Back to Journals » Journal of Inflammation Research » Volume 17
Transcription Factor IRF7 is Involved in Psoriasis Development and Response to Guselkumab Treatment
Authors Yuan X, Xin T , Yu H, Huang J, Xu Y , Ou C, Chen Y
Received 5 December 2023
Accepted for publication 13 February 2024
Published 15 February 2024 Volume 2024:17 Pages 1039—1055
DOI https://doi.org/10.2147/JIR.S450048
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Tara Strutt
Xiuqing Yuan,1,* Tiantian Xin,1,* Huanhuan Yu,1 Jian Huang,2 Yaohan Xu,1 Caixin Ou,1 Yongfeng Chen1
1Department of Dermatology, Dermatology Hospital of Southern Medical University, Guangzhou, Guangdong Province, People’s Republic of China; 2Department of Dermatology, Guangdong College of Clinical Dermatology, Anhui Medical University, Hefei, Anhui Province, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Yongfeng Chen, Dermatology Hospital of Southern Medical University, No. 2, LuJing Road, Guangzhou, Guangdong Province, 510091, People’s Republic of China, Tel +86 13802920269, Email [email protected]
Purpose: Guselkumab is a highly effective biologic agent for treating psoriasis. This study aimed to explore potential transcription factors involved in psoriasis pathogenesis and response to guselkumab treatment, aiming to provide new therapeutic strategies for psoriasis.
Patients and Methods: We analyzed gene expression and single-cell RNA-seq data from the Gene Expression Omnibus (GEO) database. Differentially expressed genes (DEGs) that upregulated in psoriasis and downregulated after guselkumab treatment were subjected to enrichment analyses. Single-cell regulatory network inference and clustering (SENIC) and regulon module analyses identified different regulon activities between the lesion and non-lesion skin of psoriasis. Cell-cell communication analysis revealed interactions among specific cell clusters. Transcription factor (TF) regulons were identified from the guselkumab-specific regulon network. Gene set enrichment analysis (GSEA) confirmed the IRF7 regulon in the validation cohort. Finally, the expression level of IRF7 was identified in plaque psoriasis before and after 12 weeks of guselkumab therapy by immunohistochemical experiment.
Results: 799 DEGs were downregulated after guselkumab treatment. Enrichment analyses highlighted the interleukin-17 (IL-17) pathway in this gene set. The M2 module exhibited the primary difference in regulon activity. Strong cell-cell interactions were observed between keratinocytes and immune cells. IRF7 regulon had significant roles in psoriasis and treatment response, as validated by GSEA analysis using the IL-17 signaling pathway as a reference. The immunohistochemical analysis unveiled substantial differences in the expression levels of IRF7 in psoriatic skin samples before and after 12 weeks of guselkumab treatment.
Conclusion: IRF7 may be the key player in psoriasis pathogenesis and the therapeutic process involving guselkumab. Targeting IRF7 might offer new therapeutic strategies for psoriasis.
Keywords: guselkumab, plaque psoriasis, IL-23/IL-17 signaling pathway, SENIC analysis, TF regulon
Introduction
Psoriasis is a chronic skin disorder distinguished by the appearance of red plaques and scaly patches arising from aberrant keratinocyte differentiation and excessive proliferation.1 It impacts an estimated global population of 125 million individuals, leading to substantial declines in both quality of life and economic burdens.2 This complex disease is influenced by immunological, genetic, and environmental factors, leading to the development of various treatment and management strategies targeting its underlying causes.3,4 Since the interleukin-23/ interleukin-17 (IL-23/IL-17) signaling pathway plays a pivotal role in its pathogenesis,5,6 monoclonal antibodies targeting this pathway have been developed and approved for clinical use, marking a paradigm shift in psoriasis management.7,8 Guselkumab, an IL-23-specific antibody, has demonstrated high efficacy in treating psoriasis.9 Although guselkumab primarily acts through the IL-23/IL-17 axis, the potential underlying mechanisms have not yet been fully explored. An in-depth exploration of guselkumab therapy from multiple perspectives could provide a comprehensive understanding of the IL-23/IL-17 pathway and help identify effective therapeutic targets for psoriasis.
Transcription factors (TFs) play a pivotal role in controlling gene expression, exerting their influence over a wide array of biological processes, including metabolism, immune response, organ development, and cellular differentiation.10 Dysregulation of TF activity can contribute to diseases like cancer, diabetes, and autoimmune disorders.11–13 Targeting TFs with drugs holds promise for therapeutically modulating gene expression, including in psoriasis treatment. The IL-23/IL-17 pathway is currently being investigated to identify potential therapeutic targets. For instance, targeting RORγT, a key regulator of Th17 cell differentiation known to induce the production of cytokines with inflammatory properties, has exhibited the potential to achieve improved effectiveness compared to inhibitors specifically targeting IL-17A.14 Inhibiting ROCK2, which hinders IRF4, a crucial transcription factor for Th17 cell differentiation and IL-17 production, may also lead to a decrease in IL-17 levels.15 These druggable targets could potentially suppress the activation of the IL-23/IL-17 pathway in treating psoriasis. However, there remain numerous unknown TFs within the IL-23/IL-17 pathway that require further exploration.
