Back to Journals » Infection and Drug Resistance » Volume 15
Three Novel Sequence Types Carbapenem-Resistant Klebsiella pneumoniae Strains ST5365, ST5587, ST5647 Isolated from Two Tertiary Teaching General Hospitals in Shanxi Province, in North China: Molecular Characteristics, Resistance and Virulence Factors
Authors Liu Y, Bai J, Kang J, Song Y, Yin D, Wang J, Li H, Duan J
Received 21 March 2022
Accepted for publication 12 May 2022
Published 18 May 2022 Volume 2022:15 Pages 2551—2563
DOI https://doi.org/10.2147/IDR.S366480
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Yujie Liu,1,* Jing Bai,1,* Jianbang Kang,2 Yan Song,2 Donghong Yin,2 Jing Wang,2 Hao Li,3 Jinju Duan2
1Department of Pharmacy, School of Pharmacy, Shanxi Medical University, Taiyuan, Shanxi, People’s Republic of China; 2Department of Pharmacy, Second Hospital of Shanxi Medical University, Taiyuan, Shanxi, People’s Republic of China; 3Department of Clinical Laboratory, First Hospital of Shanxi Medical University, Taiyuan, Shanxi, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Hao Li, Department of Clinical Laboratory, First Hospital of Shanxi Medical University, No. 85, Jiefang South Road, Taiyuan, Shanxi, People’s Republic of China, Tel +86 15340705830, Email [email protected] Jinju Duan, Department of Pharmacy, Second Hospital of Shanxi Medical University, No. 382, Wuyi Road, Xinghualing District, Taiyuan, Shanxi, People’s Republic of China, Tel +86 351 3365713, Email [email protected]
Background: Carbapenem-resistant Klebsiella pneumoniae (CRKP) represents a significant threat to public health and has already drawn worldwide attention. Hence, we aim to comprehensively analyze the case condition, as well as molecular epidemiology, resistance and virulence of three CRKP isolates with new sequence types (STs).
Methods: Three CRKP were collected from November 2019 to April 2021. The three patients’ clinical characteristics were analyzed through His system. In order to screen phenotype of metallo-carbapenemase, the modified Carbapenem Inactivation Method (mCIM) and EDTA-modified Carbapenem Inactivation Method (eCIM) were conducted. Three isolates were subjected to antimicrobial susceptibility testing (AST) using the agar dilution method or minimal broth dilution method. The string test, the sedimentation assay, biofilm formation and the serum resistance assay were performed as phenotypic experiments to assist in evaluating virulence. The presence of resistance and virulence genes were detected by Whole-Genome Sequencing (WGS). Serotypes and new STs were compared and determined by multi-locus sequence typing (MLST).
Results: Overall, all Klebsiella pneumoniae isolates were multi-resistant, but sensitive to tigecycline and colistin. Among them, all formed biofilms, strain 1 and strain 2 were classified as moderate-producers, while strain 3 as weak-producer. The results of the serum resistance assay indicated that only strain 2 was resistant. From WGS analysis, it showed that all isolates co-harbored multiple resistance genes, such as carbapenemase genes, sulfonamides, fluoroquinolones, aminoglycosides, and tetracyclines. Meanwhile, several virulence genes were also contained, including siderophores, fimbriae, capsule and lipopolysaccharides-associated genes. The serotypes of strain 1 and strain 2 manifested K35 and KL47, respectively.
Conclusion: Three novel ST5365, ST5587, ST5647 were first discovered in North China. Our study suggested that we should pay more attention to their resistance. And the results will help treat CRKP infections caused by these novel STs.
Keywords: carbapenem-resistant Klebsiella pneumoniae, CRKP, molecular characteristics, resistance, virulence factors, new sequence types
Introduction
Klebsiella pneumoniae (K. pneumoniae), belonging to the gram-negative bacillus, is an opportunistic pathogen associated with a wide range of infections, such as respiratory tract infections, bloodstream infections and urinary tract infections, which are relatively difficult to treat.1
Carbapenems are commonly used to treat severe infections in clinical setting, as well as the last resort drugs for the treatment of multi-drug resistant (MDR) bacterial infections.2 However, in recent years, the use of broad spectrum β-lactam antibiotics, especially carbapenems in clinical, are widespread. Based on statistics of Center for Antibacterial Surveillance of China since January 2021, the intensity of use of cephalosporin enzyme-containing inhibitors has risen from 4.3% in 2012 to around 6% in 2021, and the intensity of use of carbapenems tends to increase every year as well. Additionally, according to the China Antimicrobial Surveillance Network (CHINET), K. pneumoniae resistance rate to carbapenems has risen rapidly from 2.9% in 2005 to more than 25% in 2018,3 which shows the carbapenem-resistant K. pneumoniae (CRKP) strains have been dispersed in China at a boosting rate and distributed widely. That being so, CRKP represents a significant threat to public health, which comes to our urgent attention.
