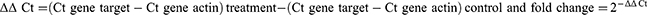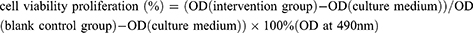Back to Journals » Journal of Inflammation Research » Volume 16
Total Flavonoids of Rhizoma Drynariae Treat Osteoarthritis by Inhibiting Arachidonic Acid Metabolites Through AMPK/NFκB Pathway
Authors Chen GY , Liu XY , Yan XE , Yu X , Liu Y, Luo J , Tao QW
Received 23 April 2023
Accepted for publication 2 September 2023
Published 20 September 2023 Volume 2023:16 Pages 4123—4140
DOI https://doi.org/10.2147/JIR.S418345
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Ning Quan
Guang-Yao Chen,1,2,* Xiao-Yu Liu,3,* Xue-Er Yan,4,* XinBo Yu,1,2 Yi Liu,5 Jing Luo,1,2 Qing-Wen Tao1,2
1Department of TCM Rheumatology, China-Japan Friendship Hospital, Beijing, People’s Republic of China; 2Beijing Key Laboratory for Immune-Mediated Inflammatory Diseases, China-Japan Friendship Hospital, Beijing, People’s Republic of China; 3School of Traditional Chinese Medicine, Beijing University of Chinese Medicine, Beijing, People’s Republic of China; 4Dongzhimen Hospital, Beijing University of Chinese Medicine, Beijing, People’s Republic of China; 5Humanities School, Beijing University of Chinese Medicine, Beijing, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Qing-Wen Tao, Email [email protected]
Objective: Previous clinical studies have found that total flavonoids of Rhizoma Drynariae (TFRD) have a good therapeutic effect on osteoarthritis (OA), but its therapeutic mechanism needs further research.
Methods: OA rat model was established by Hulth method and was intervened by TFRD. Pathological assessments were conducted to assess the protective effect of TFRD on cartilage. Serum metabolomics and network pharmacology were detected to predict the mechanism of TFRD treating OA. In further experiments, molecular biology experiment was carried out to confirm the predicted mechanisms in vivo and in vitro.
Results: TFRD can effectively reduce chondrocyte apoptosis and cartilage degeneration in OA model rats. Serum metabolomics revealed that the intervention effect may be closely related to arachidonic acid metabolism pathway. Network pharmacologic prediction showed that COX-2 was the key target of TFRD in treating OA, and its mechanism might be related with NFκB, apoptosis, AMPK and arachidonic acid metabolism pathway. In vivo experiments indicated that TFRD can inhibit the abnormal expression of COX-2 mRNA in OA model rats. In the in vitro studies, the expression of COX-2 mRNA and protein increased, AMPK phosphorylation was inhibited, and NFκB signaling pathway was activated in IL-1β-induced chondrocytes, and these changes can be reversed by TFRD. After the activation of AMPK signaling pathway or the block-down of NFκB signaling pathway, the effect of TFRD on COX-2 mRNA expression was significantly weakened.
Conclusion: TFRD can inhibit COX-2-mediated arachidonic acid metabolites, and its mechanism is closely related to AMPK/NFκB pathway, which may be a key mechanism in the treatment of OA.
Keywords: total flavonoids of Rhizoma Drynariae, osteoarthritis, network pharmacology, serum metabolomics, cyclooxygenase-2, AMPK/NFκB pathway
Introduction
Osteoarthritis (OA) is a common degenerative joint disease characterized by chronic pain and ambulation limitation, affecting about 250 million people worldwide.1 With increased aging population, the OA incidence rapidly increases and OA has become a major cause of disability.2,3 The morbidity of OA is closely related to articular cartilage, synovium, subchondral bone and surrounding muscles and ligaments, and cartilage plays a critical role in the pathogenesis of OA.4–7 The senescence of chondrocytes, joint trauma and altered structural mechanics are the most common reasons in the development of OA.1,8 In the initial phase of OA, a chronic inflammatory response in chondrocytes promotes the secretion of a mass of matrix hydrolytic enzymes, leading to matrix imbalance and cartilage degradation.9
In terms of treatment, NSAIDs possess anti-inflammatory and analgesic properties, and are recommended as a first-line treatment in most clinical guidelines.10 But the long-term of use NSAIDs is a major cause of gastrointestinal ulcers and hemorrhage.11 Glucocorticoids administered intra-articularly can provide rapid relief of inflammatory responses in joints, but repeated administration may induce a series of adverse reactions.12 Glucosamine and chondroitin sulphate have been used in the treatment of OA, but their clinical efficacy is questionable.13,14 As a result, more safe and effective drugs have remained to be developed, but in recent years a number of drug developments for OA have failed.15 The search for effective ingredients from natural compounds for the treatment of OA has become a research hotspot today.16,17 Various flavonoids extracted from plants, such as quercetin, luteolin and kaempferol, are considered candidates for disease-modifying anti-OA drugs by virtue of their anti-inflammatory and chondroprotective effects.18–22
Rhizoma Drynariae is the dried rhizome of Davallia mariesii T. Moore ex Baker, which has been applied in traditional Chinese medicine for thousands of years to treat joint pain, and flavonoids are the main active ingredients in Rhizoma Drynariae.20 In previous clinical studies, it was demonstrated that oral administration of total flavonoids from Rhizoma Drynariae (TFRD) could improve Lequesne and Visual Analogue Scale (VAS) score for joint pain in patients with OA, while it could exhibit a good safety profile.23,24 In our earlier in vitro studies, it was proved that TFRD is able to regulate the imbalance of MMPs/TIMPs caused by inflammatory factors, thus exerting a chondroprotective effect.25 However, the mechanisms of TFRD in the treatment of OA remain to be further investigated. In this study, a Hulth method was established to verify the therapeutic effect of TFRD against OA. The potential mechanism was then predicted by serum metabonomics and network pharmacology, and further validated the mechanisms by in vivo and in vitro experiments.
Materials and Methods
Experimental Animals
Six-week-old male SPF-grade Sprague-Dawley (SD) rats were purchased from Sibeifu Biotechnology Co., Ltd. (Experimental animal production licence number: SYXK (Jing) 2019-0010; Experimental animal use permit number: SCXK (Jing) 2016-0043). Rats were raised in the animal room of China-Japan Friendship Hospital, the feeding environment perfectly conforms to Laboratory Animal-Requirements of Environment and Housing Facilities (GB 14925-2010, National Laboratory Animal Standardization Technical Committee of China). The animal experiments were approved by the experimental animal care and welfare ethics committee of China-Japan Friendship Hospital (No. zryhyy21-21-05-16).
