Back to Journals » International Journal of Chronic Obstructive Pulmonary Disease » Volume 13
TNF promoter polymorphisms are associated with genetic susceptibility in COPD secondary to tobacco smoking and biomass burning
Authors Reséndiz-Hernández JM, Ambrocio-Ortiz E, Pérez-Rubio G , López-Flores LA , Abarca-Rojano E , Pavón-Romero GF , Flores-Trujillo F, Hernández-Zenteno RJ , Camarena A , Pérez-Rodríguez M, Salazar AM, Ramírez-Venegas A , Falfán-Valencia R
Received 29 July 2017
Accepted for publication 28 December 2017
Published 16 February 2018 Volume 2018:13 Pages 627—637
DOI https://doi.org/10.2147/COPD.S147688
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Richard Russell
Juan Manuel Reséndiz-Hernández,1 Enrique Ambrocio-Ortiz,1 Gloria Pérez-Rubio,1 Luis Alberto López-Flores,1 Edgar Abarca-Rojano,2 Gandhi Fernando Pavón-Romero,3 Fernando Flores-Trujillo,4 Rafael de Jesús Hernández-Zenteno,4 Ángel Camarena,1 Martha Pérez-Rodríguez,5 Ana María Salazar,6 Alejandra Ramírez-Venegas,4 Ramcés Falfán-Valencia1
1HLA Laboratory, Instituto Nacional de Enfermedades Respiratorias Ismael Cosío Villegas, Mexico City, Mexico; 2Research and Graduate Studies Section, Escuela Superior de Medicina, Instituto Politécnico Nacional, Mexico City, Mexico; 3Department of Allergy and Clinical Immunology, Instituto Nacional de Enfermedades Respiratorias Ismael Cosío Villegas, Mexico City, Mexico; 4Tobacco Smoking and COPD Research Department, Instituto Nacional de Enfermedades Respiratorias Ismael Cosío Villegas, Mexico City, Mexico; 5Unit of Medical Research in Immunology, CMN S-XXI, Instituto Mexicano del Seguro Social, Mexico City, Mexico; 6Department of Genomic Medicine and Environmental Toxicology, Instituto de Investigaciones Biomédicas, Universidad Nacional Autónoma de México, Mexico City, Mexico
Background: Smoking and smoke from biomass burning (BB) are the main environmental risk factors for COPD. Clinical differences have been described between COPD related to smoking and related to wood smoke, but no studies have shown genetic differences between patients exposed to these two risk factors.
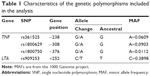
Methods: To investigate a possible association of tumor necrosis factor (TNF) promoter polymorphisms, we conducted a case–control study. A total of 1,322 subjects were included in four groups: patients with a diagnosis of COPD secondary to smoking (COPD-S, n=384), patients with COPD secondary to biomass burning (COPD-BB, n=168), smokers without COPD (SWOC, n=674), and biomass burning-exposed subjects (BBES n=96). Additionally, a group of 950 Mexican mestizos (MMs) was included as a population control. Three single nucleotide polymorphisms (SNPs) were selected in the TNF gene (rs1800629, rs361525, and rs1800750) and one SNP in the lymphotoxin alpha gene (rs909253).
Results: Statistically significant differences were found with genotype GA of the rs1800629: COPD-S vs SWOC, (p<0.001, odds ratio [OR] =2.55, 95% CI=1.53–4.27); COPD-S vs COPD-BB (p<0.01). When performing the comparison of the less severe (G1: I + II) and the more severe (G2: III + IV) levels, differences were identified in G1 (p<0.05, OR=1.94, 95% CI=1.04–3.63) and G2 (p<0.001, OR=3.68, 95% CI=1.94–3.07) compared with SWOC. Regarding genotype GA of rs361525, it has been associated when comparing COPD-BB vs BBES (p=0.0079, OR=5.99, 95% CI=1.38–53.98).
Conclusion: The heterozygous genotype GA of polymorphisms rs1800629 and rs361525 in the TNF promoter are associated with the risk of COPD.
Keywords: COPD, biomass burning, SNP, TNF
Introduction
COPD is defined as a preventable and treatable disease, which is characterized by a persistent, progressive limitation of airflow with an inflammatory response increased by particle stimulus and harmful gases in the airways. Exacerbations and comorbidities contribute to the individual severity of the disease.1 Smoking is the main environmental risk factor for COPD2 and is widely reported to increase respiratory symptoms, abnormalities in pulmonary function, and mortality.3 Air contamination by biomass burning (BB; mainly wood) has also been reported as a risk factor for COPD.4,5 Although in many aspects the COPD by biomass burning (COPD-BB) is similar to COPD by smoking (COPD-S), clinical differences have been observed. Among these, COPD-BB is most frequent in women, bronchitis is the main phenotype found and, in general, the patients have higher body mass index (BMI),6 whereas in COPD-S, the emphysema is most frequent and airflow obstruction severest.7,8 However, there are no studies showing molecular, cellular, or genetic differences between both phenotypes.