In our study, we aimed to investigate the TF regulons of guselkumab therapy for plaque psoriasis. Instead of focusing on a single or a few differentially expressed genes (DEGs) separately, we identified a set of treatment response genes to guselkumab therapy in psoriasis and explored the critical TF regulon model using single-cell RNA sequencing (scRNA-seq) data. By intersecting this critical regulon model with genes that respond after guselkumab therapy, we extracted the guselkumab therapy-related regulon network. We then divided the regulon network into multiple parts based on the TFs to form different TF Regulons and used the classic IL-17 pathway as a reference to examine their pathway activity and distribution among various cell clusters. In the validation cohort related to IL-17A inhibitory treatment for psoriasis, we evaluated the role of the IRF7 regulon in psoriasis. Our findings indicated a more significant enrichment of the IRF7 regulon compared to the IL-17 pathway during psoriasis treatment. To the best of our knowledge, this study is the first to explore the relevant TF regulon of guselkumab therapy in psoriasis at the TF regulatory level, comprehensively evaluating and validating it by referring to the IL-17 pathway. We hope that our research will enhance the comprehension of the molecular mechanism involved in guselkumab therapy for psoriasis and offer novel perspectives on targeted TF for treating psoriasis.
Materials and Methods
Data Acquisition
To investigate gene changes in plaque psoriasis skin lesions treated with guselkumab and identify relevant feature genes at the single-cell level, we utilized two datasets as the exploration cohort: GSE51440 (guselkumab-treated samples) and GSE173706 (scRNA-seq data). For the validation cohort, we searched the GEO database (http://www.ncbi.nlm.nih.gov/geo/) using the keywords “secukinumab psoriasis” or “ixekizumab psoriasis” before April 2023. We identified four expression profiling by array datasets: GSE201827, GSE137218, GSE102725, and GSE178228, which contained skin tissue treated with IL-17A inhibitors. Three datasets used secukinumab, while one used ixekizumab. All datasets were downloaded and analyzed using R software (version 4.2.2).
Differential Expression Analysis
We categorized samples into four groups for differential gene expression analysis on the GSE51440 dataset based on skin status and guselkumab treatment duration for psoriasis. These groups included non-lesion (NL) samples, as well as lesion samples collected at baseline (BL), weeks 1 (W1), and 12 (W12) after treatment. We utilized the “limma” package in R software to identify DEGs meeting specific criteria: an absolute log fold change (logFC) > 0.6 and an adjusted P value (P-adj) < 0.05. Using the “ggVennDiagram” package, we identified a set of DEGs highly expressed in BL compared with NL and down-regulated after treatment. This set was defined as the “psoriasis treatment response genes by guselkumab therapy” (PTR-GUS) set.
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) Enrichment Analyses
We performed GO and KEGG enrichment analyses to gain insights into the functions of the PTR-GUS set obtained from differential expression analysis. The GO enrichment analysis incorporated three components: biological process (BP), cellular component (CC), and molecular function (MF). The analyses were performed using the “enrichGO” and “enrichKEGG” functions in the “clusterProfiler” package (version 4.0), respectively.16 We then classified pathways as statistically significant if they had P-adj < 0.05. To visualize the results of the enrichment analyses, we utilized the “enrichplot” package to generate a tree plot.
ScRNA-Seq Data Processing
We analyzed single-cell data from plaque psoriatic (PP) and peripheral normal skin (PN) samples in the GSE173706 dataset using the “Seurat” package (version 4.0).17 The data were processed according to the official Seurat package vignette, including batch correction using the “harmony” package.18 Specifically, we performed NormalizeData, FindVariableFeatures, ScaleData, RunPCA, RunHarmony, RunUMAP, FindNeighbors, and FindClusters with default parameters. We used the first 30 Harmony dimensions as input for UMAP and a clustering resolution of 0.5 to visualize the results on the first two UMAP dimensions, resulting in 20 clusters. The identification of marker genes for individual clusters was performed using the “FindAllMarkers” function in Seurat, followed by a comparison with known signature genes of different skin cell types available in the DISCO database (https://www.immunesinglecell.org/) and widely acknowledged in scientific literature.19–21 Finally, we annotated the 20 clusters and visualized their marker genes using the “scRNAtoolVis” package in the dot plot. Additionally, we used bar plots to show the proportion of immune cell clusters in PP and PN groups.
Single-Cell Regulatory Network Inference and Clustering (SENIC) Analysis
We conducted SCENIC analysis (version 1.3.1) on the processed single-cell data to identify regulatory networks and cell clusters in psoriasis skin lesions.22 Before conducting SCENIC analysis, we integrated cells using the “MetaCell” algorithm to improve data interpretation by mitigating technical noise and batch effects and increasing statistical power.23 In the SCENIC analysis, we followed a series of steps to identify potential TF targets and assess the activity of regulons in individual cells. First, we utilized GENIE3 (version 1.20.0) to identify potential TF targets based on co-expression networks. Next, we performed TF-motif enrichment analysis and identified direct targets using RcisTarget (version 1.18.2). To conduct this analysis, we downloaded the reference genome from the cisTarget database (https://resources.aertslab.org/cistarget/). Specifically, we used the following reference datasets: hg38__refseq-r80__500bp_up_and_100bp_down_tss.mc9nr.feather and hg38__refseq-r80__10kb_up_and_down_tss.mc9nr.feather. Finally, we scored the activity of regulons in individual cells using AUCell (version 1.20.2).22 Based on the regulon activity score (RAS) matrix obtained from the SCENIC analysis, we used the UMAP algorithm to reduce dimensionality, and the first two RAS-UMAP dimensions were visualized by cell cluster or group.
