Back to Journals » Neuropsychiatric Disease and Treatment » Volume 19
Toll-Like Receptor (TLR) 1, 2, and 6 Gene Polymorphisms Support Evidence of Innate Immune Factors in Schizophrenia
Authors Sotelo-Ramírez CE , Camarena B, Sanabrais-Jiménez MA , Zaragoza-Hoyos JU , Ordoñez-Martínez B, Escamilla-Orozco RI , Gómez-González B
Received 30 May 2023
Accepted for publication 14 September 2023
Published 2 November 2023 Volume 2023:19 Pages 2353—2361
DOI https://doi.org/10.2147/NDT.S420952
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Roger Pinder
Carlo E Sotelo-Ramírez,1,2 Beatriz Camarena,2 Marco Antonio Sanabrais-Jiménez,2 Julio Uriel Zaragoza-Hoyos,2 Bruno Ordoñez-Martínez,2 Raul I Escamilla-Orozco,3 Beatriz Gómez-González4
1Posgrado en Biología Experimental, Universidad Autónoma Metropolitana-Iztapalapa, México City, México; 2Departamento de Farmacogenética, Instituto Nacional de Psiquiatría Ramon de la Fuente Muñiz, Mexico City, Mexico; 3Dirección de Servicios Clínicos, Instituto Nacional de Psiquiatría Ramón de la Fuente Muñiz, Ciudad de México, México; 4Área de Neurociencias, Departamento de Biología de la Reproducción, Universidad Autónoma Metropolitana-Iztapalapa, Ciudad de México, México
Correspondence: Beatriz Camarena, Departamento de Farmacogenética, Instituto Nacional de Psiquiatría Ramón de la Fuente Muñiz, Calz México-Xochimilco, 101, Col. San Lorenzo Huipulco, México City, 14370, Mexico, Tel +52554160-5075, Email [email protected]
Introduction: Schizophrenia is a complex psychiatric disorder with an important genetic contribution. Immunological abnormalities have been reported in schizophrenia. Toll-like receptor (TLR) genes play an important role in the activation of the innate immune response, which may help to explain the presence of inflammation in people with this disorder. The aim of this study was to analyze the association of TLR1, TLR2, and TLR6 gene polymorphisms in the etiology of schizophrenia.
Methods: We included 582 patients with schizophrenia and 525 healthy controls. Genetic analysis was performed using allelic discrimination with TaqMan probes.
Results: We observed significant differences between patients and controls in the genotype and allele frequencies of TLR1/rs4833093 (χ 2 = 17.3, p = 0.0002; χ 2 = 15.9, p = 0.0001, respectively) and TLR2/rs5743709 (χ 2 = 29.5, p = 0.00001; χ 2 = 7.785, p = 0.0053, respectively), and in the allele frequencies of TLR6/rs3775073 (χ 2 = 31.1, p = 0.00001). Finally, we found an interaction between the TLR1/rs4833093 and TLR2/rs5743709 genes, which increased the risk of developing schizophrenia (OR = 2.29, 95% CI [1.75, 3.01]).
Discussion: Our findings add to the evidence suggesting that the activation of innate immune response might play an important role in the development of schizophrenia.
Keywords: TLR1, TLR2, TLR6, gene–gene interaction, schizophrenia, immune response
Introduction
Schizophrenia is considered a debilitating, complex, and chronic disorder, characterized by the presence of positive, negative, and cognitive symptoms, that affects basic processes such as perception, emotions, and judgment.1 It is a common disorder, with a lifetime prevalence of 1%; it has been classified as the twelfth most common cause of disability in the world among psychiatric and non-psychiatric illnesses.2
The etiology of schizophrenia is unknown; however, there is evidence suggesting that immune alterations could be involved in at least a subset of patients with the disorder.3 Physiological studies have reported immune alterations in patients with the disorder, such as elevated levels of cytokines and inflammation markers, supporting the immune hypothesis.4–7 The two-hit hypothesis8 suggests that a combination of the effects of genetic susceptibility and environmental factors, such as immunological alterations during neurodevelopment, may sensitize the individual to a second stimulus, resulting in the development of schizophrenia-like disease (Figure 1).