COPD is a disease widely related to inflammation, both systemic and localized,9 and several studies have evaluated the levels of inflammation and their relation to the clinical severity of the disease.10 Some of the evaluated biomarkers include cytokines and chemokines involved in the inflammatory process, such as interleukin (IL)-1β, IL-6, IL-8, and tumor necrosis factor (TNF)-α, among others. Some studies have demonstrated relationships between increased TNF-α levels, metabolic disorders,11,12 and weakening chronic diseases, resulting in loss of weight, skeletal muscle proteins, and fat tissue.13,14 Genetic variations in the TNF promoter region have been associated with differences in its phenotypic expression and with several diseases.14,15 Regarding single nucleotide polymorphisms (SNPs), rs1800629 (G > A change in position −308) increases the transcriptional activity of the gene.15–18 Another gene reported to affect the expression of TNF-α is lymphotoxin alpha (LTA); in particular, rs909253 (G > A) in LTA has been associated with regulation of the gene.17 Genetics variations type SNP in TNF and LTA has been associated with COPD and also with some clinical phenotypes, such as lower FEV1/FVC or decline in lung function.19,20 These genes are also reportedly associated with asthma and COPD.21,22
We hypothesize that SNPs in TNF and LTA establish genetic differences between COPD related to smoking and wood smoke. The aim of this study was to determine these associations by analyzing three SNPs in the TNF gene (rs1800629, rs361525, and rs1800750) and one SNP in the LTA gene (rs909253) in Mexican mestizo (MM) population, as well as comparing the frequencies of the different analyzed SNPs with the global frequencies.
Methods
Studied population
Case and control groups
Some 1,322 subjects were included in the case–control study. These subjects attended the COPD and smoking cessation support clinics, both part of the Department of Smoking and COPD Research Department of the Instituto Nacional de Enfermedades Respiratorias Ismael Cosio Villegas (INER) at Mexico. The sample included 384 patients with a diagnosis of COPD secondary to smoking (COPD-S) and 168 with COPD secondary to biomass burning (COPD-BB). The COPD diagnosis was confirmed using pulmonary function tests, applying diagnostic criteria according to the Global Initiative for Chronic Obstructive Lung Disease (GOLD) 2011 guidelines. Individuals older than 50 years and with a tobacco index ≥5 packs/year were classified in the COPD-S group, and those with an accumulative exposure higher than 200 h/year to biomass smoke (hours a day by years exposed), never smokers were classified into the COPD-BB, this cut-off was chosen because it is the risk exposure reported in the Mexican population.22 The group of smokers without COPD (SWOC, n=674) included smokers with no evidence of pulmonary disease and with normal spirometry levels. Finally, a group of contacts exposed to wood smoke with no background of active or passive smoking was also included as biomass burning-exposed subjects (BBES, n=96). All participants completed a hereditary pathology background survey. Subjects who self-reported having chronic respiratory diseases different to COPD and/or inflammatory diseases and subjects whose ancestry were not Mexican, were excluded.
Population comparison group
A group of 950 volunteers was also included. These subjects had the following characteristics: clinically healthy, >18 years, both genders, smokers and nonsmokers, born as MMs, parents and grandparents born in Mexico, no biologically-related among themselves or with the patients or controls, and no familial history of pulmonary diseases. The participants were recruited from annual campaigns of COPD early diagnosis, performed on World COPD Day and World No-Smoking Day in the INER.
Data concerning the frequencies of alleles and genotypes for the associated polymorphisms were also included for comparison with the population data. Data were included from the National Institute of Environmental Health Sciences (NIEHS) SNPs Environmental Genome Program for the Caucasian population residing in Utah, USA (CEU), Chinese in Beijing, China (CHB), and Mexicans residing in Los Angeles, CA, USA (MXL),23 as well as from the International HapMap Project.24
All participants were previously invited to participate in the study; they signed an informed consent document and were provided with a privacy statement describing the protection of their personal data. This study was reviewed and approved by the Ethics and Science Committee in Research, with protocol number B35-09 and Institutional Review Board at National Institute of Respiratory Diseases (INER) Ismael Cosio Villegas.
SNP selection
For SNP selection, a bibliographic search was performed, which identified polymorphisms previously associated with respiratory inflammatory diseases, taking into account studies with a significance value of p<0.0519,26,27 or a minor allele frequency >5% from the 1000 Genome project. Table 1 lists the characteristics of the polymorphisms included.