Sequence types (STs) of K. pneumoniae are defined as the alleles of each strain numbered in a specified order to form their gene profiles. Multi-locus sequence typing (MLST) is a common method and widely used for identifying STs of bacteria, as well as determining the phylogenetic relationship between different STs, especially like those which are correlated to “high- risk” clone lineages, contributing to better prevention in clinic. It is now well demonstrated that STs are related to drug resistance of strains.4 As we all know, the resistance mechanism of CRKP is complex and generally may be caused by one or more factors, especially the production of carbapenemase, like ambler molecular class A (KPC), class B (VIM, IMP, NDM), and class D (OXA-48-like) types.5,6 Notably, among them, blaKPC-2 or blaNDM-1 are the predominant carbapenemase genes, which lead to outbreaks in China.7 More worrisome, aside from resistance genes, detecting a battery of virulence factors is of equal importance and has arisen considerable attention in the scientific domains. With respect to virulence, fimbriae (fimD, fimH, mrkC, mrkD), capsule-associated genes (ycfM, wabG, uge, rmpA), siderophores (entB, iucA, ybtS, fyuA, iroN), lipopolysaccharide (wbbM and wzm) are considered as four main factors,8,9 increasing the burden of the clinical treatment.
To our knowledge, the global dissemination of CRKP is largely mediated by the clonal complex CC258, which comprises ST11, ST258, and a number of closely related sequence types, like another five STs (ST270, ST340, ST379, ST407 and ST418).10,11 Apart from this, CC23 is another dominant lineage that includes many STs: like ST23, 26, 57 and 1633, of which ST23 has been proved to be associated with hypervirulence of isolates,12 as well as CC15 (ST14, ST15),13 CC147 (ST147)14 are popular in small areas. Available literature has already illustrated that in Europe, the majority of CRKP isolates typically belong to four clonal lineages: ST11, 15, 101, 258/512, and their derivatives.15 Furthermore, ST258 and its derivative ST512 are primary clones of CRKP in the Americas, while in Asia (especially in China), ST11 is particularly distributed, accounting for up to 60% of CRKP.16 Likewise, ST23, ST147, ST307 which are thought to be related to “high-risk” clones appear successively as well and have spread in different regions of China, like Beijing, Shanghai, and Zhejiang.17–19 Thus, it can be seen that the dissemination of CRKP is mostly clonal, but the STs exhibit high diversity in different regions due to the geographically specific.20 Hitherto, more than 5000 STs have been discovered, and the rate of discovery of new STs increases gradually every year. For example, in 2021, novel ST4496-KL47 CRKP isolates were discovered in Zhao et al study, which first emerged in Southeast China and were closely related to ST11-KL47 with weakened virulence.21 In another previous study, we accidentally discovered that novel ST4564 was differed from other common STs, which emerged in North China for the first time as well. Through this study, we could see that this novel ST was not related to the high- risk clones, hinting us to concentrate more on its resistance.22 Thus, it can be seen that different STs show diversity in resistance and virulence. Under the circumstance, we began to monitor and analyze the novel ST of the isolates collected. Thus, regardless of the novel STs of the CRKP isolates, comprehensive understanding via molecular epidemiology, resistance and virulence are of necessity.
A longitudinal large-scale study by Wang et al demonstrated that ST11-KPC-2 K. pneumoniae being the most common type of CRKP in China so far, especially in Northern, Eastern, Southern, Central, Northeastern and Southwestern China.23 Searching the existing literature in North China, studies on CRKP are relatively sporadical compared to other regions of China. What is more, the circumstance of discovering new sequences is even rarer. Hence, it is worth noting that three novel STs (ST5365, ST5587, ST5647) were identified in our study. Combining the case and outcome of these three patients who isolated the three CRKP strains and by whole genome sequencing (WGS) to analyze their molecular epidemiology, resistance and virulence factors will provide more information in clinical setting for further monitoring and controlling the spread of CRKP in North China.