Reagents, Kits and Antibodies
Collagenase II (C6885, Sigma, USA),0.25% trypsin-EDTA (25,200–056, Gibco, USA), penicillin streptomycin (15140122, Gibco, USA), recombinant rat IL-1β (400-01B, peprotech, USA), dexamethasone (DEX) (D4902, sigma, USA), DMEM medium (C1195500BT, Gibco, USA), fetal bovine serum (FBS) (10,099–141, Gibco, USA), Phosphate-buffered saline (SH30256.01, HyClone, USA), RIPA lysis buffer (R0010, Solarbio Life Sciences, China), Animal free blocking solution (15019S, Cell Signaling Technology, USA), Protease and phosphatase inhibitor cocktail mix (P1261, Solarbio Life Sciences, China). Polymerase chain reaction (PCR) primers for rat COX-2 and β-actin were synthesized by Tsingke Biotechnology Co., Ltd.
Column cartilage total RNA Purification Kit (101,121, TINADZ, China), PAGE Gel Fast Preparation Kit (PG112, Epizyme, China) Hematoxylin-Eosin/HE Staining Kit (G1120, Solarbio, China), Safranin O-Fast Green FCF Cartilage Stain Kit (G1371, Solarbio, China), CellTiter 96® AQueous One Solution Cell Proliferation Assay Kit (G358A, Promega, USA), Prostaglandin E2 ELISA Kit (EIA-5811, DRG, Germany), Leukotriene B4 ELISA Kit (H552-1, Nanjing Jiancheng Bioengineering Institute, China), cDNA Reverse Transcription System (A3500, Promega, USA), SYBR Green Realtime PCR Master Mix (QPK-201, Toyobo, Japan), Mitochondrial Membrane Potential Detection Kit (551302, BD, USA).
Anti-Collagen II antibody (ab34712, Abcam, UK), Anti-NFκB p65 antibody (8242T, Cell Signaling Technology, USA), Anti-Phospho-NF-κB p65 (Ser536) antibody (3033S, Cell Signaling Technology, USA), Anti-AMPKα antibody (Cell Signaling Technology, 5832S, USA), Anti-Phospho-AMPKα (Thr172) antibody (Cell Signaling Technology, 50081S, USA), Anti-IκB-α antibody (Cell Signaling Technology, 4814T, USA), Anti-Cox2 antibody (ABclonal, A1253, China), β-Actin antibody (TA-09, Zhongshan Jingqiao Biotechnology, China), horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG (ZB-530, Zhongshan Jingqiao Biotechnology, China), HRP-conjugated goat anti-rabbit IgG (ZB-2301, Zhongshan Jingqiao Biotechnology, China) and Alexa Fluor 488-conjugated Goat anti-Rabbit IgG (H+L) (ZF-0511, Zhongshan Jingqiao Biotechnology, China).
Drug Preparation
TFRD was prepared with macroporous adsorption resins described in our previous study.25 In clinical practice, the common-used dose of TFRD and glucosamine hydrochloride is 750 mg per day. This dosage was set according to an adult of 70 kg; hence, the daily dose is 10.714 mg/kg of body weight. According to the ratio of a rat to a person’s body surface area, the daily dose for an adult is 6.3 times larger than rats, and hence, the daily gavage dosage of rats is 67.5 mg/kg. The daily dosage of glucosamine hydrochloride was 1440 mg. According to the above method, the dosage of glucosamine hydrochloride in rats was 129.6 mg/kg. TFRD were prepared into a 6.75 mg/mL solution using distilled water and glucosamine hydrochloride into a 12.96 mg/mL solution, which were then stored in a refrigerator at 4°C until use.
Animal Grouping and Drug Intervention
After 7 days of acclimatization, 36 rats were randomly divided into normal group, OA model group, TFRD group and glucosamine hydrochloride group according to the random number table method. The model group, TFRD group and glucosamine hydrochloride group underwent Hulth surgery as we described previously,25 while the normal group was subjected to sham surgery without joint destruction. Three days after Hulth surgery, normal group and model group were given normal saline for gavage, while TFRD group and glucosamine hydrochloride group were intragastrically administrated with corresponding drugs. The dosage of normal saline and drugs is 0.01 mL/g according to body weight, and rats were given intragastric administration at 8:00 every day.
Sample Collection
After 28 days of intragastric administration, rats were anaesthesia by isoflurane, then blood were collected from the abdominal aorta and centrifuged to isolate serum. After centrifuged for 10 minutes at 3400 rpm at 4°C, processed specimens were immediately stored in an ultra-low-temperature freezers (−80°C) for subsequent metabolomics and ELISA detection. A bone forceps was used to remove long bones above and below the knee joint in rats, and then strip away as much of periarticular muscle tissue as possible while ensuring the integrity of the knee joint. Of these rats, three were used for pathological evaluation of cartilage. The rest six rats were used to obtain RNA of cartilage for RT-PCR analysis.
Evaluation of Cartilage Pathology
After fixation with 4% polyfluoroalkoxy for 72 h, the knee joints were decalcified with 10% EDTA-2Na solution for 8 weeks, followed by dehydration, embedding and sectioning. Knee slices of rats were subject to hematoxylin and eosin (HE) and safranin-O/fast green staining, then placed under a light microscopy to observe their pathological changes. Two researchers who were unaware of the grouping and administration of the rats performed the Mankin’s score (Table 1) and the average of the two researchers was adopted as the final result.
 |
Table 1 Mankin’s Score Evaluation |
Serum Metabolomics Detection
Liquid Chromatography and Mass Spectra (LC-MS) were used to detect the serum metabolomics of rats as follows: The serum sample was defrosted on ice, LC-ESI-MS/MS system (UPLC, ExionLC AD; MS, QTRAP System) was used for detection. The LC analytical conditions were as follows: ACQUITY UPLC HSS T3 C18 column (1.8 µm, 2.1 mm×100 mm, waters) was kept at 40°C and the flow rate was 0.4 mL/min. The injected sample volume was 2 μL for each run. The mobile phase was water-0.1% formic acid (A) and acetonitrile−0.1% formic acid (B). Gradient program was 95:5 V/V at 0 min, 10:90 V/V at 11.0 min, 10:90 V/V at 12.0 min, 95:5 V/V at 12.1 min, 95:5 V/V at 14.0 min. MS analysis were acquired on a triple quadrupole (QQQ) linear ion trap mass spectrometer (QTRAP), which equipped with an ESI Turbo Ion-Spray interface. The system operated in positive and negative ion mode and controlled by Analyst 1.6.3 software (Sciex). The ESI source operation parameters were as follows: source temperature 500 °C; ion spray voltage (IS) 5500 V (positive), −4500 V (negative); ion source gas I (GSI), gas II (GSII), curtain gas (CUR) were set at 55, 60, and 25.0 psi, respectively; the collision gas (CAD) was high.