Genotyping
DNA was obtained from peripheral blood cells obtained by venipuncture. Genomic DNA was extracted using the commercial BDtract™ Genomic DNA Isolation kit (Maxim Biotech, San Francisco, CA, USA). The genetic material was quantified using UV light absorbing spectrophotometry at 260 nm using a nanodrop device (Thermo Fisher Scientific, Waltham, MA, USA). The contamination with organic compounds and proteins was determined by establishing the ratio of 260/240 and 260/280 reads, respectively. Samples were considered free of contaminants when the ratio was 1.7–2.0 in both cases. The allelic discrimination of the SNPs was performed using commercial 20X Taqman probes (Thermo Fisher Scientific) for real-time PCR on a 7300 Real Time PCR Systems instrument (Thermo Fisher Scientific). The results were analyzed using the SDS (Sequence Detection Software) software version 1.4.
Statistical analysis
The differences between groups were evaluated by determining and comparing the allelic, genotypic, and haplotype frequencies. The statistical significance was assessed using SPSS v15.0 and Epi Info 7.1.3.0 statistical packages according to the χ2 values. The results were considered significant when the p-value after Yates correction and Cochran-Armitage trend test was <0.05. The Hardy–Weinberg equilibrium (HWE) test was performed by Pearson and Fisher’s exact test, and it was considered outside of equilibrium with p<0.05. Finally, the odds ratio (OR) was estimated with a 95% CI to assess the strength of the association.
TNF-α blood levels
According to genetic association results, 40 plasma samples were selected: 20 COPD-S and 20 COPD-BB patients. TNF-α levels were measured using the 10-plex human cytokine kit (Thermo Fisher Scientific). Data were analyzed by Mann–Whitney U test for independent groups and the correlation r2 value was calculated for GOLD grade and TNF-α.
Results
The clinical characteristics of the subjects included are listed in Table 2. The average age was ~70 years in all four groups. The gender ratio was 2:3 men for each woman in patients with COPD-S, while in the comparison group (SWOC), the ratio was 2:1 (p=0.33). In the case of COPD-BB, the ratio of genders revealed 6:6 women for each man affected, while the corresponding ratio in the group of exposed subjects without the disease (BBES) was 5:1 (p=0.46). The mean BMI was 26–28 for all groups (p>0.05, non-significant). However, in SWOC, COPD-BB, and BBES, the BMI showed the highest values. Interestingly, the lowest values for this variable were found in the COPD-S patients.
Smoking and exposure to smoke from BB are considered environmental risk variables and were evaluated by comparing groups depending on the type of exposure. For smoking, the average accumulated consumption, expressed in packs per year, was slightly higher in the group of patients with COPD related to smoking compared with the control; however, this difference was not statistically significantly (p>0.05). Regarding chronic exposure to wood smoke from BB, the COPD-BB group exhibited more years and hours of exposure (p<0.001) compared to BBES. This is reflected in the exposure index, which was higher in COPD-BB compared with BBES (p<0.001).
There were statistically significant differences in the pulmonary function variable between cases and controls.
Genotyping
Four SNPs were genotyped in two genes: three in TNF (rs361525, rs1800629, and rs1800750) and one in LTA (rs909253). The results of the allelic and genotype frequencies are shown in Tables 3 and 4. HWE was checked for all the SNP markers from all groups. The control groups (SWOC and BBES) were in HWE, although the rs1800629 in group SWOC had a slight deviation by the Pearson test (p=0.0185); however, in Fisher’s exact test, the p-value was 0.0688. All the markers included in the study are shown in a ternary plot (Figure S1).
To determine the possible association with the polymorphisms analyzed between cases and control groups, the allelic and genotype frequencies were compared between groups using the χ2 test. No statistically significant difference was found in genotypes from rs909253 in LTA or the rs1800750 in TNF.
COPD-S to smoking vs SWOC
A statistically significant difference for genotype GA of the rs1800629 in TNF was found between COPD-S vs SWOC (p=0.00013, OR=2.54, 95% CI=1.53–4.26). According to this result, allele A of this SNP was associated with susceptibility to COPD related to smoking (p=0.001, OR=2.17, 95% CI=1.33–3.53).
COPD secondary to wood smoke vs subjects exposed to wood smoke without COPD
The comparison performed between COPD related to COPD-BB and nonsmoking BBES showed a statistically significant difference in the frequency of genotype GA of rs631525 (p=0.0079, OR=5.99, 95% CI=1.38–53.98). Regarding the allelic frequencies of the same polymorphism, allele A was increased in the case group compared with the control (p=0.009, OR=5.69, 95% CI=1.31–24.71). Given its low frequency in BBES, allele A was assessed using Fisher’s exact test.
