C). In KEGG, five pathways (hsa04512, hsa05205, hsa04510, hsa05410, hsa05414) were significantly enriched, and as displayed in top 3 (Figure S1D–F), these data indicated that DETFTGs would make a difference to migration/invasion in LGG.
The Cluster1/2 is Clinical Prognosis-Related Followed by hTFs Annotation
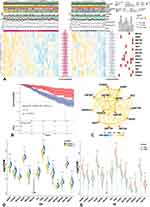
To evaluate whether the results of unsupervised learning were clinically significant, we performed Wilcoxon-test to compare the distribution of clinical features (grade, age, IDH status, 1p19q co-deletion status, MGMT promoter methylated status, KPS, histology, immune score, stromal score) between Cluster1/2 cohorts (Figure 2A). Our findings indicated that grade (p < 0.05), age (p < 0.001), 1p19q co-deletion status (p < 0.05), KPS (p < 0.01), histology (p < 0.01), immune score (p < 0.001), and stromal score (p < 0.001) were significantly clinical signatures. Besides, Cluster1 demonstrated that these findings were consistent with poor prognosis (Figure 2B) and poor clinical signature.
The functional annotations of the 14-hub TFs (hTFs) were conducted by DAIVD and Cistrome databases (Figure 2A). Co-relation network (Figure 2C) was also performed and indicated that most hTFs had positive interactions, while the only 2-negative relationships were between TAZ-FOXN3 and TAZ-ZFP91. Meanwhile, the network also demonstrated that the very stronger interactions were between ADNP-KMT2D/ZNF597-ADNP/YBX1-HDGF. The mRNA expressions of hTFs between Cluster1/2 and LGG/GTEx were conducted (Figure 2D and E), and the obtained results were consistent with lower levels (p < 0.05).
Prognostic Predict Model of kTFs and Its Validation
We used mRNA expression of the hTFs to establish a prognosis risk-signature model via lasso algorithm in the training cohort (TCGA-LGG). The best λ was defined when partial likelihood deviance was at the minimum value (Figure 3A-I~II). Meanwhile, only 6-TFs out of 14-hTFs were screening by the lasso algorithm, and they were marked as the kTFs. A prognostic-related risk-score of each sample was calculated by the coefficient of kTFs with the lasso algorithm. Risk groups (high/low) were generated by the median value of the risk-score. Expression levels of kTFs between high/low-risk groups demonstrated significantly higher expression in the high-risk group, except for ZNF423 (Figure 3B). The ROC curve showed that this risk model could satisfactorily predict 1-year (AUC = 0.75), and 3-year (AUC = 0.722) survival rates (Figure 3A-III). Furthermore, we also compared OS between high/low-risk groups (Figure 3A-IV), and found that OS of the high-risk group was significantly shorter (p = 0.0014).
The distribution of risk-score between Cluster1/2 and grades was performed by t-test (Figure 3C-I~II). Our data suggested that a higher risk-score was significantly (p < 0.05) consistent with higher-grade and Cluster1 (poorer-survival). These results also indicated that more than half of Cluster1 samples were in a high-risk group, which suggested that cluster algorithm based on hTFs’ NES revealed consistent findings with lasso algorithm based on kTFs’ expression values.
To confirm whether risk-score could better predict the prognosis of primary-LGG patients, we performed uni-Cox regression with risk-score and clinical-pathological variates (grade, IDH mutation status, TERT promoter status, 1p19q co-deletion status, MGMT promoter methylation status). As a result (Figure 3D), higher grade, IDH wildtype, 1p19q non-codeletion, MGMT promoter unmethylation and higher risk-score were harmful to prognosis, with hazard ratios (HR)>1 and p-values <0.001. Next, we selected risk-score and significant clinical-pathological indicators (IDH, 1p19q, MGMT, grade) to build a prognostic-predicted nomogram via multi-Cox algorithm (Figure 3E). Meanwhile, the multivariable regression calibration curves showed satisfactory precision of regression models in 1/3/5-year survival (Figure 3F-I~III).
We also validated the results in the external independent validation cohort of CGGA. Risk-score in the validation cohort was calculated via kTFs coefficients followed by ROC and KM analysis. As the outcomes of ROC curve, robust evidence correlated the risk-score with prognosis (Figure 3G-I), where AUC in 1/3/5 years was 0.905/0.681/0.697, respectively. KM analysis between high/low-risk groups also showed significant differences (p = 0.00028) in OS (Figure 3G-II). Then, we performed uni-Cox algorithm with risk-score and clinical-pathological variates, which revealed consistent outcomes (Figure 3H) with training cohort, except MGMT (p> 0.05). Nevertheless, a few missing data of MGMT in the CGGA cohort might cause this bias. Besides, MGMT promoter methylation status systemically demonstrated coincidence with patients’ OS.11,12 Then, we used the same clinical-pathological variates in the training cohort and risk-score to build the prognostic predicted nomogram (Figure 3I). Similar results were found in the external independent validation cohort, where calibration curves (Figure 3J-I~III) indicated robust precision of risk-score in the nomogram.