Family, twin, and adoption studies have shown genetic and environmental components of schizophrenia, estimating a heritability of 80–85%. Interestingly, genome-wide association studies (GWAS) of the disorder have identified several genes from the immune system.9–12 An additional GWAS analysis of high-risk genes, including distal genes and those associated with weak GWAS signals, identified overrepresented biologically plausible pathways, showing that the signaling pathway of toll-like receptors (TLR) was overrepresented in schizophrenia.12
TLRs are thus possible agents involved in immunological alterations at the level of the central nervous system. Studies carried out in animal models show schizophrenia-like symptoms, including hyperlocomotion, anxiolytic-like behavior, social withdrawal, and cognitive impairment, in TLR2 knockout mouse brains.1,13 Specifically, the TLR1 and TLR6 genes on chromosome 4p14 and the TLR2 gene on 4q31.3 have shown lower levels of gene expression in patients with schizophrenia than controls, suggesting a relationship between TLRs and schizophrenia; however, the causes of these alterations remain unknown.14,15 For this reason, it is important to study TLR1, TLR2, and TLR6 and their association with the etiology of schizophrenia.
Specifically, TLR1, TLR2, and TLR6 belong to the group of receptors located on the cell surface and can form heterodimers, which allow the identification of a wide range of ligands, such as damaged cells, regions of the cell wall of exogenous microorganisms, and the microbiota.16 In addition, it has been reported that TLRs protect the intestine from potential damage from host bacteria.17 Also, the intestinal integrity can be affected by changes in the microbiota composition, which can result in bacterial translocation and cytokine synthesis in the systemic circulation; therefore, slowdowns in different neurobiological pathways can increase the risk of developing schizophrenia.18
The aim of this study was thus to analyze the association between the TLR1, TLR2, and TLR6 gene polymorphisms and schizophrenia.
Materials and Methods
Sample Selection
This study included 582 Mexican patients, recruited from a schizophrenia clinic from the Instituto Nacional de Psiquiatria Ramon de la Fuente Muñiz, who met the DSM-5 criteria for schizophrenia according to an evaluation with the Spanish version of the Mini International Neuropsychiatric Interview (MINI-5). The study also included a control group of 525 healthy volunteers, also evaluated with the MINI-5 scale, without any diagnosis of mental illness or family history of mental disorders. All participants were Mexican mestizos with a family background of at least three generations born in Mexico, 18 years of age or older, with no chronic immunological diseases. The investigation was carried out in accordance with the last version of the Declaration of Helsinki. All patients provided written informed consent at time of recruitment and were obtained after the nature of the procedures had been fully explained. The study design was reviewed and approved by the Research Ethics Committee of the Institute (CEI/C/017/2016).
SNP Selection
Three polymorphisms, one each from TLR1, TLR2, and TLR6, were chosen in accordance with data published in PubMed, the UCSC Genome Browser (human, GRCh/hg38), and the Genetic Association Database.19 We selected polymorphisms with functionality based on nucleotide changes, location within the gene (regulatory region), or previous association with RNA expression levels, inflammatory processes, or schizophrenia and other psychiatric disorders (Table 1).
 |
Table 1 TLRs Gene Polymorphisms Analyzed in the Study |
Genotyping
Genomic DNA was extracted from peripheral blood samples using the Flexigene DNA kit (Qiagen, Minneapolis, MN). Genotyping was analyzed with custom TaqMan assays for rs4833095 (C_44103606_10), rs5743596 (C_30687147_10), rs4833093 (C_44102593_10), rs3804099 (C_22274563_10), rs7656411 (C_29420880_10), rs5743709 (C_25607738_10), rs5743810 (C_1180648_20), rs3775073 (C_25809256_30), and rs5743827 (C_1180651_20); it was performed with the allele-specific TaqMan assay method, using the ABI Prism®7500 Sequence Detection System according to the manufacturer’s protocols (Applied Biosystems Inc., Foster City, CA). The final volume of the reaction was 7 µL, consisting of 100 ng of genomic DNA, 1X TaqMan Universal Master Mix, and 0.71x SNP Genotyping Assay Mix (Applied Biosystems Inc.). After denaturing at 95°C for 1 min, 40 cycles of PCR were performed, denaturing at 95°C for 15s and annealing at 60°C for 1 min.