COPD related to smoking vs COPD related to wood smoke
To determine whether there were differences between COPD-S and COPD-BB, their frequencies were compared. A statistically significant difference was found for genotype GA of rs1800629 (p=0.017), which was increased in the COPD-S group compared with COPD-BB.
GOLD severity analysis
The association of different genotypes with the severity of the disease was evaluated using logistic regression, including as covariables the GOLD level and the pulmonary function measurements. No statistically significant differences were found (p>0.05). To determine the strength of the association of the minor allele, we used the Kruskal–Wallis test in an additive recessive model (GA + AA vs GG, for both SNPs associated) and found no association with the covariables included.
A contrast between the less severe (G1: I + II) and severe (G2: III + IV) levels in patients with COPD related to smoking compared with the SWOC group yielded a statistically significant difference in genotype GA of the rs1800629 comparing G1 (p=0.0441, OR=1.94, 95% CI=1.04–3.63) and G2 (p=0.00018, OR=3.68, 95% CI=1.94–3.07) against the SWOC group. In the case of patients with COPD related to wood smoke, genotype GA of the rs361525 was increased in group G1 compared with BBES (p=0.0305, OR=5.09, 95% CI=1.07–24.26). The frequencies are included in Table 5.
Haplotypes
The haplotype analysis was performed using the Haploview v4.2 software; no haplotype was found in association with the polymorphisms analyzed; only between rs361525 and rs909253 was r2=0.62.
TNF-α blood levels
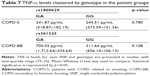
According to our genetic association results in the TNF gene, 40 plasma samples were selected in base of the followed characteristics: 20 COPD-S patients, 10 homozygous (GG) and 10 heterozygous for the rs1800629 and 20 COPD-BB were chosen: 10 homozygous (GG) and 10 heterozygous for the rs361525.
No significant differences were found when compared COPD-S vs COPD-BB cytokine levels (Table 6). For the association of TNF-α with COPD severity, all the subjects were classified by GOLD grade and correlation coefficient was calculated. No correlation between TNF-α and GOLD grade was found. TNF-α levels were compared for each associated genotype in rs1800629 and rs361525, respectively, and no statistical significant differences were found (Table 7). Interestingly, TNF-α plasma levels in COPD-BB heterozygous (GA) compared with homozygous was increased 2-fold (700.55 vs 311.64 pg/mL, p=0.108).
Population comparison
For rs361525, we considered values from the NIEHS SNPs Environmental Genome Program,23 whereas for rs1800629, data were collected from the same populations available in Phases II + III, release 28 of the International HapMap Project.24 Table 8 presents the genotype and allele frequencies for both SNPs in a Caucasian population (CEU), a Chinese population (CHB), and four Mexican populations (MXL, MM, SWOC, and BBES).
  | Table 8 Genotype and allele frequency of rs361525 and rs1800629 among the populations |
Allele G and genotype GG for both SNPs were the most frequent in the populations studied. For the rs361525 GA, the highest frequency was found in the Caucasian population (>14%). In the Mexican population, there was a difference in the distribution of this heterozygous genotype, exceeding 9% in MMs of our population control group, while in smokers with no COPD, the frequency was 7.7%. According to the data available for Mexican-Americans, <4% of this population had genotype GA. Subjects exposed to wood smoke showed the lowest frequency in Mexicans (2.1%). Genotype AA was only identified in MMs (~1.5%) and was absent in the remaining populations included in the study.
Genotype GA of rs1800629 had a heterogeneous distribution between the populations studied; almost one-third of the Caucasian population (31%) has this genotype compared with 15.3% in the Chinese population. The Mexican populations showed the lowest frequencies (~12%) in individuals with Mexican ancestry residing in Los Angeles, CA, USA. This percentage decreased to 7.3% in MMs and to 4.3% and 3.1% in Mexicans from the SWOC and BBES groups, respectively. The homozygous AA genotype was only identified in <2% in CEU and <1% in CHB. In Mexicans, its frequency was very low (0.3%–0.7%).
Figure 1 shows the frequency of genotype GA for both SNPs in all populations. The frequency of genotypes associated in the control groups (SWOC and BBES) and MMs was compared with that corresponding to the CEU, CHB, and MXL populations included in this analysis. rs361525 GA was significantly decreased in MMs vs CEU (p=0.0181) and, ultimately, was increased compared with BBES (MMs vs BBES, p=0.0314). A similar finding was observed with rs1800629 GA, which was decreased in MMs compared to CEU and CHB (p<0.001 for both comparisons). However, this frequency was higher compared with the SWOC group (7.3% vs 4.6%, p=0.036).