These results indicated that risk-score was an independent predictor of prognosis in primary-LGG, and the risk-score-based nomogram could also provide a robust prognostic prediction in LGG.
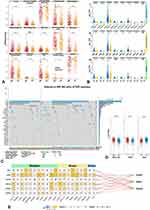
More Immunocyte Infiltrating and Higher Immune Checkpoints (ICPs) Expression in a High-Risk Group
We performed 3-independent algorithms (xCell/CIBERSORT/TIMER) to calculate the infiltration score of immunocytes and studied the distinction between high/low-risk groups. Interestingly, there were 14 (xCell, Figure 4A), 9 (CIBERSORT, Figure S2B), and 6 (TIMER, Figure S2A) kinds of terms showing significant differences. However, all results were consistent with higher-infiltrating scores in the high-risk group. Of note, macrophage, mast cell, monocyte, and myeloid dendritic cell (mDC) with more characteristics of immunosuppression showed the higher-infiltrating score (Figures 4A and S2A). The results of xCell (Figure 4A), monocyte, macrophage, mast cell, activated-mDC, and T helper 2 cells (Th2) showed significantly higher composition in the high-risk group. Still, “T cell NK” showed significantly higher infiltration in low-risk group. In TIMER (Figure S2A), macrophage, mDC, and neutrophil showed higher infiltration in the high-risk group. Moreover, in CIBERSORT (Figure S2B), macrophage M2, activated-mast cell, neutrophil, and resting-memory CD4+ T cell were higher-enriched in the high-risk group. Meanwhile, resting-NK cells were also higher-enriched in the high-risk group. These findings indicated that immune cells with more skewed functions of immunosuppression were more abundant in a high-risk group, whereas immune cells with more functions of anti-tumor immunity were less present. However, some anti-tumor immunocytes were also higher-significantly enriched in the high-risk group (“macrophage M1” in Figures 4A and S2B, “CD8+ T cell” in Figure S2A and B). Interestingly, their infiltration scores were at a lower level than others. Besides, it is well known that there is still a small number of anti-tumors immunocytes in tumor tissue, and the level of anti-tumor immunocytes in LGG is not as exhausted as in GBM.
To thoroughly investigate the immune status levels between high/low-risk groups, we analyzed the expression of ICPs in grade and Cluster1/2 (t-test). The high-risk group revealed results (Figure 4B-II) consistent with a higher level of expression in most ICPs (p < 0.05), except TIGIT. Meanwhile, expressions of ICPs in Cluster1 (poorer prognosis) and Grade-III were associated with higher levels (Figure 4B-I/III).
The above findings suggested that patients in the high-risk group may receive higher-level of immunosuppression and could be regarded as having “hot-tumor”.
Higher-Level of TMB in High-Risk Group and kTFs Associated with m6A Gene
Next, we analyzed the somatic mutation in LGG. As shown in Figure 4C, findings indicated that the low-risk group, Cluster2, and Grade-II were consistent with higher mutation frequency in IDH1, which represented a better prognosis. TMB was calculated and transformed to logarithmic form for normalization (Figure 4D). Poorer-prognosis groups (high-risk group, Cluster1 and Grade-III) demonstrated that these results were consistent with higher levels of TMB (t-test, p < 0.0001). Risk groups based on kTFs were a prognostic predictor, represented the immunosuppression level, and reflected the ICPs, especially in CD274/PDCD1/PDCD1LG2, which are the most widely studied. Therefore, patients in the high-risk group may benefit from ICPs inhibitor or other immunotherapies.