After the data preprocessing, unsupervised PCA (principal compound analysis) was performed by statistics function prcomp within R software. VIP values were extracted from OPLS-DA result, which was generated using R package MetaboAnalystR. Significantly regulated metabolites between groups were determined by P<0.05, VIP ≥ 1 and absolute fold change (FC) ≥ 1.2. Identified metabolites were annotated using KEGG compound database (http://www.kegg.jp/kegg/compound/), significantly enriched pathways are identified with a hypergeometric test’s p-value for a given list of metabolites. Metaboanalyst (http://www.Metaboanalyst.ca/) was used to perform metabolic pathway enrichment analysis for differential metabolites, and Homo sapiens was selected as the pathway library.
ELISA
The concentrations of leukotriene B4 and PGE2 in the rat serum were determined by a commercial ELISA kit (competitive method) according to the manufacturers’ instructions. The OD value of each standard and sample pore was detected at 450 nm wavelength by a microplate reader, with a reference wavelength of 650 nm. A model of standard curve was constructed by Curve Expert 1.4 software, and the concentrations of each sample were calculated by OD value.
Network Pharmacology Prediction
The TCMSP database26 was used to obtain all the chemical compounds in Rhizoma Drynariae. Then, chemical compounds were input into PubChem database27 to obtain their structural formula, from which flavonoid compounds were screened. The criteria of drug-likeness (DL)≥0.18 and oral bioavailability (OB)≥30% were employed to identify active compounds.28 In the meantime, the flavonoid ingredients that failed to meet the above standards, if proven therapeutic effects on OA in the previous literature, were also included in the investigated compounds. The compounds obtained in the above two ways were combined as the key compounds of TFRD in the treatment of OA, and were applied to the subsequent TFRD quality control by an Ultra High Performance Liquid Chromatography coupled to a Mass Spectrometer (UHPLC/MS).
The gene targets corresponding to the key compounds of TFRD treating OA were obtained from the TCMSP database. Meanwhile, with “osteoarthritis” as the keyword, targets were identified from Genecard29 and OMIM database,30 the results of the two databases were merged as OA-related gene targets. The intersection targets between TFRD and OA were defined as potential targets of TFRD.
Compound-target (C-T) network of TFRD for OA was established by Cytoscape software and key compounds and targets were identified based on the number of nodes. KEGG analysis of potential targets of TFRD for the treatment of OA was performed using the Metascape database31 to identify the key pathways in the treatment of OA with TFRD. PPI analysis of potential targets of TFRD for the treatment of OA was performed using the String database32 to identify key TFRD targets in the treatment of OA.
RT-PCR
Column cartilage total RNA Purification Kit was adopted to isolate RNA from cartilage according to manufacturers’ instructions. A Nanodrop spectrophotometer (Thermo Scientific, USA) was used to detect RNA concentration and quality. 20 μL mixed solution (1 μg total RNA, 4 μg MgCl2, 2 μL reverse transcription buffer, 2 μL dNTP mixture, 0.5 μL recombinant RNasin ribonuclease inhibitor, 0.63 μL AMV reverse transcriptase, 1 μL Oligo (dT) 15 primer and nuclease-free water) was used for the construction of a transcription system (A3500, Promega, USA). The mixture was heated at 42°C for 15 min, then heated at 95°C for 5 min to obtain cDNA.
A 20 µL of reaction system (10 µL SYBR Green real-time PCR master mix, 2 µL cDNA, 0.8 µL gene forward and reverse primer (10 µmol/L) and 7.2 µL distilled water) was generated. Then, real-time PCR was carried out on a QuantStudio Real-Time PCR cycler. The protocol was as follows: Preheated at 95°C for 60s, then heated at 95°C for 15s, 60°C for 15s, and 72°C for 45s for 40 cycles. The relative expression was analyzed based on the 2−ΔΔCt method using the following equations:
Chondrocyte Isolation, Culture and Identification
Five-day-old male SPF-grade SD sucking rats were purchased from Sibeifu Biotechnology Co., Ltd. The sucking rats were euthanized with carbon dioxide and subsequently surface-sterilized by soaked in 75% alcohol for 10 minutes. Keen cartilage of rats was isolated with scalpels, washed with PBS twice, and digested by 0.25% trypsin-EDTA for 1 hour at 37°C. Then, 0.2% type II collagenase was used to decompose the cartilage for another 6 hours. After complete digestion, digestive fluids and remnants of cartilage were filtered and triturated through nylon screen (100 μm). The liquid was centrifuged and resuspended into DEME complete mediums (89% DEME basic medium, 10% FBS and 1% Penicillin-Streptomycin). Chondrocytes were seeded in poly-lysine-coated dishes and cultured at 37°C in a 5% CO2 humidified incubator. To preserve the chondrocyte phenotype, the experiments selected the third generation of chondrocytes. Prior to experiments, immunofluorescence was employed to verify whether the isolated cells expressed positive for type II collagen and the flow cytometry calculate the positive rate.
Cell Viability Assays and Drug Intervention
1×104 chondrocytes and 100 µL of complete DEME mediums were planted in a 96-well plates. Different concentrations of TFRD was added to the plates. The substrate was aspirated after a 12-hour incubation, then the cells were washed 3 times with PBS. 100 µl of DEME medium was added and cells were incubated for 4 h, 20 µL MTS was added to each well and incubated for another 2 h. The cell viability of the intervention group was then calculated according to the following formula:
The cells were planted in 6-well plates and 48-well plates (for immunofluorescence). The cells were pre-treated with TFRD for 1 h, then 10 ng/mL IL-1β was added for inflammatory stimulation. Cells were rinsed with PBS for 3 times, then total proteins were extracted for subsequent Western Blotting detection.