Regulon Modules Analysis Based on Connection Specificity Index (CSI) Matrix
We identified regulon modules in psoriasis skin lesions using the context-dependent CSI method that measures associations between TFs and their target genes.24 Initially, we calculated the Pearson correlation coefficient (PCC) of activity scores across various pairs of regulons. Subsequently, we defined the CSI between two fixed regulons, A and B, as the fraction of regulons whose PCC with A and B was lower than that between A and B. We then applied hierarchical clustering with Euclidean distance to the resulting CSI matrix to identify distinct regulon modules. Furthermore, we constructed a regulon association network using a threshold of CSI > 0.8 to explore the relationships between different regulons. The result was visualized by the “ggplot2” package. We prioritized the regulon module with the most remarkable inter-group differences for further analysis, which may be crucial to psoriasis pathogenesis.
Inference of Cell-Cell Communication of Specific Cell Clusters
To understand the cell-cell communication networks underlying psoriasis etiology, we focused on the M2 regulon module that showed the most significant inter-group differences, with the keratinocyte subgroups KC.cycling and KC.bas.1 exhibiting the most significant changes. To investigate how these differences may affect other cell types in the lesions, we utilized the “CellChat” package to infer the cell communication network of these keratinocytes based on quantitative inference from scRNA-seq data.25 All data analysis parameters were set according to the official documentation available on Github (https://github.com/sqjin/CellChat).
Construction of Gene Regulatory Network (GRN) Specific to the PTR-GUS Set
To unravel the intricate gene regulatory networks underlying psoriasis, we used SCENIC analysis and regulon module analysis to identify the regulon module with the most remarkable inter-group differences based on scRNA-seq data. We then compared the genes in the PTR-GUS set with those in the identified module to investigate any shared features between them. After extracting the common genes and identifying the TFs that regulate them with high confidence, we constructed a GRN specific to the PTR-GUS set using the “igraph” package.
Scoring TF Regulons in the GRN Specific to the PTR-GUS Set
To further analyze the GRN specific to the PTR-GUS set identified from single-cell RNA sequencing data, we partitioned it into multiple units based on TFs. We defined a TF regulon as any TF with more than 30 target genes. We then used the AUCell algorithm to score the TF regulons in the scRNA-seq data.22 To enhance the interpretation of these scores, we incorporated the IL-17 signaling pathway associated with psoriasis pathogenesis as a reference gene set. Specifically, we extracted the IL-17 signaling pathway from the KEGG gene set available through the Enrichr library (https://maayanlab.cloud/Enrichr/) and integrated it into the TF regulons.26
Verification of IRF7 Regulon Associated with PTR-GUS Set
To validate the IRF7 regulon linked to the PTR-GUS set identified in the exploration cohort, we applied consistent data preprocessing and differential analysis methods to distinct datasets within the validation cohort. Firstly, we utilized the “ggvenn” package to identify the common genes between the IRF7 regulon and the IL17 pathway. Additionally, we employed the “clusterProfiler” package (version 4.0) for GSEA analysis, considering different grouping conditions.16 The generation of gene lists, based on logFC values, served as inputs for subsequent GSEA. In order to enhance the interpretability of the enrichment outcomes, we incorporated the IL-17 signaling pathway, known to be associated with psoriasis pathogenesis, as reference gene sets for GSEA analysis. The GSEA results were visualized using the “ggplot2” package.
Immunohistochemical Validation of IRF7 Regulon Expression in Psoriatic Skin and Effect of Guselkumab Treatment
Immunohistochemical experiments were performed using paraffin-embedded skin samples obtained from three plaque psoriasis patients before and after 12 weeks of guselkumab treatment. The skin tissues were sectioned to a thickness of 3 um. Hydrogen peroxide blockage and antigen retrieval steps were performed following deparaffinization and rehydration. Subsequently, the sections were incubated with an IRF7 antibody (1:200 dilution, Proteintech, USA) for 30 minutes. A goat anti-rabbit IgG peroxidase conjugate (Proteintech, USA) was then applied as the secondary antibody for 20 minutes at 37°C. Finally, the slides were observed under a bright-field microscope at magnifications of 10x. The study received ethical approval from the Research Ethics Committee of Dermatology Hospital, Southern Medical University.
Results
Selection of Datasets and Samples
With the criteria mentioned above, we obtained six datasets to perform subsequent analyses. Detailed information regarding the extracted samples can be found in Table 1.
 |
Table 1 Summary of Six GEO Datasets Involved in the Exploration and Validation Cohort |
Identification of PTR-GUS Set
We detected 2138 differentially expressed genes (DEGs) between the BL and NL groups in the GSE51440 dataset. Among these, 1321 genes were found to be upregulated, while 817 genes were downregulated based on the predefined criteria. In comparison, for the W12 group relative to the BL group, we identified 1867 DEGs, including 844 upregulated genes and 1023 downregulated genes. However, no DEGs were found between the BL and W1 groups, possibly due to the short duration of guselkumab therapy. We created volcano plots (Figure 1A and B) to show the results of differential analyses with positive findings. Using a Venn diagram, we identified 799 DEGs (Figure 1C) named the PTR-GUS set.
Functional Enrichment Pathways of PTR-GUS Set
We performed functional enrichment analysis for genes in the PTR-GUS set using GO and KEGG analyses. We identified 572 significant GO pathways and 13 enriched KEGG pathways (Supplementary Table 1). Tree plots were used to visualize the top 30 enriched GO pathways and all KEGG pathways (Figure 1D and E). Enriched GO pathways included defense against viruses and bacteria, mitotic cell division, and epidermis development. Similar findings were observed in the KEGG analysis, which revealed a significant enrichment of immune-related pathways, such as the IL-17 signaling pathway. Additionally, infectious disease-related pathways, metabolic pathways, and cell cycle pathways were also identified as enriched pathways. These results further supported the involvement of immune processes, infection response, and cellular metabolism in the pathogenesis of the condition.