Statistical Analysis
The power analysis was with QUANTO v.1.2.4 software,20 with a power of 0.9 to detect a twofold increased risk, assuming an additive genetic model, a risk allele frequency of 0.22, an α level of 0.05, and a control–case ratio of 1:1. Genotype and allele frequency analyses were performed with a chi-square test using Epidat version 3.1.21 Bonferroni’s correction for multiple testing was applied considering the analysis of nine SNPs in the study (0.05/9), adjusting to p < 0.0055. A linkage disequilibrium (LD) matrix based on the D’ parameter in the patients and controls was estimated using Haploview version 4.2.22 THESIAS software was used to analyze the haplotype effect. The results were expressed as a haplotypic odds ratio in reference to the most frequent haplotype.23
Gene–gene (GXG) interaction was analyzed using Multifactor Dimensionality Reduction (MDR) software version 3.0.2 and MDR Permutation Testing software version 1.0 beta.24,25 This program is used to detect high-order nonlinear or non-additive interactions in case–control studies with small sample size to improve estimation of the effect of multiple genetic loci in the development of a disease.23–25 The interaction models are evaluated using testing balanced accuracy (TBA), which measures how often individuals are correctly classified with respect to their case/control status, and cross-validation consistency (CVC), which evaluates the consistency with which individuals are classified. The best final model is defined for values of TBA of 0.55 to 0.69, a CVC of 10, which is considered strong evidence of a multifactor interaction, and a significant p-value derived from 1000 permutations.24,26 The MDR software avoids problems in the use of parametric statistics to analyze high-order interactions, minimizes false-positive results due to multiple testing, and does not assume a genetic model.26
Results
Demographic and Clinical Characteristics
We include a total sample of 1106 subjects, 581 cases and 525 controls. In the cases sample, 221 (38%) were females and 360 males (62%); and in the control group, 322 (61.3%) were females and 203 (38.7%) males. The mean age and SD in cases was 40.61±2.4 and in the control group was 32.13±11.6. Most of the patients were single (90%), unemployed (72%), and males (63%). Average age and years of education were 40.6±12.4 and 10.25±0.13 years, respectively.
Association Analysis
We observed Hardy Weinberg equilibrium (HWE) for the SNPs TLR1/ rs4833095, TLR1/rs5743596, TLR1/rs4833093, TLR2/rs3804099, TLR2/rs5743709, and TLR6/rs5743810 in controls (p>0.05). On the other hand, we did not observe HWE for the SNPs TLR2/rs7656411, TLR6/rs3775073, and TLR6/rs5743827 in controls (p<0.05). Regarding the patients, the TLR2/rs3804099 and TLR2/rs7656411 polymorphisms were found in HWE (p≥0.05); we did not find HWE for the variants TLR1/rs4833095, TLR1/rs5743596, TLR1/4833093, TLR2/rs5743709, TLR6/rs3775073, TLR6/rs5743810, and TLR6/rs5743827 (p<0.05).
Genotype and allele distributions of TLR1, TLR2, and TLR6 gene polymorphisms are shown in Table 2. There were no differences between the TLR1/rs4833095, TLR1/rs5743596, and TLR2/rs3804099 gene polymorphisms between patients with schizophrenia and controls (Table 2). However, we observed significant differences between patients and controls in genotype and allele frequencies of TLR1/rs4833093 (χ2 = 17.3, p = 0.0002; χ2 = 15.9, p = 0.0001; OR = 1.5, 95% CI [1.26, 1.99], respectively), of TLR2/rs5743709 (χ2 = 29.5, p = 0.00001; χ2 = 7.785, p = 0.0053; OR = 0.75, 95% CI [0.61, 0.91], respectively), and in the allele frequencies of TLR6/rs3775073 (χ2 = 31.1, p = 0.00001; OR = 0.82, 95% CI [0.69, 0.98]) (Table 2).
 |
Table 2 Genotype and Allele Frequencies of SNPs Analyzed in Patients and Controls |
Haplotype Analysis
Since the variants TLR2/rs7656411, TLR6/rs3775073, and TLR6/rs5743827 were not found in HWE, they were excluded from further analysis. Analysis of the TLR1 gene indicates that the polymorphic variants are not found in linkage disequilibrium (D’ = 0.762, r2 = 0.095; D’ = 0.674, r2 = 0.04; D’ = 0.745, r2 = 0.299), which indicates that these variants are subject to recombination processes (Figure 2A). Similarly, polymorphic variants of the TLR2 gene were not found in LD (D’ = 0.48, r2 = 0.163), so it was not possible to carry out the haplotype frequency analysis (Figure 2B).