While kTFs participated in transcription, it remained unclear whether they could be involved in epigenetic modification. We next performed correlation analysis with m6A regulatory genes, as methylation was the most common epigenetic modification. The 20-m6A genes were divided into 3 parts by function: Readers, Writers, and Erasers. The Pearson’s correlation matrix (Figure 4E) showed that ADNP had the most significant relationship with m6A regulatory genes and an almost positive relation between Writer’s and Eraser’s genes (p < 0.001), followed by KMT2D. Mantel’s test revealed that ADNP also had a stronger positive relation to Writer and Reader (Figure 4E, p < 0.01, r > 0.6). Besides, KMT2D had a stronger positive relation to Writer (Figure 4E, p < 0.01, r > 0.6). A previous study found that ADNP were functioning in the form of transcription complex,13 which was similar to the Write process. Meanwhile, KMT2D encoded protein functioned as methyltransferase, and it also served in a protein complex.14,15 However, substantial systematic research is needed to explain how ADNP and KMT2D interacted with the m6A process. In conclusion, kTFs showed positive relation to m6A regulatory genes, especially in processes involving Reader and Writer’s genes.
Discussion
Previous studies on LGG often focused on single-gene level as the starting point, thus potentially neglecting the biological effect to some degree. Research starting with the gene sets could promote exploring and developing the less-bias predictable prognostic model. Transcription factors are the crucial proteins regulating gene expression and key functional molecules in the signaling pathway. Therefore, this study adopted a comprehensive bio-information approach beginning with TFs target gene sets to develop and validate the immune-related prognostic predict model via integrating multiple-dimension data including NES, clinical information, mRNA expression, and somatic mutation levels (Figure S3).
Global TFTGS score was first calculated by ssGSEA. Bio-function-based survival analyses helped to explore screening OS-related TFTGS. Therefore, our findings suggested that the hTFs were not only OS-related but had fewer bias features with prognostic and biological meaning. In this way, enrichment results confidently supported the evidence that the DETFTGs were involved in tumor invasion and migration (Figure S1D~F). Besides, enriched pathways were associated with molecular function in GO/GSEA analysis (Figure S1A-C). Immunocyte infiltration and TBM analysis indicated the “hot-tumor” status and TBM ratio in a high-risk group. Finally, kTFs were related to m6A regulatory genes. Our approach generated a robust prognosis-model-based nomogram that contributed to the treatment of LGG.
Adopting a bio-information approach based on NES of TFTGS could yield less-biased global information, considering that TF has a crucial role in gene expression. As the lasso algorithm was screened, kTFs were engaged for calculating risk-score. The relation of prognosis with immunocytes was getting accumulated research. Gjorgjevski16 and Szulzewsky’s17 works, for instance, found that macrophage polarization and their bio-markers would be developed to potential targets for future anti-glioma therapy. We also analyzed the immune microenvironment based on our novel predicted model. However, tumor is not only a group of malignant tumor cells but a complex environment with many kinds of cells. Therefore, based on works like Gjorgjevski16 and Szulzewsky,17 we performed comprehensive methods (eg, deconvolution algorithm) to assess the status of infiltration of up to 22-kinds of immunocytes in different predicted groups. This would be an extension to the previous research. As the result, the high-risk group revealed a higher expression level with almost all ICPs (Figure 4B-II), which suggested that patients in the high-risk group could benefit from adopting ICPs inhibitor. According to this finding, novel drugs can be designed for targeting HAVCR2 and PDCD1LG2. Immune score from xCell showed that the high-risk group was consistent with a higher score, which suggested that patients in the high-risk group were with “hot-tumor”. “Hot-tumor” is likely to trigger a stronger immune response and usually responds well to immunotherapy. Besides, the high-risk group demonstrated findings (Figure 4D) consistent with high levels TMB. Higher TMB is usually associated with higher levels of neoantigens that can be recognized by the immune system.18 Therefore, “hot-tumor” (high-risk group) showed a higher level of macrophage M1 infiltrating (Figures 4A and S2B). Our analysis provided novel insights that could be used to develop new immunotherapeutic strategies for personalized treatment.
In conclusion, we presented a comprehensive analysis with systematically and closely combining algorithm and bio-function that could be used to predict prognosis in lower-grade glioma patients. We trained a novel independent prognostic indicator and developed an easy-to-use nomogram for clinicians to evaluate the prognosis of LGG patients, having the potential to inform treatments and therapeutic strategies for this difficult condition.
Abbreviations
DETFTGs, differential expressed transcription factor target genes; hTFs, hub transcription factors; kTFs, key transcription factors; NES, normalized enrichment score; TF, transcription factor; TFTGS, TF target gene sets; TMB, tumor mutational burden; TPM, transcripts per kilobase million.
Acknowledgments
We thank the teams of the TCGA and CGGA. Peidong Liu, Ruojie Wu, and Jinhao Zhang are co-first authors for this study.