Cell Clusters Annotation of scRNA-Seq Dataset
We utilized the Seurat and Harmony packages to process the scRNA-seq dataset. By comparing cell markers for each cluster with commonly used skin cell markers from databases and literature, we identified 20 distinct cell populations (Figure 2A). Among these populations, keratinocytes (KC) exist in several states, including KC.bas.1–3, KC.cycling, KC.spin.1–3, and KC.gran. These different states represent the maturation progression of keratinocytes, starting from basal cells and progressing through cycling, spinous, and ultimately granular cells. In addition to keratinocytes, various immune cells, including macrophages or dendritic cells (IMM.Macro/DC), natural killer cells or T cells (IMM.NK/T), and mast cells (IMM.Mast) contribute to the immune response in the skin, performing functions such as antigen presentation, killing infected cells, and participating in allergic reactions and inflammation. Furthermore, endothelial cells (EC) such as EC.vascular.1–2, EC.lympha, and EC.tip are responsible for blood or lymphatic vessel formation and maintenance in the skin. Pericytes provide support to endothelial cells, while fibroblasts (Fibroblast.1–2) synthesize and organize the extracellular matrix. Glial cells, closely related to fibroblasts, contribute to skin homeostasis. Lastly, melanocytes produce melanin for skin pigmentation and protection against UV radiation. We then visualized cell-specific markers for these 20 cell clusters using a dot plot (Figure 2B). Additionally, percentage plots showed a significantly higher abundance of immune cells in PP compared with the PN group, particularly in the IMM.NK/T cell cluster (Figure 2C). This finding was consistent with the widely accepted understanding that psoriasis is an immune-mediated disease.
Identification of Differential Regulation Between Groups
Using the SENIC analysis, we derived a RAS matrix that was subsequently employed to conduct an unsupervised clustering analysis of keratinocyte clusters by applying the UMAP algorithm. Distinctive variances were identified in the regulon activity of keratinocyte clusters between the PP and PN groups, while other cell types showed no notable distinctions (Figure 3A).
Significant Differences in the M2 Regulon Module Between Groups
Using SENIC and regulon module analysis, we found that the regulon activity of the M2 module was significantly higher in PP compared with the PN group. Specifically, keratinocyte clusters, particularly KC.cycling and KC.bas.1 clusters showed a notably higher regulon activity within the M2 module in PP than in the PN group. Additionally, immune cell clusters such as IMM.NKT and IMM.Macro/DC had slightly higher regulon activity in the PP group (Figure 3B).
Abnormal Keratinocyte-Immune Cell Interactions Drive Excessive Immune Activation
CellChat analysis revealed strong associations between the KC.cycling and KC.bas.1 keratinocyte subpopulations and IMM.NKT, IMM.Macro/DC, and EC.lympha clusters (Figure 3C). Moreover, migration inhibitory factor (MIF) signaling pathways were more pronounced in KC.cycling and KC.bas.1 clusters in the PP group compared with the PN group. Interestingly, only the KC.bas.1 cluster in the PP group exhibited secretion of tumor necrosis factor (TNF) pathway-related substances (Figure 3D and E). These observed differences in cell-cell interactions and signaling pathways may contribute to excessive immune activation and the aggregation of immune cells in psoriasis lesions.
Discovery of a PTR-GUS Set Specific GRN
By intersecting the PTR-GUS set with genes from the M2 regulon module, we identified a GRN specific to the PTR-GUS set (Figure 4). The GRN included several TFs, such as IRF7, STAT1, ETV4, ELF1, and TFDPI, which directly regulated dozens of genes. Additionally, other TFs, like HTATIP2, TBX1, and MYBL2, directly regulated fewer genes. The above TFs interacted with each other through several co-regulated genes. However, some transcription factors, such as IRF4 and SREBF1, were found to individually regulate CD274 and KRT6A, respectively. These findings provided insights into the involvement of specific TFs in psoriasis development and the potential effects of guselkumab treatment at the gene regulatory level.
Four TF Regulons Outperformed the IL-17 Pathway in Assessing Psoriasis
Using the aforementioned criteria, we identified five TFs (IRF7, STAT1, ELF1, ETV4, and TFDPI) and their target genes to construct the TF regulon set. Combining these regulons with the IL-17 pathway, we employed the AUCell algorithm for scoring. Our analysis revealed that, except for the TFDP regulon, IRF7, STAT1, ELF1, and ETV4 regulon exhibited more significant intergroup differences compared with the IL-17 pathway (Figure 5A–F). Notably, the enrichment level of the IRF7 regulon demonstrated the most pronounced disparities among the IL-17 pathway and other TF regulons. These findings suggested that the TF regulons may offer a more effective assessment of psoriasis than the conventional IL-17 pathway.