Western Blot
After intervention, the cell culture supernatant was discarded. Chondrocytes were rinsed twice by PBS. Each well was added with 250 µL RIPA lysate, gently blown for 5–10 times and lysed on ice for 25 min. The lysate was centrifuged (4°C, 10,000 r/min, 5 min) and the supernatants were collected. Total protein concentration was detected by a bicinchoninic acid (BCA) assay kit. Samples were adjusted to the same protein concentration by RIPA lysate, and 5 × loading buffer was added, boiled for 5 min and centrifuged (4°C, 10,000 r/min, 5 min).
10% SDS-PAGE gel was configured by a PAGE Gel Fast Preparation Kit base on the kit’s instructions. 40 µg proteins were added into each well of the SDS-PAGE gel, labeled with marker protein. Electrophoresis was carried out at 150 V until the bromophenol blue reached the bottom of the gel. The PVDF membrane was activated by methanol for 40s before transfer. A mini gel holder cassette, the filtrate paper, PVDF membrane and SDS-PAGE gel were prepared to make a sandwich transfer system and electrophoresis for 55 min at 70 V. Proteins in gels were transferred to PVDF membranes, and then incubated by blocking solution for 1 h at room temperature. PVDF membranes were incubated with primary antibodies at 4°C overnight and with secondary antibodies at room temperature for 1.5 hours in succession. The samples were finally detected by chemiluminescence system using ECL luminous fluid.
Immunofluorescence
The cells were washed twice with PBS after the intervention and then fixation was carried out using 4% paraformaldehyde. 0.1% Triton X-100 was used to penetrate the cytomembrane, and 1 × animal free blocking solution was utilized to block cells for 1 h and the appropriate concentration of primary antibody was added and incubated at 4°C overnight. After discarding the primary antibody, Alexa Fluor 488-conjugated Goat anti-Rabbit IgG (H+L) was added and incubated at room temperature for 1 h, then the plate was covered with fluorescent mounting medium with DAPI (4,6-diamidino-2-phenylindole). An inverted fluorescence microscope (Zeiss, Germany) was used to observe the location of target protein and DAPI.
Flow Cytometry
Flow cytometry was used for the identification of chondrocyte purity, chondrocytes were subjected to digestion of EDTA-free trypsin and then fixed, and the membrane was broken. Type II collagen antibodies (1:100 dilution) was added into dishes for 1 h incubation. Finally, these cells were washed with 1×Buffer, followed by the addition of Alexa Fluor 488-conjugated Goat anti-Rabbit IgG (H+L) to cells (1:50 dilution) and a 30 min incubation under a light-free circumstance. For detection, 1× Assay Buffer was used to wash chondrocytes twice, and 0.5 mL 1× Assay Buffer was added to suspend the cells. The prepared cells were assayed by a flow cytometer (BD FACSCantoII, USA).
Statistical Analysis
The Statistical Package for Social Sciences (SPSS) version 19 was used for data analysis. Continuous variables are presented as the mean values ± standard deviation (SD). Student’s t-test was used to assess the differences between the two groups. A p-value < 0.05 was considered to indicate a statistically significant difference. Correlations were performed by Pearson correlation analysis, the coefficient R > 0.4 and p-value < 0.05 was considered to be statistically significant.
Results
Effect of TFRD on Cartilage Pathology in OA Rats
The OA rats were established by Hulth surgery, and intervened by TFRD and the positive control drug glucosamine hydrochloride, then the rat knee joints were used for cartilage pathological assessment. The rats did not show obvious emaciation or discomfort during the experiment, and the rats did not die during the experiment.
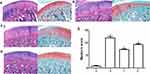
Compared to the normal group of cartilage, HE staining revealed vacuole-like changes in cartilage tissue in OA rats, suggesting chondrocyte apoptosis. At the same time, the staining degree of safranin-O obviously decreased, suggesting the reduction of cartilage matrix (Figure 1A and B). HE staining revealed that TFRD could significantly relieve the vacuole-like change and deletion of safranin-O staining in OA model rats (Figure 1C). Positive control drug glucosamine hydrochloride can relieve the deletion of safranin-O staining (Figure 1D). Mankin’s scoring of cartilage tissue indicated that TFRD and positive control drug glucosamine hydrochloride had an ameliorative effect on cartilage degeneration (Figure 1E). Pathological sections of the whole joint see Supplementary Material.
Effect of TFRD on Serum Metabolomics in OA Rats
The serum of blank control group, Hulth model group and TFRD treated group was detected by untargeted metabolomics, and the differential metabolites between groups were screened for further KEGG pathway analysis. Results showed that 906 metabolites were detected in rat serum by Liquid Chromatography and Mass Spectra (LC-MS). PCA Score plot and OPLS-da plot results showed a clear separation of metabolome profiles in blank control group, Hulth model group and TFRD treated group, suggesting differences in metabolomics results among groups (Figure 2A and B).
Compared with the blank control group, 80 metabolites in the model group were changed (59 up-regulated and 21 down-regulated, Figure 2C. 1). Compared with the Hulth model group, 20 metabolites in TFRD treated group were changed (9 up-regulated and 11 down-regulated, Figure 2C. 2). KEGG enrichment analysis was then performed for the differential metabolites of blank control group vs Hulth model group, and Hulth model group vs TFRD treated group (Figure 2D). Among all the corresponding pathways, arachidonic acid pathway ranked first. The above results indicated arachidonic acid pathway is the key of TFRD treating OA. In metabolites associated with arachidonic acid pathway, (±)15-HETE, leukotriene B4, prostaglandin E2, prostaglandin J2, prostaglandin B2 and prostaglandin A2 increased after OA modeling, while reduced glutathione decreased. Leukotriene B4, prostaglandin B2 and prostaglandin A2 in TFRD group decreased after TFRD intervention, while arachidonic acid elevated.