Gene–Gene Interaction
Gene–gene interaction analysis showed the best interaction model for TLR1 and TLR2 genes with TBA of 0.5824, a maximum CVC of 10/10, p < 0.0001, and a 1000-fold permutation test. This result showed a significant interaction between two polymorphic variants of the TLR1 and TLR2 genes, reflecting an increased risk in the Mexican population of developing schizophrenia (OR = 2.3, 95% CI [1.75, 3.01] (Table 2)).
Discussion
This study investigated the effect of three candidate genes involved in initiating the innate immune response in a sample of Mexican patients with schizophrenia. The demographic characteristics showed that those with schizophrenia were more likely to be male, have a lower educational level, be single, and be unemployed, which is consistent with data reported in the literature.27–29
To our knowledge, this is the first genetic study of TLR1, TLR2, and TLR6 and schizophrenia in Mexican population. Several genome-wide association studies in patients with schizophrenia have shown associations with chromosomal regions containing genes involved in the immune system and inflammation,11,12,30–32 however only one has shown an association between genes encoding proteins in the signaling cascade initiated by the TLRs.12
We find an association of TLR1/rs4833093, TLR2/rs5743709, and TLR6/rs3775073 with schizophrenia. Carriers of the G allele of the TLR1/rs4833093 gene polymorphism have a risk of developing schizophrenia that is 1.5 times that of a control group. The G allele of TLR2/rs5743709 and T allele of TLR6/rs3775073 confer protective effects from schizophrenia.
The TLR1/rs4833093 gene polymorphism is located near a regulatory region, which could be altering the expression of the gene,19 supporting findings of a decreased expression of TLR1, TLR2, and TLR6 genes in patients with schizophrenia, as compared to healthy controls.15 Studies have found that mRNA stability is affected by the number of optimal and non-optimal codons;33,34 TLR2 and TLR6 variants could thus be related to RNA stability conferring a protective effect from schizophrenia.
In addition, we found no association between the TLR1/rs4833095, TLR1/rs5743596, or TLR2/rs3804099 polymorphisms and schizophrenia, consistent with a study showing no association between TLR1/4833095 and schizophrenia in the North Indian population. That study observed an association between TLR2/rs3804099 and an increased risk of developing schizophrenia.35 Oliveira et al reported associations between TLR2/rs4696480, TLR2/rs3804099, TLR4/rs1927914, and TLR4/rs11536891 and bipolar disorder, suggesting a pleiotropic effect of TLR genes in psychotic spectrum disorders.36
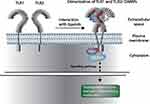
The TLR1, TLR2, and TLR6 gene polymorphisms analyzed in the present study were not found in LD, suggesting that these variants are subject to a recombination process. We also found an epistatic effect between TLR1/rs4833093 and TLR2/rs5743709, conferring an increased risk for developing schizophrenia. Interestingly, members of the same TLR subfamily tend to form heterodimers to detect their ligands; in particular, TLR2 forms dimers with TLR1 to recognize various pathogen-associated molecular patterns (PAMP).37 The immune activation of TLR in early developmental stages increases the risk of developing such neuropsychiatric disorders.38 A hypothesis has been formulated about how inflammation may be influencing neurobiological alterations in schizophrenia. This theory is based on immune activation by translocation of bacteria and the synthesis of cytokines in the intestine binding to vagus nerve receptors and reaching the hypothalamus through retrograde neuronal transport in the CNS, which activates microglia, leading to the synthesis of cytokines within the brain. These messengers activate indoleamine 2.3-dioxygenase (IDO1), which is the enzyme in charge of metabolizing tryptophan through the kynurenine pathway, leading to an increase in kynurenic acid, which is associated with alterations in glutamate neurotransmission, which has been associated with the symptomatology of schizophrenia.4 Therefore, the effect of the interaction of the TLR1 and TLR2 variants may result in alterations in neural TLR activation, altering brain function and increasing the risk. The role of TLR activation in individuals with psychotic disorders, however, remains unknown. It has been suggested that damage-associated molecular patterns (DAMP) are involved in the pathophysiology of schizophrenia, but the precise mechanism remains unclear. We hypothesize that the presence of polymorphisms in TLR1 and TLR2 genes can modify the affinity between DAMPs and adapter proteins responsible for activating the signaling cascade of the innate immune response (Figure 3). Preclinical evidence suggests that perinatal infections can trigger an immune and inflammatory response, followed by oxido-nitrosative stress that can lead to behavioral abnormalities in children.38,39 It would also be interesting to study proteins that participate in the signaling cascade of TLR1, TLR2, and TLR6 receptors, such as myeloid differentiation primary response 88 (MyD88), which has been associated with the etiology of schizophrenia in the Spanish population.13 There were certain limitations to our study. First, we evaluated only three polymorphisms in each TLR1, TLR2, and TLR6 gene; therefore, the analysis of additional SNPs located in exons or regulatory regions could be important to understand the role of TLR in schizophrenia. Second, we did not match the sex between patients and control group, which could yield important information. Third, our small sample size could not be effective in detecting small genetic effects of TLR genes in the etiology of schizophrenia. Fourth, we did not observe HWE in some SNPs analyzed reducing the genetic information in the study. We did not find HWE in the patients group. Interestingly, it has been suggested that a HW disequilibrium might be explained by genetic association showing a link between the locus studied and the disease under study.40
It is important to note that because schizophrenia is a multifactorial disorder, with a high comorbidity with other psychiatric disorders, it is difficult to elucidate its etiology.41 Genetic factors have a major impact on its development; however, gene–environment interaction studies must be carried out to fully characterize its pathophysiology.
Conclusion
Our findings showed an association between TLR1, TLR2, and TLR6 genes and schizophrenia. This suggests that the activation of innate immune response might play an important role in the development of this disorder; however, replication of this study in a larger sample size may help to clarify the role of innate immune genes in the etiology of schizophrenia.
Abbreviations
TLR, Toll-like receptor; GWAS, genome-wide association studies; MINI-5, Mini International Neuropsychiatric Interview; LD, linkage disequilibrium; GXG, Gene–gene interaction; MDR, Multifactor Dimensionality Reduction; TBA, testing balanced accuracy; CVC, cross-validation consistency; OR, odd radio; PAMP, pathogen-associated molecular patterns; DAMP, damage-associated molecular patterns; MyD88, myeloid differentiation primary response 88; CNS, central nervous system; TIR, Toll-interleukin-1; TIRAP, TIR domain-containing adaptor protein.
Acknowledgments
The study was supported by Instituto Nacional de Psiquiatría Ramon de la Fuente Muñiz. CSR was supported by Consejo Nacional de Ciencia y Tecnologia Scholarship (No. 926391). The authors thank all the participants who agreed to participate in the study.
Disclosure
The authors report no conflicts of interest in this work.