A Gradual Decrease in the Enrichment Score of IRF7 Regulon During IL-23/IL-17 Inhibitor Therapy for Psoriasis
Based on AUCell analysis, we observed that the IRF7 regulon displayed the most substantial intergroup disparities when compared to the IL-17 pathway as a reference. Therefore, we evaluated the potential correlation between the IRF7 regulon and the different states of biologics therapy for psoriasis at various time points. Similarly, we applied the IL17 pathway as a reference. The IRF7 regulon and the IL17 pathway exhibited significant differences in their gene composition, as evidenced by the Venn diagram, which shows only three common genes between them (Figure 6A). Prior to treatment, the IRF7 regulon exhibited a significantly higher positive enrichment score in lesional tissue than in non-lesional tissue, as evidenced by its comparison with the IL17 pathway gene set. In the initial stage of treatment, there was a slight decrease in the enrichment score of the IRF7 regulon. At the same time, the IL-17 signaling pathway showed a significant negative enrichment level in the lesions after the first week of treatment compared to the baseline. However, as the treatment duration increased, this trend reversed, with the enrichment score of the IRF7 regulon becoming slightly more negative than that of the IL-17 pathway (Figure 6B–F). These findings supported the involvement of the IRF7 regulon in psoriasis development and suggested its potential as a treatment target, as the enrichment level of the regulon gradually decreased with disease improvement.
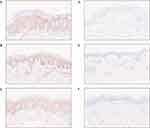
Downregulation of IRF7 Expression in Psoriatic Skin Following Treatment
Based on previous analysis, the IRF7 regulon was found to have significantly higher expression levels in psoriasis compared to healthy skin. Immunohistochemistry experiments were conducted to validate this observation. The results showed a significant reduction in IRF7 expression in psoriatic skin after 12 weeks of guselkumab treatment compared to its high expression levels prior to treatment (Figure 7A–F). This finding aligned with the observations from the AUCell analysis. Notably, apart from keratinocytes, inflammatory cells within the dermal stroma of psoriatic skin also exhibited high levels of IRF7 expression prior to treatment.
Discussion
Psoriasis is a chronic immune-mediated skin condition characterized by erythematous plaques and scaling, where immune dysfunction plays a central role in its pathophysiology.1 The IL-23/IL-17 signaling pathway has emerged as a critical pathway leading to psoriasis, and various therapeutic targets have been developed, including TNF-α, IL-17A, and IL-23 monoclonal antibodies.5,27 Anti-cytokine antibody therapy has shown superior clinical remission rates compared with traditional treatments, highlighting the importance of cytokines such as TNF, IL-23, and IL-17 in psoriasis.28
It is widely accepted that IL-23 inhibitors act upstream by blocking the interaction between IL-23 and its receptor on Th17 cells that produce proinflammatory cytokines such as IL-17. This blockade effectively prevents the activation and proliferation of these cells, leading to a reduction in downstream inflammation.5,29 Notably, the activities of transcription factors play a pivotal role in these processes. Several studies have provided evidence in support of this aspect. For example, STAT3, a key transcription factor associated with inflammatory responses, has been found to be activated by IL-23 in psoriatic lesions. This immune activation prompts the generation of proinflammatory cytokines and chemokines, ultimately exerting a substantial impact on the pathogenesis of psoriasis.30,31 Other transcription factors, such as NF-kB and AP-1, have also been implicated in regulating psoriasis-related genes and may be influenced by IL-23 signaling.32,33 Despite these findings, specific transcription factors associated with the progression of psoriasis through IL-23 still require further exploration and understanding. In our study, we mainly utilized bioinformatics methods to analyze response genes in psoriasis following anti-IL23 therapy. Our aim was to gain insights into the potential involvement of transcription factors in the pathogenesis and response to IL-23 inhibitor therapy for psoriasis.
The PTR-GUS set demonstrated enrichment in the IL-17 signaling pathway, as well as pathways related to cell cycle and keratinocyte differentiation. These results supported the notion that the customized gene set accurately reflects the recognized pathogenesis of psoriasis. The scenic regulon analysis identified the M2 module as the primary differentiating factor in regulon activity between PP and PN.
Particularly keratinocyte clusters KC.cycling and KC.bas.1 showed significant differences in regulon activity, while no notable variations were observed among other cell groups. Epidermal keratinocytes play a crucial role in innate immunity by influencing the recruitment and modulation of T cells in the skin, contributing to disease onset.6 We hypothesized that these two keratinocyte subpopulations with distinct regulon activity differences might affect other cells through cell communication. To investigate this hypothesis, we performed cell-cell communication analysis, revealing close interactions between these keratinocyte populations and immune cell clusters IMM.NKT and IMM.Macro/DC. Further exploration unveiled the enrichment of inflammation-related pathways, including the TNF signaling pathway exclusively enriched in PP and higher activation of the MIF signaling pathway in PP. The significance of the TNF signaling pathway in psoriasis is widely acknowledged, and TNF-targeting biologics have demonstrated efficacy in treating psoriasis.2 Similarly, previous studies have highlighted the chemotactic functions of MIF and its role as a significant regulatory factor in recruiting inflammatory cells.34 MIF exerts its biological effects through binding to receptors CD74, CD44, CXCR4, and so forth in a paracrine manner.35 Elevated levels of MIF in psoriasis patients can activate mitogen- and stress-activated kinase 1 (MSK1) and other pathways, promoting keratinocyte proliferation.36 These findings potentially explained the higher proportion of immune cells and the more pronounced inflammatory response observed in keratinocytes from PP compared with PN.
Our analysis demonstrated that both the regulon activity of the M2 module and gene expression of the PTR-GUS set were significantly elevated in lesion skin of psoriasis. To create a guselkumab-specific regulon network, we identified genes from the PTR-GUS set within the M2 module. This network consisted of TFs STAT1, IRF7, ELF1, and ETV4 and their regulated genes. Using the AUCell algorithm, we assessed the potential significance of these TF regulons concerning psoriasis. Remarkably, we observed statistical differences between PP and PN, with the activities of these four regulons, particularly IRF7, showing higher enrichment scores compared to the classical IL-17 pathway. Our validation cohort further confirmed the roles of the IRF7 regulon in psoriasis. Notably, the enrichment score of IRF7 regulon displayed similar temporal changes to those of the IL-17 pathway and was even more associated with psoriasis, suggesting its potential role as a biomarker for psoriasis. Furthermore, except for being highly expressed in keratinocytes of psoriatic skin, IRF7 was predominantly expressed in inflammatory cells. Hence, targeting IRF7 may offer a more cell-specific therapeutic approach compared to targeting IL-23 or IL-17 cytokines.