Discussion
COPD is a disease with a widely documented inflammatory component; altered levels of different inflammatory biomarkers have been described, and there are different hypotheses regarding its initiation and progression, including those related to immunological, biochemical, and genetic events. Additionally, different polymorphisms whose protein products are associated with inflammation have been reported. In our study, an association was found between COPD and some polymorphisms in the promoter of TNF gene. The GA genotype of rs1800629 is associated with the susceptibility of COPD secondary to smoking, while the GA genotype of the rs361525 is associated to COPD-BB. Furthermore, genotype GA of rs1800629 was identified to be more frequent in the group with COPD-S compared with COPD-BB.
Regarding rs1800629, which represents the substitution of guanine by adenine (G > A), it has been reported that allele A (minor allele) increases the gene’s transcriptional rate16,38,39 and has been associated with inflammatory autoimmune diseases.28–31
In a Turkish population, rs1800629 was not found to be associated with the GOLD grade; however, the GA genotype was associated with exacerbations and hospitalizations.32 In another study performed in the population of Helsinki, the GA genotype of the same polymorphism was associated with paraseptal emphysema, which is a COPD clinical phenotype.25
Regarding genetic associations and COPD, there are studies showing relevance for the TNF-308G/A SNP in the population of Taiwan, in which genotype GA was associated with chronic bronchitis and an increased inflammatory response.33 This result was included in a meta-analysis performed by Zhang et al in 2011,34 in which genotype GA in position −308 was shown to be associated with higher risk in Asian populations but not in Caucasian populations. This association remains even when the populations are stratified by smoking habits.34 However, these results have not been repeated in other populations.
Another polymorphism that has been associated in COPD is rs361525 GA, which was found to increase protein expression in lung secretions and bioactivity with neutrophil inflammation, increasing their chemotaxis, in addition to clinical features of the disease, including its progression.18 In our study, we found that rs361525 GA is increased in patients with COPD related to wood smoke in the GOLD grades, with low severity levels compared with the control group.
Previously, in our work team, employing sequencing analysis, we have identified several SNPs associated to COPD in Mexican population;35 however, this is the first report of a genetic association study in COPD secondary to BB or wood smoke.
On the other hand, high levels of TNF-α have been associated with SNPs in LTA.36 In our study, we analyzed rs909253 of the LTA gene, but we did not find differences in the allele genotype frequencies or haplotypes of this variant between the groups studied; this polymorphism has been associated with decreased risk to COPD in the Russian population;37 however, its frequency is low in the MM population and probably this behavior influences the fact that we have not able to identify haplotypes with SNP in the TNF promoter gene.
In general, these types of analysis have been performed in Caucasian populations, without population validations, particularly in those with ancestral contributions from diverse origins, which is the case of the MM populations. In our study, we compared the frequencies of the alleles and genotypes associated with susceptibility and higher severity of COPD in the MM populations and the Caucasian, Chinese, and Mexican-American population data. We observed that the GA genotype was found in a lower proportion in the BBES group compared with the rest of the populations. A similar result was reported by Meenagh et al in 2002,38 in which they found polymorphisms in genes that encode different cytokines, the GA genotype was found at a lower frequency (5%) compared with the Caucasian, Asian, and African populations. Interestingly, in this report, this genotype was less frequent in the BBES group compared with the SWOC group, possibly because the group with exposure to BB smoke came from rural areas with higher proportions of native ancestral contributions, while the smoker group individuals were mainly from urban areas. Nonetheless, there was no statistically significant difference between the groups. At the population level, our results agree with recently reported findings in Mexican populations, where a higher frequency of the GA genotype of rs1800629 is found in mestizo populations compared with native populations.39
Another interesting result was that the lowest average BMI was found in the group of patients with COPD related to smoking. BMI is an independent prognostic factor in COPD,40 as it has been reported as a risk factor for death.41 In 2012, a study was performed in a Taiwanese population, in which the TNF-863 (rs1800630) was associated with an increased BMI;42 however, Eagan et al found increased levels of TNF-α were associated with a loss of lean body mass;43 nevertheless, there are no studies in our population regarding TNF polymorphism and COPD clinical phenotype related to BMI. In our results, we had patients with exceptionally high BMI; however, there are no statistical differences between the study groups.
There are no studies of genetic associations and COPD secondary to smoke from BB. However, Kurmi et al reported that wood smoke was a potential oxidizing agent and they demonstrated that ascorbate oxidation and glutathione reduction were induced by a mixture of biomass and wood.44 Previously, Ramírez-Venegas et al8 in 2006 described clinical differences between patients with COPD related to BB vs smoking, such as a higher partial carbon dioxide saturation (PaCO2) and a high prevalence of bronchitis among patients with COPD related to wood smoke.