References
1. McCutcheon RA, Reis Marques T, Howes OD. Schizophrenia-An Overview. JAMA Psychiatry. 2020;77(2):201–210. doi:10.1001/jamapsychiatry.2019.3360
2. Charlson FJ, Ferrari AJ, Santomauro DF, et al. Global epidemiology and burden of schizophrenia: findings from the global burden of disease study 2016. Schizophr Bull. 2018;44(6):1195–1203. doi:10.1093/schbul/sby058
3. Benros ME, Nielsen PR, Nordentoft M, et al. Autoimmune diseases and severe infections as risk factors for schizophrenia: a 30-year population-based register study. Am J Psychiatry. 2011;168(12):1303–1310. doi:10.1176/appi.ajp.2011.11030516
4. Khandaker GM, Cousins L, Deakin J, Lennox BR, Yolken R, Jones PB. Inflammation and immunity in schizophrenia: implications for pathophysiology and treatment. Lancet Psychiatry. 2015;2(3):258–270. doi:10.1016/S2215-0366(14)00122-9
5. Wang AK, Miller BJ. Meta-analysis of cerebrospinal fluid cytokine and tryptophan catabolite alterations in psychiatric patients: comparisons between schizophrenia, bipolar disorder, and depression. Schizophr Bull. 2018;44(1):75–83. doi:10.1093/schbul/sbx035
6. Khandaker GM, Dantzer R, Jones PB. Immunopsychiatry: important facts. Psychol Med. 2017;47(13):2229–2237. doi:10.1017/S0033291717000745
7. Feigenson KA, Kusnecov AW, Silverstein SM. Inflammation and the two-hit hypothesis of schizophrenia. Neurosci Biobehav Rev. 2014;38:72–93. doi:10.1016/j.neubiorev.2013.11.006
8. Lee KW, Woon PS, Teo YY, Sim K. Genome wide association studies (GWAS) and copy number variation (CNV) studies of the major psychoses: what have we learnt? Neurosci Biobehav Rev. 2012;36(1):556–571. doi:10.1016/j.neubiorev.2011.09.001
9. Lee SH, DeCandia TR, Ripke S, Yang J. Estimating the proportion of variation in susceptibility to schizophrenia captured by common SNPs. Nat Genet. 2012;44(3):247–250. doi:10.1038/ng.1108
10. Prata DP, Costa-Neves B, Cosme G, Vassos E. Unravelling the genetic basis of schizophrenia and bipolar disorder with GWAS: a systematic review. J Psychiatr Res. 2019;114:178–207. doi:10.1016/j.jpsychires.2019.04.007
11. Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511(7510):421–427. doi:10.1038/nature13595
12. Lin JR, Cai Y, Zhang Q, et al. Integrated post-GWAS analysis sheds new light on the disease mechanisms of schizophrenia. Genetics. 2016;204(4):1587–1600. doi:10.1534/genetics.116.187195
13. García-Bueno B, Gassó P, MacDowell KS, et al. Evidence of activation of the Toll-like receptor-4 proinflammatory pathway in patients with schizophrenia. J Psychiatry Neurosci. 2016;41(3):E46–E55. doi:10.1503/jpn.150195
14. Kéri S, Szabó C, Kelemen O. Uniting the neurodevelopmental and immunological hypotheses: neuregulin 1 receptor ErbB and Toll-like receptor activation in first-episode schizophrenia. Sci Rep. 2017;7(1):4147. doi:10.1038/s41598-017-03736-3
15. Kozłowska E, Agier J, Wysokiński A, et al. The expression of toll-like receptors in peripheral blood mononuclear cells is altered in schizophrenia. Psychiatry Res. 2019;272:540–550. doi:10.1016/j.psychres.2018.12.138
16. Chen CY, Shih YC, Hung YF, Hsueh YP. Beyond defense: regulation of neuronal morphogenesis and brain functions via Toll-like receptors. J Biomed Sci. 2019;26(1):90. doi:10.1186/s12929-019-0584-z
17. Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118(2):229–241. doi:10.1016/j.cell.2004.07.002
18. Caso JR, Balanzá-Martínez V, Palomo T, García-Bueno B. The microbiota and gut-brain axis: contributions to the immunopathogenesis of schizophrenia. Curr Pharm Des. 2016;22(40):6122–6133. doi:10.2174/1381612822666160906160911
19. Kent WJ, Sugnet CW, Furey TS, et al. The human genome browser at UCSC. Genome Res. 2002;12(6):996–1006. doi:10.1101/gr.229102
20. Gaudeman WJ, Morrison JM Quanto 1.2.4: a computer program for power and sample size calculations for genetic-epidemiology studies. University of Southern California; 2019. Available from: http://biostats.usc.edu/Quanto.html.