ELISA assay was applied to validate the content of leukotriene B4 and prostaglandin E2 in the serum, thus to verify the serum metabolomics results. The ELISA results of leukotriene B4 and prostaglandin E2 in each sample were fitted to the original peak values of metabolomics, and a good linear relationship can be observed between the two methods (Figure 2E). These results provide a reliable basis for the serum metabolomics detection.
Network Pharmacology Prediction of TFRD Treating OA
With “Rhizoma Drynariae” as the keyword, 71 compounds were found by TCMSP database, 41 were identified as flavonoids, and 10 met the standard (OB≥30% and DL≥0.18), which were (2R)-5,7-dihydroxy-2-(4-hydroxyphenyl) chroman-4-one, Aureusidin, Eriodyctiol (flavanone), kaempferol, naringenin, (+)-catechin, eriodictyol, luteolin, davallioside A_qt and xanthogalenol. Through literature search, 4 flavonoid compounds (Astragalin,33 (-)-epicatechin,34 Procyanidin B2,35 and Procyanidin D36) did not meet the criteria but were proved to be important components in the treatment by literature, so they were also included. The 14 active compounds of TFRD were identified with UHPLC-MS (Figure 3). Detailed information was presented as Table 2.
 |
Table 2 Chemical Identification of Total Flavonoids of Rhizoma Drynariae |
Two hundred and seventy-five targets corresponding to the compounds of TFRD were screened out from TCMSP; 3013 OA-related genes were identified from GeneCards database (Relevance≥4), and there were 94 TFRD-OA intersection genes. Based on the above result, a C-T network was established, among which 167 nodes (155 targets, 12 compounds) and 273 edges exists (Figure 4A). Results showed that PTGS2 (COX-2) had the highest number of connections with targets, suggesting that it was the potential key targets of TFRD against OA (Figure 4B). The TFRD-OA intersection genes were then imported into Metascape database for KEGG pathway enrichment analysis, and the top 20 KEGG pathways were screened out in order of enrichment significance (Figure 4C). Combined with literature analysis, NF-κB signaling pathway,37 multi-species apoptosis pathway,38 AMPK signaling pathway39 and arachidonic acid metabolism pathway40 play a pivotal role in the treatment of OA, thus may be the critical mechanism in TFRD treating OA.
Effect of TFRD on Cartilage COX-2 mRNA in OA Rats
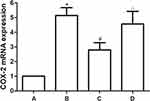
The expression of COX-2 mRNA in OA rat knee cartilage was detected by RT-PCR. Results showed that the mRNA level of COX-2 increased significantly after OA modeling, which can be reversed by TFRD intervention. The positive control drug glucosamine hydrochloride had no significant effect on the expression of COX-2 mRNA in cartilage of OA rats (Figure 5).
Chondrocyte Identification
The synthesis and secretion of type II collagen are the characteristics of chondrocytes, and type II collagen staining was visualized in cells isolated from knee cartilage of rats by immunofluorescence. Flow cytometry was then used to detect the proportion of type II collagen-positive cells. SW1353 cells, a type of chondrosarcoma cells with multiple phenotype of chondrocytes, were often used in OA-related research. However, SW1353 cells do not secret type II collagen, thus were used as the negative control.
Immunofluorescence results indicated that type II collagen staining was positive in most cells isolated from knee cartilage of rats (Figure 6A–C). Flow cytometry results suggested 98.2% of the isolated chondrocytes were positive for type II collagen, which confirmed the high purity of the isolated chondrocytes and were applicable for subsequent research (Figure 6D). The flow cytometry results of SW1353 cells showed no type II collagen-positive cells (Figure 6E).
Effects of TFRD on Chondrocyte Proliferation
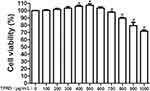
Chondrocytes were treated with 100, 200, 300, 400, 500, 600, 700, 800, 900 and 1000 μg/mL TFRD. Results showed that low concentration of TFRD (400 and 500 μg/mL) could promote the proliferation of chondrocytes, and 700, 800, 900 and 1000 μg/mL TFRD could significantly inhibit the proliferation of chondrocytes (Figure 7). Therefore, 600 μg/mL TFRD was selected as the maximum concentration in the follow-up intervention experiment, half of the maximum concentration (300 μg/mL) was selected as the medium-dose intervention concentration, and one quarter (150 μg/mL) was selected as the low-dose intervention concentration.
Effects of TFRD on AMPK/NFκB Signaling Pathway in Chondrocytes
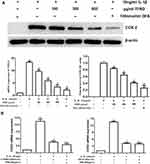
Chondrocytes were pretreated with different concentrations of TFRD or positive control drug dexamethasone for 1 h, then stimulated with 10 ng/mL IL-1β for 1 h or 12 h. After stimulated with IL-1β for 1 h, the content of IKB-α in cell lysates decreased apparently, the level of NFκB p65 had no significant change, and the level of p-NFκB p65 increased apparently. In further experiments, the cell nuclei were extracted using a Nuclear Extraction kit. Results showed that the expression of NFκB p65 in the nucleus significantly increased. The above results suggested that IL-1β stimulation can abnormally activate NFκB signaling pathway. After stimulated with IL-1β for 12 h. The phosphorylation of AMPK in chondrocytes significantly decreased, suggesting that AMPK signaling pathway was inhibited. The above changes can be reversed by TFRD and the positive control drug dexamethasone, and TFRD showed a dose-dependent effect (Figure 8).
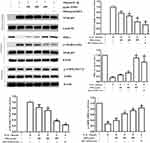 |
Figure 8 Effect of TFRD on AMPK/NFκB signaling pathway of IL-1β-stimulated chondrocytes. *P<0.05 compared with the blank control group, #P<0.05 compared with the IL-1β-stimulated group. |
Effect of TFRD on COX-2 of Chondrocytes in vitro
The expression of COX-2 mRNA and protein in chondrocytes stimulated by IL-1β was significantly increased. TFRD inhibited the expression of COX-2 mRNA and protein in a dose-dependent manner (Figure 9A). In further experiments, 200 μmol/L A-76966241 and 20 μmol/L JSH-2342 were used to activate AMPK signaling pathway and inhibit NFκB pathway, respectively. Results showed that COX-2 mRNA expression significantly decreased after the above intervention, and on the basis of this intervention, the effect of TFRD on COX-2 was significantly weakened (Figure 9B).