The results of several in vivo and in vitro studies indicate that the increase in the production of TNF-α leads to an increase in inflammation and to a pro-oxidative response, which is important in the disease’s pathogenesis. TNF-α regulates inflammation and is thought to play a key role in the respiratory and systemic characteristics of COPD.45–47 Several studies have found increased TNF-α concentrations in the serum and in pulmonary fluids, and more specifically, these increased levels have been found during infection exacerbations.30,31 In our analysis, in the determination of TNF-α levels, we did not find statistical differences; however, among COPD-BB patients harboring heterozygous rs361525 (GA) had 2-fold increased plasma levels in comparison to those homozygous for common allele (GG). TNF-α has also been associated with COPD systemic manifestations such as decreased BMI48 and abnormal energy expenditures at rest.49 The clinical manifestations, such as BMI, severity of the disease, and its progression, are heterogeneous and may be the result of several pathogenic processes involving abnormalities in different biological pathways such as the oxidative stress, apoptotic control disequilibrium, and changes in the immune system (inflammation).14 These pathways, either individually or collectively, may be responsible for the phenotypic differences of the disease.
Finally, regarding why two different SNPs in the same gene are associated with COPD secondary to different ambiental risk factors could be related to genetic composition in mestizo populations, as both associated SNPs have been previously found related in an independent way.
In the present work, we studied the involvement of individual SNPs in the promoter region of TNF and LTA. A limitation of our work is that rs1800629 has a deviation of the HWE. Although it is true that this point is critical in these kinds of studies, the allelic/genotype frequency of this polymorphism is very similar to other reports in the MM population. Another limitation was that we were not able to classify patients regarding clinical phenotype (ie, bronchitis, emphysema), because not all patients had imaging tests, mainly CT scans. It would be desirable to conduct research including clinical, histological, and radiological variables, in addition to wider regions of these genes, especially those involved in genetic regulation, to identify groups of variants contributing to COPD establishment and/or progression.
Currently, there are no studies evaluating the participation of genetic factors related to inflammation that include both main environmental risk factors for COPD. Hence, the present investigation constitutes a source of information for the design of further studies on this issue. Identifying genetic susceptibility in COPD secondary to different ambiental factors, particularly in the inflammation way, could lead to better pharmacological interventions, where genomic and personalized medicine have a principal role.
Acknowledgment
Partial data of this manuscript were presented in abstract form in the European Respiratory Society (ERS) 2015 international congress from September 26–30, 2015.50
Disclosure
The authors report no conflicts of interest in this work.
References
GOLD. Global Strategy for the Diagnosis, Management and Prevention of COPD. 2017 [webpage on the Internet]. Available from: http://goldcopd.org/gold-2017-global-strategy-diagnosis-management-prevention-copd/. Accessed July 28, 2017. | ||
Nardini S, Annesi-Maesano I, Del Donno M, et al. The AIMAR recommendations for early diagnosis of chronic obstructive respiratory disease based on the WHO/GARD model. Multidiscip Respir Med. 2014;9(1):46. | ||
Anthonisen NR, Skeans MA, Wise RA, et al. The effects of a smoking cessation intervention on 14.5-year mortality: a randomized clinical trial. Ann Intern Med. 2005;142(4):233–239. | ||
Albalak R, Frisancho AR, Keeler GJ. Domestic biomass fuel combustion and chronic bronchitis in two rural Bolivian villages. Thorax. 1999;54(11):1004–1008. | ||
Sezer H, Akkurt I, Guler N, Marakoğlu K, Berk S. A case-control study on the effect of exposure to different substances on the development of COPD. Ann Epidemiol. 2006;16(1):59–62. | ||
Ramírez-Venegas A, Sansores RH, Quintana-Carrillo RH, et al. FEV1 decline in patients with chronic obstructive pulmonary disease associated with biomass exposure. Am J Respir Crit Care Med. 2014;190(9):996–1002. | ||