Materials and Methods
Collection of Bacterial Strains and Clinical Information
Three CRKP isolates were collected from two tertiary teaching general hospitals in Shanxi Province in North China, from November 2019 to April 2021. Among these, two of them were derived from The First Hospital of Shanxi Medical University, while the remaining one originated from The Second Hospital of Shanxi Medical University, and all the K. pneumoniae isolates were confirmed by matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF-MS).
All information came from the hospital’s electronic medical record database, exempting patients from informed consent. The following data were obtained via the laboratory information system: demographic characteristics (patient’s name, gender and age), bacteria species, department, complications, clinical diagnosis, and length of hospital stays.
Antimicrobial Susceptibility Testing (AST)
All bacterial isolates were subjected to antibiotic sensitivity tests (AST) using the agar dilution method or minimal broth dilution method as recommended by the Clinical and Laboratory Standards Institute 2020 (CLSI 2020).9 The antimicrobial agents tested include cefoperazone/sulbactam (CSL), imipenem (IPM), meropenem (MEM), levofloxacin (LEV), aztreonam (ATM), trimethoprim/sulfamethoxazole (SXT), amikacin (AMK), tigecycline (TGC) and colistin (COL). In our study, the first five antibiotics used the agar dilution method, while the latter four ones used the minimal broth dilution method in accordance with the CLSI 2020. The AST results were interpreted by measuring the minimum inhibitory concentrations (MICs) which were determined to be the lowest concentration of antibiotics at which the strains were incubated at 37°C for 24 h with no visible growth.24 All results were interpreted by the CLSI 2020 (CSL >64 μg/mL, IPM ≥8 μg/mL, MEM≥ 8 μg/mL, LEV ≥2 μg/mL, ATM ≥16 μg/mL, SXT ≥4/76 μg/mL, AMK ≥64 μg/mL, COL ≥4 μg/mL) except the tigecycline following the FDA guidelines (MIC ≤2 μg/mL).
Escherichia coli ATCC 25922 was used as a quality control strain.
Phenotypic Detection of Carbapenemase and Hypermucoviscousity
Recommended by the CLSI2020, in this study, phenotypic screening for preliminarily determining whether the strain produced metallo-carbapenemase was performed in accordance with the modified Carbapenem Inactivation Method (mCIM) and EDTA-Modified Carbapenem Inactivation Method (eCIM).25 To be brief, a single colony of isolates was inoculated into a tube containing 2 mL trypticase soy broth (TSB) and a tube containing 2 mL TSB with 5mM EDTA, respectively, and then stir the colony immediately to dissolve enough to be invisible. A disk containing 10 μg meropenem was placed in each tube and incubated at 37°C for 4 h. After that, these disks were taken out and placed on MH agar plates that were inoculated with a lawn of the meropenem-susceptible Escherichia coli ATCC25922 (0.5 McFarland standard). The results were interpreted according to CLSI 2020.26
Furthermore, two different methods were used to test for initially screened hypermucoviscous phenotype of the three isolates: i) The string test was widely used and performed as previously described. Briefly, isolates were streaked on blood agar plates and cultured overnight. The single colony was stretched with an inoculation loop to measure the visible string, and a string longer than 5 mm was considered to the results positive.27 ii) The sedimentation assay was performed as described by Wakler et al,28 based on the principle that hypermucoviscous strains cannot sediment sufficiently, leading to the supernatant turbid. In brief, cultures of K. pneumoniae were grown overnight in 5 mL LB medium at 37°C. Then, these cultures were sedimented at 2500 g for 5 min. The optical density at 600 nm (OD600) of the top 500 μL of supernatant was determined by a microplate reader. The results were expressed as a ratio of the OD600 of the supernatant and the OD600 of the starting culture.29
The Biofilm Formation Assay
The biofilm was detected by crystal violet staining as previously described.30 As for the biofilm formation assay, specifically, the bacterial suspension (0.5 McFarland standard) was diluted from the overnight culture, and then took 10 µL and 190 µL LB broth into the 96-well plates, incubating for 24 h. Each well was equipped with three multiple ones. Afterwards, we took out the 96-well plate, sucked off the supernatant and cleaned it with sterilized distilled water for 3 times. Next, 200 µL of 99% methanol was added to each well for 15 minutes. Additionally, the biofilm was stained with 200 μL 1% crystal violet staining solution for 20 about minutes, and each well was washed thrice with the sterilized distilled water. After that, added 200 μL of 95% v/v ethanol for 10 minutes to solubilize the stained biofilms, and then transferred it to a new 96-well plate, and the OD570 were measured with a microplate reader. The results were interpreted based on the previous studies.31
The Serum Resistance Assay
The serum bactericidal assay was performed as described in Podschun’s study with modifications.32 In brief, the bacteria were first streaked on blood agar culture-medium (BAP) plates and incubated overnight to collect a single colony for inoculation in 5 mL brain heart infusion (BHI) broth for 4–6 h at 37°C, 200rpm until the optical density at 630 nm (OD630) was measured to be about 0.6. Then, bacteria were diluted to 106 cells/nil in physiologic saline. After that, the 50 µL of bacterial suspensions were combined with 150 µL of human serum in a 2 mL centrifuge tube, mixed and incubated for 0, 1, 2, and 3 h, respectively, at 37°C, 200rpm. Moreover, 5 µL of bacterial solution was taken at each time point and diluted 100 times with normal saline. With that, 50 µL were taken and inoculated on the MH agar plate for counting. In addition, each strain was tested at least three times, and the results were interpreted as the percentage of inoculum in terms of viable counts were graded from 1 to 6 in accordance with previous studies.