21. Hervada VX, Santiago IP, Vazquez F, et al. Epidad 3.0 Programa para análisis epidemiológicos de datos tabulados. Rev Esp Salud Publica. 2004;78(2):277–280. doi:10.1590/S1135-57272004000200013
22. Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi:10.1093/bioinformatics/bth457
23. Tregouet DA, Garelle V. A new JAVA interface implementation of THESIAS: testing haplotype effects in association studies. Bioinformatics. 2007;23(8):1038–1039. doi:10.1093/bioinformatics/btm058
24. Hahn LW, Ritchie MD, Moore JH. Multifactor dimensionality reduction software for detecting gene-gene and gene-environment interactions. Bioinformatics. 2003;19(3):376–382. doi:10.1093/bioinformatics/btf869
25. Ritchie MD, Hahn LW, Moore JH. Power of multifactor dimensionality reduction for detecting gene-gene interactions in the presence of genotyping error, missing data, phenocopy, and genetic heterogeneity. Genet Epidemiol. 2003;24(2):150–157. doi:10.1002/gepi.10218
26. Moore JH MDR 101—Part 4—results. Epistasis blog from the computational genetics laboratory at the University of Pennsylvania; 2019. Available from: http://www.epistasisblog.org/2006/12/.
27. Castle DJ, Murray RM. The neurodevelopmental basis of sex differences in schizophrenia. Psychol Med. 1991;21(3):565–575. doi:10.1017/s0033291700022194
28. Canuso CM, Pandina G. Gender and schizophrenia. Psychopharmacol Bull. 2007;40(4):178–190.
29. Kahn RS, Sommer IE, Murray RM, et al. Schizophrenia. Nat Rev Dis Primers. 2015;1:15067. doi:10.1038/nrdp.2015.67
30. Avramopoulos D, Pearce BD, McGrath J, et al. Infection and inflammation in schizophrenia and bipolar disorder: a genome wide study for interactions with genetic variation. PLoS One. 2015;10(3):e0116696. doi:10.1371/journal.pone.0116696
31. Jia P, Wang L, Meltzer HY, Zhao Z. Common variants conferring risk of schizophrenia: a pathway analysis of GWAS data. Schizophr Res. 2010;122(1–3):38–42. doi:10.1016/j.schres.2010.07.001
32. Purcell SM, Wray NR; International Schizophrenia Consortium. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460(7256):748–752. doi:10.1038/nature08185
33. Presnyak V, Alhusaini N, Chen YH, et al. Codon optimality is a major determinant of mRNA stability. Cell. 2015;160(6):1111–1124. doi:10.1016/j.cell.2015.02.029
34. Forrest ME, Pinkard O, Martin S, et al. Codon and amino acid content are associated with mRNA stability in mammalian cells. PLoS One. 2020;15(2):e0228730. doi:10.1371/journal.pone.0228730
35. Sharma I, Priya I, Sharma S, et al. Association of toll-like receptor 2 gene polymorphism (rs3804099) with susceptibility to Schizophrenia risk in the Dogra population of Jammu region, North India. Eur J Psychiatry. 2022;36(2):106–113. doi:10.1016/j.ejpsy.2022.02.001
36. Oliveira J, Etain B, Lajnef M, et al. Combined effect of TLR2 gene polymorphism and early life stress on the age at onset of bipolar disorders. PLoS One. 2015;10(3):e0119702. doi:10.1371/journal.pone.0119702
37. Al-Haddad BJS, Jacobsson B, Chabra S, et al. Long-term risk of neuropsychiatric disease after exposure to infection in utero. JAMA Psychiatry. 2019;76(6):594–602. doi:10.1001/jamapsychiatry.2019.0029
38. Venkatasubramanian G, Debnath M. The TRIPS (Toll-like receptors in immuno-inflammatory pathogenesis) Hypothesis: a novel postulate to understand schizophrenia. Prog Neuropsychopharmacol Biol Psychiatry. 2013;44:301–311. doi:10.1016/j.pnpbp.2013.04.001
39. Abrahams VM, Bole-Aldo P, Kim YM, et al. Divergent trophoblast responses to bacterial products mediated by TLRs. J Immunol. 2004;173(7):4286–4296. doi:10.4049/jimmunol.173.7.4286
40. Royo JL. Hardy Weinberg equilibrium disturbances in case-control studies lead to non-conclusive results. Cell J. 2021;22(4):575. doi:10.22074/cellj.2021.7195
41. Owen MJ, Sawa A, Mortensen PB. Schizophrenia. Lancet. 2016;388(10039):86–97. doi:10.1016/S0140-6736(15)01121-6
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.