Rivera RM, Cosio MG, Ghezzo H, Salazar M, Pérez-Padilla R. Comparison of lung morphology in COPD secondary to cigarette and biomass smoke. Int J Tuberc Lung Dis. 2008;12(8):972–977. | ||
Ramírez-Venegas A, Sansores RH, Pérez-Padilla R, et al. Survival of patients with chronic obstructive pulmonary disease due to biomass smoke and tobacco. Am J Respir Crit Care Med. 2006;173(4):393–397. | ||
Barnes PJ, Chowdhury B, Kharitonov SA, et al. Pulmonary biomarkers in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2006;174(1):6–14. | ||
Sin DD, Vestbo J. Biomarkers in chronic obstructive pulmonary disease. Proc Am Thorac Soc. 2009;6(6):543–545. | ||
Oudijk EJD, Lammers JWJ, Koenderman L. Systemic inflammation in chronic obstructive pulmonary disease. Eur Respir J Suppl. 2003;46:5s–13s. | ||
Debigaré R, Marquis K, Côté CH, et al. Catabolic/anabolic balance and muscle wasting in patients with COPD. Chest. 2003;124(1):83–89. | ||
Tanni SE, Pelegrino NR, Angeleli AY, Correa C, Godoy I. Smoking status and tumor necrosis factor-alpha mediated systemic inflammation in COPD patients. J Inflamm (Lond). 2010;7(1):29. | ||
Han MK, Agusti A, Calverley PM, et al. Chronic obstructive pulmonary disease phenotypes: the future of COPD. Am J Respir Crit Care Med. 2010;182(5):598–604. | ||
Wilson AG, Symons JA, McDowell TL, McDevitt HO, Duff GW. Effects of a polymorphism in the human tumor necrosis factor alpha promoter on transcriptional activation. Proc Natl Acad Sci U S A. 1997;94(7):3195–3199. | ||
Wu WS, McClain KL. DNA polymorphisms and mutations of the tumor necrosis factor-alpha (TNF-alpha) promoter in Langerhans cell histiocytosis (LCH). J Interferon Cytokine Res. 1997;17(10):631–635. | ||
Messer G, Spengler U, Jung MC, et al. Polymorphic structure of the tumor necrosis factor (TNF) locus: an NcoI polymorphism in the first intron of the human TNF-beta gene correlates with a variant amino acid in position 26 and a reduced level of TNF-beta production. J Exp Med. 1991;173(1):209–219. | ||
Sapey E, Wood AM, Ahmad A, Stockley RA. Tumor necrosis factor-{alpha} rs361525 polymorphism is associated with increased local production and downstream inflammation in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2010;182(2):192–199. | ||
Sandford AJ, Chagani T, Weir TD, Connett JE, Anthonisen NR, Paré PD. Susceptibility genes for rapid decline of lung function in the lung health study. Am J Respir Crit Care Med. 2001;163(2):469–473. | ||
Migita O, Noguchi E, Koga M, et al. Haplotype analysis of a 100 kb region spanning TNF-LTA identifies a polymorphism in the LTA promoter region that is associated with atopic asthma susceptibility in Japan. Clin Exp Allergy. 2005;35(6):790–796. | ||
Hogg JC, Chu F, Utokaparch S, et al. The nature of small-airway obstruction in chronic obstructive pulmonary disease. N Engl J Med. 2004;350(26):2645–2653. | ||
Pérez-Padilla R, Regalado J, Vedal S, et al. Exposure to biomass smoke and chronic airway disease in Mexican women. A case-control study. Am J Respir Crit Care Med. 1996;154(3 Pt 1):701–706. | ||
National Institute of Environmental Health Sciences (NIEHS). SNPs Program. Washington, DC; 2015. [homepage on the Internet]. Available from: http://egp.gs.washington.edu/. Accessed July 28, 2017. | ||
International HapMap 3 Consortium, Altshuler DM, Gibbs RA, Peltonen L, et al. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467(7311):52–58. | ||
Kukkonen MK, Tiili E, Vehmas T, Oksa P, Piirilä P, Hirvonen A. Association of genes of protease-antiprotease balance pathway to lung function and emphysema subtypes. BMC Pulm Med. 2013;13(1):36. | ||
Morales-García G, Falfán-Valencia R, García-Ramírez RA, et al. Pandemic influenza A/H1N1 virus infection and TNF, LTA, IL1B, IL6, IL8, and CCL polymorphisms in Mexican population: a case-control study. BMC Infect Dis. 2012;12(1):299. | ||
Wilson AG, di Giovine FS, Blakemore AI, Duff GW. Single base polymorphism in the human tumour necrosis factor alpha (TNF alpha) gene detectable by NcoI restriction of PCR product. Hum Mol Genet. 1992;1(5):353. | ||
D’Alfonso S, Richiardi PM. A polymorphic variation in a putative regulation box of the TNFA promoter region. Immunogenetics. 1994;39(2):150–154. | ||
Sun R, Huang Y, Zhang H, Liu R. MMP-2, TNF-α and NLRP1 polymorphisms in Chinese patients with ankylosing spondylitis and rheumatoid arthritis. Mol Biol Rep. 2013;40(11):6303–6308. | ||
Lee YH, Bae S-C, Choi SJ, Ji JD, Song GG. Genome-wide pathway analysis of genome-wide association studies on systemic lupus erythematosus and rheumatoid arthritis. Mol Biol Rep. 2012;39(12):10627–10635. | ||