IRF7 is a member of the interferon regulatory factor family and shares DNA-binding domain homology with IRF3. It plays a significant role in producing type I interferons (IFN-I), which activate the JAK-STAT pathway and induce the expression of immune response-related genes by creating proinflammatory factors.37,38 Integrated bioinformatic analysis by Tang et al identified IRF7 as a critical potential gene for psoriasis treatment.39 Raposo et al also reported a substantial upregulation of IRF7 in psoriatic lesions compared to non-lesion skin, a finding corroborated by our immunohistochemistry experiment that demonstrated a positive correlation between IRF7 overexpression and psoriasis.40 Following 12 weeks of guselkumab treatment, a significant reduction in IRF7 expression was observed in the lesion skin of psoriasis patients. Additionally, our in silico experiments using a validation cohort revealed an association between the IRF7 regulon and psoriasis and its correlation with disease improvement through anti-IL-23/IL-17 therapy. These findings emphasized the significance of elevated IRF7 expression in psoriasis, which was subsequently downregulated following effective treatment. This observation implied the potential contribution of IRF7 to the development and progression of the disease.
In the broader context, research has illuminated the essential role of transcription factor IRF-7 in activating the IFN induction pathway in plasmacytoid dendritic cells (pDC).41,42 Mirroring this, the participation of IRF7, alongside IRF3, in the NF-kB signaling pathway-mediated production of IL-23 by dendritic cells has been identified as a potential contributor to the increased proliferation of epidermal cells in psoriasis.43 Intriguingly, SIDT1-deficient mice exhibited reduced severity in an imiquimod-induced acute psoriasis-like model, attributed to decreased production of IFN-I and IFN-dependent chemokines. Further mechanistic insights indicated that SIDT1 facilitated the nuclear migration of IRF7 in pDC, emphasizing the critical role of IRF7 in psoriasis.44 Our immunohistochemistry experiment also revealed high expression of IRF7 in inflammatory cells within psoriatic skin prior to treatment. These findings highlighted the essential role of IRF7 in the development of psoriasis, as it governed the production of proinflammatory factors and chemokines while also promoting abnormal proliferation of epidermal cells.
Our study has certain limitations that should be acknowledged. Firstly, we focused only on genes overexpressed in psoriasis and showed a decreased response after treatment, neglecting genes downregulated in psoriasis and exhibiting an increased response after treatment. Secondly, our research solely relied on gene expression changes in datasets without considering other factors, such as environmental or genetic factors, that might influence disease development. Thirdly, the sample size used in our exploration cohort was relatively small due to the availability of only one dataset containing skin tissue from patients undergoing guselkumab therapy for psoriasis. More samples would be required to validate the reliability of our findings, if possible. Lastly, our study mainly relied on secondary analysis of publicly available databases, and we only validated the expression of IRF7 in skin tissue from limited patients before and after guselkumab treatment. Investigating the role and mechanism of IRF7 regulon in psoriasis using experimental approaches will be the primary focus in future work.
Conclusions
Through our analysis of the regulon network in patients undergoing guselkumab therapy for psoriasis, we have identified the involvement of the IRF7 regulon in the development of psoriasis and the treatment process with guselkumab. Our findings suggest that targeting IRF7 might offer a novel therapeutic strategy for treating psoriasis, presenting an alternative to the current cytokine-targeted therapies.
Abbreviations
GEO, Gene Expression Omnibus; scRNA-seq, single-cell RNA sequencing; DEGs, Differentially expressed genes; SENIC, Single-cell regulatory network inference and clustering; TF, Transcription factor; GSEA, Gene set enrichment analysis; IL-17, interleukin-17; IL-23, interleukin-23; NL, non-lesion; BL, lesion samples collected at baseline; logFC, log fold change; P-adj, adjusted P value; PTR-GUS, psoriasis treatment response genes by guselkumab therapy, GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; BP, biological process; CC, cellular component; MF, molecular function; PP, plaque psoriatic skin; PN, peripheral normal skin; RAS, regulon activity score; CSI, Connection Specificity Index; PCC, Pearson correlation coefficient; GRN, gene regulatory network; KC, keratinocytes; KC.bas, keratinocyte basal cells; KC.cycling, Keratinocyte cycling cells; KC.spin, keratinocyte spinous cells; KC.gran, keratinocyte granular cells; IMM, immune cells; IMM.Macro/DC, macrophages or dendritic cells; IMM.NK/T, natural killer cells or T cells; IMM.Mast, mast cells; EC, endothelial cells; TNF, tumor necrosis factor; MSK1, mitogen- and stress-activated kinase 1; MIF, migration inhibitory factor; IFN-I, type I interferons; pDC, plasmacytoid dendritic cells.
Data Sharing Statement
Upon a reasonable request, individuals can obtain the datasets and R code utilized in this study directly from the corresponding author.
Ethics Approval and Informed Consent
The current study received ethical approval from the Research Ethics Committee of the Dermatology Hospital affiliated with Southern Medical University in Guangzhou, China (IRB#2023024). The anonymized processing of patient data allowed for the waiver of informed consent. The study was conducted following the principles outlined in the Declaration of Helsinki.