Discussion
Our previous study found that TFRD can protect IL-1β from the imbalance between MMPs/TIMP in SW1353 cells in vitro, suggesting that TFRD can play a therapeutic role in OA by regulating the decomposition and synthesis balance of cartilage matrix.25 We have also found in other studies that TFRD can play a role in the treatment of rheumatoid arthritis by inhibiting the inflammatory response.43 In this study, pathological assessment indicated that TFRD had chondro-protective effect in OA rat model.
In further experiments, we observed the changes of serum metabolites in OA rats after TFRD intervention. The potential mechanism of TFRD treating OA was then predicted by serum metabonomics and network pharmacology. Metabonomics results suggested that arachidonic acid metabolism pathway was the key pathway for TFRD treating OA. The subsequent network pharmacology predicted that COX-2 and arachidonic acid metabolism pathway are the pivotal target and pathway of TFRD against OA. TFRD may also be closely related to NFκB signaling pathway, apoptosis pathway and AMPK signaling pathway.
COX-2 plays an important role in the occurrence and development of OA and is a key target for OA treatment.44 During the inflammatory response, COX-2 catalyzes the conversion of arachidonic acid into prostaglandins, which are then released to promote the inflammatory response.45 The increase of COX-2 mRNA in chondrocytes of OA model rats was accompanied by the increase of prostaglandin E2, prostaglandin J2, prostaglandin B2 and prostaglandin A2 in serum, which reveals the activation of COX-2-mediated arachidonic acid pathway. After TFRD treatment, COX-2 mRNA in chondrocytes decreased, and prostaglandin B2 and prostaglandin A2 decreased in serum, while arachidonic acid elevated. These results suggest that TFRD inhibits the decomposition of arachidonic acid into prostaglandins by inhibiting the expression of COX-2 in cartilage, which may be the key mechanism of TFRD’s protective effect on cartilage.
In inflammatory response, the transcription and synthesis of COX-2 are mainly regulated by NFκB pathway.46 Under normal conditions, NFκB p65·NFκB p55·IKB-α is present in the cytoplasm as a trimer, and inflammatory processes activate IKK-α ubiquitination, leading to phosphorylation and degradation of IKB-α.47 After the IKB-α degradation, the NFκB p65·NFκB p55 dimer is released and transferred to the nucleus (nuclear heterotopia of NFκB).48 Upon entry into the nucleus, NFκB p65·NFκB p55 is phosphorylated and activated, binding to specific DNA sequences to stimulate transcription of relevant inflammatory mediators, including COX-2.49
In vitro experiments, we stimulated chondrocytes with IL-1β to establish OA model. Results showed that the mRNA and protein contents of COX-2 in chondrocytes significantly increased after IL-1β stimulation, while IKB-α decreased, the phosphorylation level of NFκB p65 increased in chondrocytes, and NFκB p65 increased in the nucleus, suggesting the abnormal activation of NFκB pathway. TFRD can inhibit COX-2 expression and NFκB pathway to a certain extent. In further experiments, JSH-23 (an inhibitor of NFκB pathway) was applied to block NFκB signaling pathway, and TFRD was used to intervene chondrocytes. It was revealed that the inhibition of COX-2 was significantly weakened by TFRD, suggesting that TFRD mainly regulates the synthesis of COX-2 through NFκB pathway.
AMPK signaling pathway involved in a variety of energy-related biological processes, including cell apoptosis and inflammatory responses.50–52 AMPK regulates IKK-α ubiquitination through SIRTs, thereby affecting the phosphorylation and degradation of IKB-α, thus regulates NFκB pathway.53 After IL-1β stimulation, the phosphorylation of AMPK decreased, suggesting that AMPK signaling pathway was inhibited. This change can be reversed by TFRD, indicating that TFRD can activate AMPK signaling pathway by promoting AMPK phosphorylation, and may inhibit inflammatory factor-mediated chondrocyte apoptosis through this pathway. In further experiments, A-769662 was used to activate AMPK signaling pathway, and TFRD was used to intervene chondrocytes on this basis. The results showed that the inhibition of COX-2 was significantly reduced. These results suggest that AMPK signaling plays a key role in TFRD regulation of COX-2.
Conclusion
In conclusion, through network pharmacology, serum metabolomics, in vivo and in vitro experiments, this study revealed that it is a key mechanism for TFRD treating OA by inhibiting the chondrocyte COX-2 expression through AMPK/NFκB signaling pathway, thus blocking the downstream arachidonic acid metabolic pathway.
Ethics Approval and Consent to Participate
The animal experiments were approved by the experimental animal care and welfare ethics committee of China-Japan Friendship Hospital (No. zryhyy21-21-05-16).
Funding
This study was supported by National High Level Hospital Clinical Research Funding (2023-NHLHCRF-YXHZ-ZRMS-07, 2022-NHLHCRF-LX-02-0103), National Natural Science Foundation of China (No. 81804042,81673941), National Famous Traditional Chinese Medicine Experts inheritance studio of Xiaoping Yan (2019-PP-001), National Regional Chinese Medicine Diagnostic and Treatment Centre for Rheumatic Diseases Project (2019-ZX-006), National Key Clinical Specialty Capacity Building Project (2011-ZDZK-001).
Disclosure
Guang-Yao Chen, Xiao-Yu Liu and Xue-Er Yan are co-first authors in this work. The authors declare that there are no conflicts of interest in this work.