Toonen EJM, Barrera P, Fransen J, et al. Meta-analysis identified the TNFA −308G > A promoter polymorphism as a risk factor for disease severity in patients with rheumatoid arthritis. Arthritis Res Ther. 2012;14(6):R264. | ||
Özdoğan N, Tutar N, Demir R, Saatçi Ç, Kanbay A, Büyükoğlan H. Is TNF-α gene polymorphism related to pulmonary functions and prognosis as determined by FEV1, BMI, COPD exacerbation and hospitalization in patients with smoking-related COPD in a Turkish population? Rev Port Pneumol. 2014;20(6):305–310. | ||
Huang SL, Su CH, Chang SC. Tumor necrosis factor-alpha gene polymorphism in chronic bronchitis. Am J Respir Crit Care Med. 1997;156(5):1436–1439. | ||
Zhang S, Wang C, Xi B, Li X. Association between the tumour necrosis factor-α-308G/A polymorphism and chronic obstructive pulmonary disease: an update. Respirology. 2011;16(1):107–115. | ||
Reséndiz-Hernández JM, Sansores R, Hernández-Zenteno R de J, et al. Identification of genetic variants in the TNF promoter associated with COPD secondary to tobacco smoking and its severity. Int J Chron Obstruct Pulmon Dis. 2015;10(1):1241–1251. | ||
Tomasdottir H, Hjartarson H, Ricksten A, Wasslavik C, Bengtsson A, Ricksten SE. Tumor necrosis factor gene polymorphism is associated with enhanced systemic inflammatory response and increased cardiopulmonary morbidity after cardiac surgery. Anesth Analg. 2003;97(4):944–949, table of contents. | ||
Danilko KV, Korytina GF, Akhmidishina LZ, Ianbaeva DG, Zagidullin SZ, Victorova TV. [Association of cytokines genes (ILL, IL1RN, TNF, LTA, IL6, IL8, IL0) polymorphic markers with chronic obstructive pulmonary disease]. Mol Biol (Mosk). 2007;41(1):26–36. | ||
Meenagh A, Williams F, Ross OA, et al. Frequency of cytokine polymorphisms in populations from western Europe, Africa, Asia, the Middle East and South America. Hum Immunol. 2002;63(11):1055–1061. | ||
Castro-Martínez XH, Leal-Cortés C, Flores-Martínez SE, et al. Tumor necrosis factor haplotype diversity in Mestizo and native populations of Mexico. Tissue Antigens. 2014;83(4):247–259. | ||
Schols AM, Slangen J, Volovics L, Wouters EF. Weight loss is a reversible factor in the prognosis of chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 1998;157(6 Pt 1):1791–1797. | ||
Celli BR, Cote CG, Marin JM, et al. The body-mass index, airflow obstruction, dyspnea, and exercise capacity index in chronic obstructive pulmonary disease. N Engl J Med. 2004;350(10):1005–1012. | ||
Chen YC, Liu SF, Chin CH, et al. Association of tumor necrosis factor-alpha-863C/A gene polymorphism with chronic obstructive pulmonary disease. Lung. 2010;188(4):339–347. | ||
Eagan TML, Gabazza EC, D’Alessandro-Gabazza C, et al. TNF-α is associated with loss of lean body mass only in already cachectic COPD patients. Respir Res. 2012;13(1):48. | ||
Kurmi OP, Dunster C, Ayres JG, Kelly FJ. Oxidative potential of smoke from burning wood and mixed biomass fuels. Free Radic Res. 2013;47(10):829–835. | ||
Mukhopadhyay S, Hoidal JR, Mukherjee TK. Role of TNFalpha in pulmonary pathophysiology. Respir Res. 2006;7(1):125. | ||
Sapey E, Ahmad A, Bayley D, et al. Imbalances between interleukin-1 and tumor necrosis factor agonists and antagonists in stable COPD. J Clin Immunol. 2009;29(4):508–516. | ||
Aaron SD, Angel JB, Lunau M, et al. Granulocyte inflammatory markers and airway infection during acute exacerbation of chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2001;163(2):349–355. | ||
Görek Dilektaşli A, Ulubay G, Bayraktar N, Eminsoy I, Oner Eyüboğlu F. The effects of cachexia and related components on pulmonary functions in patients with COPD. Tuberk Toraks. 2009;57(3):298–305. | ||
Nguyen LT, Bedu M, Caillaud D, et al. Increased resting energy expenditure is related to plasma TNF-alpha concentration in stable COPD patients. Clin Nutr. 1999;18(5):269–274. | ||
Falfán-Valencia R, Reséndiz-Hernández J, Sansores R, Ramírez-Venegas A. TNF promoter polymorphisms are associated with susceptibility and increased severity in secondary to tobacco and biomass smoke COPD in Mexican population. In: 3.1 Molecular Pathology and Functional Genomics. European Respiratory Society. 2015;46:PA4887. |
Supplementary material
  | Figure S1 De Finetti diagram for all markers ternary plot shows rs1800629 in the smokers without COPD group. (A) Marker in Hardy–Weinberg disequilibrium (p=0.018). |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.