Each isolate was classified as highly sensitive (grade 1 or 2), intermediately sensitive (grade 3 or 4), or resistant (grade 5 or 6).27
Whole-Genome Sequencing (WGS)
Genomic DNA was extracted using the Ezup Column Bacteria Genomic DNA Purification Kit and then conducted quality inspection by Software FastQ (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/), subsequently sequenced on the Illumina HiSeq platform. Then, Software SPAdes (http://cab.spbu.ru/software/spades/) were used to assemble the next-generation sequencing data, and Software Prinses-G (https://updeplasrv1.epfl.ch/prinses/) was used for sequence correction. Using BLAST, the gene sequences respectively with the CARD (https://card.mcmaster.ca/) and VFDB (http://www.mgc.ac.cn/VFs/), comparing the database of bacterial drug resistance genes and virulence gene screening and annotation.
Multi-Locus Sequence Typing (MLST) and Serotypes
Multi-locus sequence typing (MLST) was performed to explore the characteristics of the clinical K. pneumoniae isolates. Briefly, seven K. pneumoniae housekeeping genes (infB, tonB, pgi, gapA, phoE, rpoB and mdh), as well as serotypes determined via wzi locus were amplified and sequenced. Whereafter, the positive products were sequenced by Sangon Biotech, and the results were submitted to K. pneumoniae MLST database ((http://bigsdb.web.pasteur.fr/klebsiella/klebsiella.html).) for comparison. The primers were shown in Supplementary Table S1. In the previous study, some researchers have demonstrated that ST profiles sharing 100% genetic identity in at least 6 of 7 MLST loci were grouped into a clonal complex (CC).22 So that, we used the software goeBURST 1.2.1 version (http://www.phyloviz.net/goeburst/) to identify the molecular epidemiological relationships of the CC.
Results
Clinical Characteristics of the Isolates
Three patients infected with CRKP were finally involved in our research. Clinical data revealed that there were two males of the three patients, who all shared a common clinical diagnosis (lung infection), while one female received a case of severe pneumonia. Additionally, all of them have been administered appropriate antibiotics for treatment during the isolation of strains (Table 1).
 |
Table 1 Clinical Characteristics of the Three Strains |
Among them, one patient whose sputum sample cultured strain 1 was a 53-year-old man hospitalized in the ICU of The Second Hospital of Shanxi Medical University, and totally in hospital for 164 days. Initially, he was admitted to hospital and diagnosed with cervical spinal cord injury with lung infection and electrolyte disturbances on 3 November, 2020. After 15 days, CRKP was isolated from his sputum culture. Subsequently, this was followed by the discovery of Acinetobacter baumannii and Pseudomonas aeruginosa (P. aeruginosa) in sputum specimens, P. aeruginosa in urine specimens, and Enterococcus faecium in catheter urine. After anti-infection treatment for a period of time, he finally got better before discharging from hospital. Additionally, the other two strains from sputum samples were isolated from a 57-year-old man (strain 2) and an 81-year-old woman (strain 3), receiving treatments in the plastic burn surgery and ICU, respectively, at First Hospital of Shanxi Medical University. The former patient was admitted to hospital and diagnosed with a lung infection along with hypokalemia and hypoproteinemia on 21 November, 2019. CRKP was isolated from his sputum culture after 7 days. Soon afterwards, Acinetobacter baumannii was also found in sputum samples, as well as P. aeruginosa and Staphylococcus haemolyticus in urine samples. At last, he also got better and was discharged from the hospital after 56 days. The latter patient was admitted to hospital on 26 March, 2020 and diagnosed with severe pneumoniae and electrolyte disturbances accompanied by a history of diabetes. CRKP was isolated from sputum culture in 15 days. After that, Staphylococcus aureus and Haemophilus influenzae were detected in sputum samples, and Escherichia coli was detected in urine sample. Ultimately, after 21 days of hospitalization, she was discharged in a slightly better condition but remained critical.