Funding
This work was supported by funding from the National Natural Science Foundation of China (grant numbers: 81872063).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Venneti S, Huse JT. The evolving molecular genetics of low-grade glioma. Adv Anat Pathol. 2015;22(2):94–101. doi:10.1097/pap.0000000000000049
2. Ohgaki H, Kleihues P. The definition of primary and secondary glioblastoma. Clin Cancer Res. 2013;19(4):764–772. doi:10.1158/1078-0432.Ccr-12-3002
3. Hai L, Zhang C, Li T, et al. Notch1 is a prognostic factor that is distinctly activated in the classical and proneural subtype of glioblastoma and that promotes glioma cell survival via the NF-κB(p65) pathway. Cell Death Dis. 2018;9(2):158. doi:10.1038/s41419-017-0119-z
4. Tao Z, Li T, Ma H, et al. Autophagy suppresses self-renewal ability and tumorigenicity of glioma-initiating cells and promotes Notch1 degradation. Cell Death Dis. 2018;9(11):1063. doi:10.1038/s41419-018-0957-3
5. Yi L, Zhou X, Li T, et al. Notch1 signaling pathway promotes invasion, self-renewal and growth of glioma initiating cells via modulating chemokine system CXCL12/CXCR4. J Exp Clin Cancer Res. 2019;38(1):339. doi:10.1186/s13046-019-1319-4
6. Tong L, Li J, Li Q, et al. ACT001 reduces the expression of PD-L1 by inhibiting the phosphorylation of STAT3 in glioblastoma. Theranostics. 2020;10(13):5943–5956. doi:10.7150/thno.41498
7. Lim SO, Li CW, Xia W, et al. Deubiquitination and stabilization of PD-L1 by CSN5. Cancer Cell. 2016;30(6):925–939. doi:10.1016/j.ccell.2016.10.010
8. Li Q, Sun Y, Liu B, et al. ACT001 modulates the NF-κB/MnSOD/ROS axis by targeting IKKβ to inhibit glioblastoma cell growth. J Mol Med. 2020;98(2):263–277. doi:10.1007/s00109-019-01839-0
9. Kolmykov S, Yevshin I, Kulyashov M, et al. GTRD: an integrated view of transcription regulation. Nucleic Acids Res. 2021;49(D1):D104–D111. doi:10.1093/nar/gkaa1057
10. Li Y, Xiao J, Bai J, et al. Molecular characterization and clinical relevance of m(6)A regulators across 33 cancer types. Mol Cancer. 2019;18(1):137. doi:10.1186/s12943-019-1066-3
11. Mathur R, Zhang Y, Grimmer MR, et al. MGMT promoter methylation level in newly diagnosed low-grade glioma is a predictor of hypermutation at recurrence. Neuro Oncol. 2020;22(11):1580–1590. doi:10.1093/neuonc/noaa059
12. Bell EH, Zhang P, Fisher BJ, et al. Association of MGMT promoter methylation status with survival outcomes in patients with high-risk glioma treated with radiotherapy and temozolomide: an analysis from the NRG oncology/RTOG 0424 trial. JAMA Oncol. 2018;4(10):1405–1409. doi:10.1001/jamaoncol.2018.1977
13. Vandeweyer G, Helsmoortel C, Van Dijck A, et al. The transcriptional regulator ADNP links the BAF (SWI/SNF) complexes with autism. Am J Med Genet C Semin Med Genet. 2014;166c(3):315–326. doi:10.1002/ajmg.c.31413
14. Tekendo-Ngongang C, Kruszka P, Martinez AF, Muenke M. Novel heterozygous variants in KMT2D associated with holoprosencephaly. Clin Genet. 2019;96(3):266–270. doi:10.1111/cge.13598
15. Froimchuk E, Jang Y, Ge K. Histone H3 lysine 4 methyltransferase KMT2D. Gene. 2017;627:337–342. doi:10.1016/j.gene.2017.06.056
16. Gjorgjevski M, Hannen R, Carl B, et al. Molecular profiling of the tumor microenvironment in glioblastoma patients: correlation of microglia/macrophage polarization state with metalloprotease expression profiles and survival. Biosci Rep. 2019;39(6). doi:10.1042/bsr20182361
17. Szulzewsky F, Pelz A, Feng X, et al. Glioma-associated microglia/macrophages display an expression profile different from M1 and M2 polarization and highly express Gpnmb and Spp1. PLoS One. 2015;10(2):e0116644. doi:10.1371/journal.pone.0116644
18. Maleki Vareki S. High and low mutational burden tumors versus immunologically hot and cold tumors and response to immune checkpoint inhibitors. J Immunother Cancer. 2018;6(1):157. doi:10.1186/s40425-018-0479-7
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.