References
1. Hunter DJ, Bierma-Zeinstra S. Osteoarthritis. Lancet. 2019;393(10182):1745–1759. doi:10.1016/S0140-6736(19)30417-9
2. Kim HA. Osteoarthritis - insights from recent research. J Rheum Dis. 2022;29(3):132–139. doi:10.4078/jrd.2022.29.3.132
3. Du X, Liu ZY, Tao XX, et al. Research progress on the pathogenesis of knee osteoarthritis. Orthop Surg. 2023;15(9):2213–2224. doi:10.1111/os.13809
4. Zhou J, Huang J, Li Z, et al. Identification of aging-related biomarkers and immune infiltration characteristics in osteoarthritis based on bioinformatics analysis and machine learning. Front Immunol. 2023;14:1168780. doi:10.3389/fimmu.2023.1168780
5. Cao X, Cui Z, Ding Z, et al. An osteoarthritis subtype characterized by synovial lipid metabolism disorder and fibroblast-like synoviocyte dysfunction. J Orthop Transl. 2022;33:142–152. doi:10.1016/j.jot.2022.02.007
6. Zhao K, Ruan J, Nie L, Ye X, Li J. Effects of synovial macrophages in osteoarthritis. Front Immunol. 2023;14:1164137. doi:10.3389/fimmu.2023.1164137
7. Hu W, Chen Y, Dou C, Dong S. Microenvironment in subchondral bone: predominant regulator for the treatment of osteoarthritis. Ann Rheum Dis. 2021;80(4):413–422. doi:10.1136/annrheumdis-2020-218089
8. Dilley JE, Bello MA, Roman N, McKinley T, Sankar U. Post-traumatic osteoarthritis: a review of pathogenic mechanisms and novel targets for mitigation. Bone Rep. 2023;18:101658. doi:10.1016/j.bonr.2023.101658
9. Pulik Ł, Łęgosz P, Motyl G. Matrix metalloproteinases in rheumatoid arthritis and osteoarthritis: a state of the art review. Reumatologia. 2023;61(3):191–201. doi:10.5114/reum/168503
10. Primorac D, Molnar V, Matišić V, et al. comprehensive review of knee osteoarthritis pharmacological treatment and the latest professional societies’ guidelines. Pharmaceuticals. 2021;14(3):205. doi:10.3390/ph14030205
11. Bindu S, Mazumder S, Bandyopadhyay U. Non-steroidal anti-inflammatory drugs (NSAIDs) and organ damage: a current perspective. Biochem Pharmacol. 2020;180:114147. doi:10.1016/j.bcp.2020.114147
12. Saltychev M, Mattie R, McCormick Z, Laimi K. The magnitude and duration of the effect of intra-articular corticosteroid injections on pain severity in knee osteoarthritis: a systematic review and meta-analysis. Am J Phys Med Rehabil. 2020;99(7):617–625. doi:10.1097/PHM.0000000000001384
13. Clegg DO, Reda DJ, Harris CL, et al. Glucosamine, chondroitin sulfate, and the two in combination for painful knee osteoarthritis. N Engl J Med. 2006;354(8):795–808. doi:10.1056/NEJMoa052771
14. Vo NX, Le NNH, Chu TDP, et al. Effectiveness and safety of glucosamine in osteoarthritis: a systematic review. Pharmacy. 2023;11(4):117. doi:10.3390/pharmacy11040117
15. Spil W, Kubassova O, Boesen M, Bay-Jensen A, Mobasheri A. Osteoarthritis phenotypes and novel therapeutic targets. Biochem Pharmacol. 2019;165:41–48. doi:10.1016/j.bcp.2019.02.037
16. Wang H, Yan Y, Pathak JL, et al. Quercetin prevents osteoarthritis progression possibly via regulation of local and systemic inflammatory cascades. J Cell Mol Med. 2023;27(4):515–528. doi:10.1111/jcmm.17672
17. Pérez-Lozano ML, Cesaro A, Mazor M, et al. Emerging natural-product-based treatments for the management of osteoarthritis. Antioxidants. 2021;10(2):265. doi:10.3390/antiox10020265
18. Zhang J, Yin J, Zhao D, et al. Therapeutic effect and mechanism of action of quercetin in a rat model of osteoarthritis. J Int Med Res. 2020;48(3):1–9. doi:10.1177/0300060519873461
19. Wang X, Xie W, Bi Y, et al. Quercetin suppresses apoptosis of chondrocytes induced by IL-1β via inactivation of p38 MAPK signaling pathway. Exp Ther Med. 2021;21(5):468. doi:10.3892/etm.2021.9899
20. Chen G, Liu X, Chen J, et al. Prediction of rhizoma drynariae targets in the treatment of osteoarthritis based on network pharmacology and experimental verification. Evid Based Complement Alternat Med. 2021;2021:5233462. doi:10.1155/2021/5233462
21. Kang B, Ryu J, Lee C, Hwang S. luteolin inhibits the activity, secretion and gene expression of MMP-3 in cultured articular chondrocytes and production of MMP-3 in the rat knee. Biomol Ther. 2014;22(3):239–245. doi:10.4062/biomolther.2014.020
22. Hämäläinen M, Nieminen R, Vuorela P, Heinonen M, Moilanen E. Anti-inflammatory effects of flavonoids: genistein, kaempferol, quercetin, and daidzein inhibit STAT-1 and NF-kappaB activations, whereas flavone, isorhamnetin, naringenin, and pelargonidin inhibit only NF-kappaB activation along with their inhibitory effect on iNOS expression and NO production in activated macrophages. Mediators Inflamm. 2007;2007:1–10. doi:10.1155/2007/45673
23. Cao J, Chen D, Zhang Z. Clinical study on the treatment of osteoarthritis with glucosamine potassium sulfate in combination with strong bone capsule. J Clin Ratio Drug Use. 2014;7(19):30–31. doi:10.15887/j.cnki.13-1389/r.2014.19.092
24. Zhou R, Chen C, He L, Wang C. Clinical observation of oral osteopractic total flavonoids in the patients with knee osteoarthritis. China Med Herald. 2011;8(02):77–78.