Acknowledgments
We sincerely appreciate the authors of GSE51440, GSE173706, GSE201827, GSE137218, GSE102725, and GSE178228 for sharing their data, which greatly contributed to the success of our study. Special thanks go to Jarning Gau and Shipeng Guo from the GZDlab organization for their invaluable technical suggestions. We also acknowledge the hard work of our researchers and the valuable feedback provided by the reviewers.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This research received no external funding.
Disclosure
The authors declare no competing interests related to this study.
References
1. Griffiths CEM, Armstrong AW, Gudjonsson JE, Barker JNWN. Psoriasis. Lancet. 2021;397(10281):1301–1315. doi:10.1016/s0140-6736(20)32549-6
2. Armstrong AW, Read C. Pathophysiology, clinical presentation, and treatment of psoriasis: a review. JAMA. 2020;323(19):1945–1960. doi:10.1001/jama.2020.4006
3. Rendon A, Schäkel K. Psoriasis pathogenesis and treatment. Int J Mol Sci. 2019;20(6):1475. doi:10.3390/ijms20061475
4. Ko SH, Chi CC, Yeh ML, Wang SH, Tsai YS, Hsu MY. Lifestyle changes for treating psoriasis. Cochrane Database Syst Rev. 2019;7(7):CD011972. doi:10.1002/14651858.CD011972.pub2
5. Ghoreschi K, Balato A, Enerbäck C, Sabat R. Therapeutics targeting the IL-23 and IL-17 pathway in psoriasis. Lancet. 2021;397(10275):754–766. doi:10.1016/S0140-6736(21)00184-7
6. Lowes MA, Suárez-Fariñas M, Krueger JG. Immunology of psoriasis. Annu Rev Immunol. 2014;32:227–255. doi:10.1146/annurev-immunol-032713-120225
7. Conrad C, Gilliet M. Psoriasis: from pathogenesis to targeted therapies. Clin Rev Allergy Immunol. 2018;54(1):102–113. doi:10.1007/s12016-018-8668-1
8. Armstrong AW, Puig L, Joshi A, et al. Comparison of biologics and oral treatments for plaque psoriasis: a meta-analysis. JAMA Dermatol. 2020;156(3):258–269. doi:10.1001/jamadermatol.2019.4029
9. Reich K, Armstrong AW, Langley RG, et al. Guselkumab versus secukinumab for the treatment of moderate-to-severe psoriasis (ECLIPSE): results from a Phase 3, randomised controlled trial. Lancet. 2019;394(10201):831–839. doi:10.1016/S0140-6736(19)31773-8
10. Lambert SA, Jolma A, Campitelli LF, et al. The human transcription factors. Cell. 2018;172(4):650–665. doi:10.1016/j.cell.2018.01.029
11. Calissi G, Lam EWF, Link W. Therapeutic strategies targeting FOXO transcription factors. Nat Rev Drug Discov. 2021;20(1):21–38. doi:10.1038/s41573-020-0088-2
12. Zhang Z, Xu L, Xu X. The role of transcription factor 7-like 2 in metabolic disorders. Obes Rev. 2021;22(5):e13166. doi:10.1111/obr.13166
13. Kanno Y, Vahedi G, Hirahara K, Singleton K, O’Shea JJ. Transcriptional and epigenetic control of T helper cell specification: molecular mechanisms underlying commitment and plasticity. Annu Rev Immunol. 2012;30:707–731. doi:10.1146/annurev-immunol-020711-075058
14. Gege C. RORγt inhibitors as potential back-ups for the Phase II candidate VTP-43742 from Vitae Pharmaceuticals: patent evaluation of WO2016061160 and US20160122345. Expert Opin Ther Pat. 2017;27(1):1–8. doi:10.1080/13543776.2017.1262350
15. Biswas PS, Gupta S, Chang E, et al. Phosphorylation of IRF4 by ROCK2 regulates IL-17 and IL-21 production and the development of autoimmunity in mice. J Clin Invest. 2010;120(9):3280–3295. doi:10.1172/JCI42856
16. Wu T, Hu E, Xu S, et al. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation. 2021;2(3):100141. doi:10.1016/j.xinn.2021.100141
17. Satija R, Farrell JA, Gennert D, Schier AF, Regev A. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol. 2015;33(5):495–502. doi:10.1038/nbt.3192
18. Korsunsky I, Millard N, Fan J, et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Methods. 2019;16(12):1289–1296. doi:10.1038/s41592-019-0619-0
19. Li M, Zhang X, Ang KS, et al. DISCO: a database of deeply integrated human Single-Cell Omics data. Nucleic Acids Res. 2022;50(D1):D596–D602. doi:10.1093/nar/gkab1020
20. Zou Z, Long X, Zhao Q, et al. A single-cell transcriptomic atlas of human skin aging. Dev Cell. 2021;56(3):383–397.e8. doi:10.1016/j.devcel.2020.11.002
21. Merleev A, Ji-Xu A, Toussi A, et al. Proprotein convertase subtilisin/kexin type 9 is a psoriasis-susceptibility locus that is negatively related to IL36G. JCI Insight. 2022;7(16):e141193. doi:10.1172/jci.insight.141193
22. Aibar S, González-Blas CB, Moerman T, et al. SCENIC: single-cell regulatory network inference and clustering. Nat Methods. 2017;14(11):1083–1086. doi:10.1038/nmeth.4463