25. Chen G, Chen J, Liu X, Xu Y, Tao Q, Yong YK. Total flavonoids of rhizoma drynariae restore the MMP/TIMP balance in models of osteoarthritis by inhibiting the activation of the NF-κB and PI3K/AKT pathways. Evid Based Complement Altern Med. 2021;2021(2):1–14. doi:10.1155/2021/6634837
26. Ru J, Li P, Wang J, et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J Cheminform. 2014;6(1):13. doi:10.1186/1758-2946-6-13
27. Kim S, Thiessen PA, Bolton EE, et al. PubChem substance and compound databases. Nucleic Acids Res. 2016;44(D1):D1202–D1213. doi:10.1093/nar/gkv951
28. Wang K, Gao Y, Lu C, et al. Uncovering the complexity mechanism of different formulas treatment for rheumatoid arthritis based on a novel network pharmacology model. Front Pharmacol. 2020;11:1035. doi:10.3389/fphar.2020.01035
29. Marilyn S, Irina D, Alexander J, et al. GeneCards version 3: the human gene integrator. Database. 2010;2010:baq020. doi:10.1093/database/baq020
30. Hamosh A, Amberger J, Bocchini C, Scott A. Online Mendelian Inheritance in Man (OMIM): victor McKusick’s magnum opus. Original Article. 2021;185(11):3259–3265. doi:10.1002/ajmg.a.62407
31. Zhou Y, Zhou B, Pache L, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10(1):1523. doi:10.1038/s41467-019-09234-6
32. Damian S, Gable AL, Nastou KC, et al. The STRING database in 2021: customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2020;D1:605–612. doi:10.1093/nar/gkaa1074
33. Ma Z, Piao T, Wang Y, Liu J. Astragalin inhibits IL-1β-induced inflammatory mediators production in human osteoarthritis chondrocyte by inhibiting NF-κB and MAPK activation. Int Immunopharmacol. 2015;25(1):83–87. doi:10.1016/j.intimp.2015.01.018
34. Wan N, Tantowi N, Seng FL, Mohamed S. Epicatechin and scopoletin rich Morinda citrifolia (Noni) leaf extract supplementation, mitigated osteoarthritis via anti-inflammatory, anti-oxidative, and anti-protease pathways. J Food Biochem. 2019;43(3):1–12. doi:10.1111/jfbc.12755
35. Wang A, Leong DJ, He Z, et al. Procyanidins mitigate osteoarthritis pathogenesis by, at least in part, suppressing vascular endothelial growth factor signaling. Int J Mol Sci. 2016;17(12):2065. doi:10.3390/ijms17122065
36. Masuda I, Koike M, Nakashima S, et al. Apple procyanidins promote mitochondrial biogenesis and proteoglycan biosynthesis in chondrocytes. Sci Rep. 2018;8(1):7229. doi:10.1038/s41598-018-25348-1
37. Lepetsos P, Papavassiliou KA, Papavassiliou AG. Redox and NF-κB signaling in osteoarthritis. Free Radic Biol Med. 2019;132:90–100. doi:10.1016/j.freeradbiomed.2018.09.025
38. Hwang HS, Kim HA. Chondrocyte apoptosis in the pathogenesis of osteoarthritis. Int J Mol Sci. 2015;16(11):26035–26054. doi:10.3390/ijms161125943
39. Wang J, Li J, Song D, Ni J, Yan M. AMPK: implications in osteoarthritis and therapeutic targets. Am J Transl Res. 2020;12(12):7670–7681.
40. Li Z, Ma D, Peng L, Li Y, Liao Z, Yu T. Compatibility of Achyranthes bidentata components in reducing inflammatory response through Arachidonic acid pathway for treatment of Osteoarthritis. Bioengineered. 2022;13(1):1746–1757. doi:10.1080/21655979.2021.2020394
41. Sanders MJ, Ali ZS, Hegarty BD, Heath R, Snowden MA, Carling D. Defining the mechanism of activation of AMP-activated protein kinase by the small molecule A-769662, a member of the thienopyridone family. J Biol Chem. 2007;282(45):32539–32548. doi:10.1074/jbc.M706543200
42. Fan P, Abderrahman B, Chai TS, Yerrum S, Jordan VC. Targeting peroxisome proliferator-activated receptor γ to increase estrogen-induced apoptosis in estrogen-deprived breast cancer cells. Mol Cancer Ther. 2018;17(12):2732–2745. doi:10.1158/1535-7163.MCT-18-0088
43. Chen GY, Luo J, Liu Y, Yu XB, Liu XY, Tao QW. Network pharmacology analysis and experimental validation to investigate the mechanism of total flavonoids of rhizoma drynariae in treating rheumatoid arthritis. Drug Des Devel Ther. 2022;16:1743–1766. doi:10.2147/DDDT.S354946
44. Su W, Liu G, Mohajer B, et al. Senescent preosteoclast secretome promotes metabolic syndrome associated osteoarthritis through cyclooxygenase 2. eLife. 2022;11:e79773. doi:10.7554/eLife.79773
45. Jin K, Qian C, Lin J, Liu B. Cyclooxygenase-2-Prostaglandin E2 pathway: a key player in tumor-associated immune cells. Front Oncol. 2023;13:1099811. doi:10.3389/fonc.2023.1099811
46. Sun P, Quan J, Wang S, Zhuang M, Liu Z, Guan X. lncRNA-PACER upregulates COX-2 and PGE2 through the NF-κB pathway to promote the proliferation and invasion of colorectal-cancer cells. Gastroenterol Rep. 2020;9(3):257–268. doi:10.1093/gastro/goaa060
47. Yu H, Lin L, Zhang Z, Zhang H, Hu H. Targeting NF-κB pathway for the therapy of diseases: mechanism and clinical study. Signal Transduct Target Ther. 2020;5(1):209. doi:10.1038/s41392-020-00312-6
48. Zhou Y, Cui C, Ma X, Luo W, Zheng SG, Qiu W. Nuclear factor κB (NF-κB)-mediated inflammation in multiple sclerosis. Front Immunol. 2020;11:391. doi:10.3389/fimmu.2020.00391
49. George M, Lang M, Gali CC, et al. Liver X receptor activation attenuates oxysterol-induced inflammatory responses in fetoplacental endothelial cells. Cells. 2023;12(8):1186. doi:10.3390/cells12081186
50. Trefts E, Shaw RJ. AMPK: restoring metabolic homeostasis over space and time. Mol Cell. 2021;81(18):3677–3690. doi:10.1016/j.molcel.2021.08.015
51. Ge Y, Zhou M, Chen C, Wu X, Wang X. Role of AMPK mediated pathways in autophagy and aging. Biochimie. 2022;195:100–113. doi:10.1016/j.biochi.2021.11.008
52. Hsu CC, Peng D, Cai Z, Lin HK. AMPK signaling and its targeting in cancer progression and treatment. Semin Cancer Biol. 2022;85:52–68. doi:10.1016/j.semcancer.2021.04.006
53. Zheng Z, Bian Y, Zhang Y, Ren G, Li G. Metformin activates AMPK/SIRT1/NF-κB pathway and induces mitochondrial dysfunction to drive caspase3/GSDME-mediated cancer cell pyroptosis. Cell Cycle. 2020;19(10):1089–1104. doi:10.1080/15384101.2020.1743911
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.