23. Baran Y, Bercovich A, Sebe-Pedros A, et al. MetaCell: analysis of single-cell RNA-seq data using K-nn graph partitions. Genome Biol. 2019;20(1):206. doi:10.1186/s13059-019-1812-2
24. Fuxman Bass JI, Diallo A, Nelson J, Soto JM, Myers CL, Walhout AJM. Using networks to measure similarity between genes: association index selection. Nat Methods. 2013;10(12):1169–1176. doi:10.1038/nmeth.2728
25. Jin S, Guerrero-Juarez CF, Zhang L, et al. Inference and analysis of cell-cell communication using CellChat. Nat Commun. 2021;12(1):1088. doi:10.1038/s41467-021-21246-9
26. Kuleshov MV, Jones MR, Rouillard AD, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44(W1):W90–W97. doi:10.1093/nar/gkw377
27. Deng Y, Chang C, Lu Q. The inflammatory response in psoriasis: a comprehensive review. Clin Rev Allergy Immunol. 2016;50(3):377–389. doi:10.1007/s12016-016-8535-x
28. Kim J, Krueger JG. Highly effective new treatments for psoriasis target the IL-23/Type 17 T cell autoimmune axis. Annu Rev Med. 2017;68:255–269. doi:10.1146/annurev-med-042915-103905
29. Gaffen SL, Jain R, Garg AV, Cua DJ. The IL-23-IL-17 immune axis: from mechanisms to therapeutic testing. Nat Rev Immunol. 2014;14(9):585–600. doi:10.1038/nri3707
30. Hou Y, Zhu L, Tian H, et al. IL-23-induced macrophage polarization and its pathological roles in mice with imiquimod-induced psoriasis. Protein Cell. 2018;9(12):1027–1038. doi:10.1007/s13238-018-0505-z
31. Ravipati A, Nolan S, Alphonse M, et al. IL-6R/Signal transducer and activator of transcription 3 signaling in keratinocytes rather than in T cells induces psoriasis-like dermatitis in mice. J Invest Dermatol. 2022;142(4):1126–1135.e4. doi:10.1016/j.jid.2021.09.012
32. Albanesi C, Madonna S, Gisondi P, Girolomoni G. The Interplay between keratinocytes and immune cells in the pathogenesis of psoriasis. Front Immunol. 2018;9:1549. doi:10.3389/fimmu.2018.01549
33. Novoszel P, Holcmann M, Stulnig G, et al. Psoriatic skin inflammation is promoted by c-Jun/AP-1-dependent CCL2 and IL-23 expression in dendritic cells. EMBO Mol Med. 2021;13(4):e12409. doi:10.15252/emmm.202012409
34. Bernhagen J, Krohn R, Lue H, et al. MIF is a noncognate ligand of CXC chemokine receptors in inflammatory and atherogenic cell recruitment. Nat Med. 2007;13(5):587–596. doi:10.1038/nm1567
35. Jankauskas SS, Wong DWL, Bucala R, Djudjaj S, Boor P. Evolving complexity of MIF signaling. Cell Signalling. 2019;57:76–88. doi:10.1016/j.cellsig.2019.01.006
36. Gesser B, Rasmussen MK, Raaby L, et al. Dimethylfumarate inhibits MIF-induced proliferation of keratinocytes by inhibiting MSK1 and RSK1 activation and by inducing nuclear p-c-Jun (S63) and p-p53 (S15) expression. Inflamm Res. 2011;60(7):643–653. doi:10.1007/s00011-011-0316-7
37. Negishi H, Taniguchi T, Yanai H. The Interferon (IFN) Class of Cytokines and the IFN Regulatory Factor (IRF) transcription factor family. Cold Spring Harb Perspect Biol. 2018;10(11):a028423. doi:10.1101/cshperspect.a028423
38. Taniguchi T, Ogasawara K, Takaoka A, Tanaka N. IRF family of transcription factors as regulators of host defense. Annu Rev Immunol. 2001;19:623–655. doi:10.1146/annurev.immunol.19.1.623
39. Tang S, Jiang W, Xu P, et al. Integrated bioinformatic analysis of key biomarkers and signalling pathways in psoriasis. Scott Med J. 2022;67(1):7–17. doi:10.1177/00369330221078993
40. Raposo RA, Gupta R, Abdel-Mohsen M, et al. Antiviral gene expression in psoriasis. J Eur Acad Dermatol Venereol. 2015;29(10):1951–1957. doi:10.1111/jdv.13091
41. Guo H, Zhang J, Zhang X, et al. SCARB2/LIMP-2 regulates IFN production of plasmacytoid dendritic cells by mediating endosomal translocation of TLR9 and nuclear translocation of IRF7. J Immunol. 2015;194(10):4737–4749. doi:10.4049/jimmunol.1402312
42. Honda K, Ohba Y, Yanai H, et al. Spatiotemporal regulation of MyD88-IRF-7 signalling for robust type-I interferon induction. Nature. 2005;434(7036):1035–1040. doi:10.1038/nature03547
43. Zhu H, Lou F, Yin Q, et al. RIG-I antiviral signaling drives interleukin-23 production and psoriasis-like skin disease. EMBO Mol Med. 2017;9(5):589–604. doi:10.15252/emmm.201607027
44. Morell M, Varela N, Castillejo-López C, et al. SIDT1 plays a key role in type I IFN responses to nucleic acids in plasmacytoid dendritic cells and mediates the pathogenesis of an imiquimod-induced psoriasis model. EBioMedicine. 2022;76:103808. doi:10.1016/j.ebiom.2021.103808
 © 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.