The AST Results and Phenotypic Screening
In this study, the susceptibility tests of eight kinds of antibiotics were carried out for the three isolates collected, including β- lactams and β- lactamase inhibitor combination, carbapenems, quinolones, monobactams, sulfonamides, aminoglycosides, glycylcycline and polypeptides. The results indicated that the three CRKP strains had varying degrees of resistance to different classes of antibiotics, nonetheless, they were all sensitive to tigecycline and colistin. Specifically, all the three isolates showed resistance to the first five antibiotics. Of note, strain 1 was sensitive to amikacin, and strain 2 was sensitive to trimethoprim/sulfamethoxazole, while strain 3 showed resistance to both two antibiotics (Table 2).
 |
Table 2 The Antibiotic Sensitivity Test Results of the Three Strains |
Additional phenotypic experiment was done to confirm whether to produce metal β-lactamase. The results of mCIM/eCIM method were that strain 1 showed positive, while the other two performed negative.
Phenotypic Assessment of Virulence
In order to preliminarily evaluate the virulence of these three CRKP isolates, we comprehensively carried out the string test, the sedimentation assay, biofilm formation and the serum resistance assay. Overall, we performed the string test and the sedimentation assay as initial assessments of hypermucoviscousity. The string test turned out that the three CRKP isolates all presented negative. And in the meantime, the results of the sedimentation assay showed the same as well.
Simultaneously, the biofilm formation experiment and the serum resistance assay were also performed to help assess virulence. The upshots were that all of the three strains formed biofilms. Moreover, strain 1 and strain 2 were classified as moderate-producers, and strain 3 as weak-producer (Figure 1). On the other hand, the serum resistance assay indicated that the levels of drug resistance in serum are quite different. Strain 1 (grade 2) and strain 3 (grade 3) both showed sensitivity, while strain 2 showed resistance (grade 6) (Figure 2).
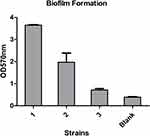 |
Figure 1 Biofilm formation of these three strains. |
 |
Figure 2 The results of serum resistance assay of these three strains. |
Resistance and Virulence Genes Detection by WGS
To confirm the resistance pattern, we identified the related resistance genes of the three strains. From the WGS analysis, we found that the strain 1 co-harbored blaNDM-1 and blaKPC-2, as well as carried several other resistance genes, like β- lactams genes (blaCTX-M, blaTEM, blaSHV, blaSME, blaLEN, blaOXY and blaCARB), sulfonamides (sul1, sul2, sul3), fluoroquinolones (qnrS, qnrB, qnrA), aminoglycosides (APH (6)- Id, APH (3”)- Ia, APH (3”)- Ib, APH (3”)- Ic, AAC (3)- IIa, aadA16, aadA6), tetracyclines (tetR, tetA, tetB, tet31). Strain 2 co-existed blaKPC-2 and blaIMI, likewise, it carried β- lactams genes (blaSHV, blaTEM, blaCTX-M, blaLEN, blaOXY, blaSME, blaCARB and blaLRA), fluoroquinolones (norB), aminoglycosides (acrD), tetracyclines (tetD). Strain 3 mainly carried class D carbapenemase genes, like blaOXA-23-like, blaOXA-24-like, blaOXA-51-like, blaOXA-58-like, and AmpC enzyme blaCMY was detected as well. In addition, resistance genes like sulfonamides (sul1, sul2, sul3), aminoglycosides (aadA, APH (6)- Id, APH (3’)- Ia, APH (3’)- Ib, AAC (6’)- Ib), tetracyclines (tetR, tetA, tetB, tet31) were also found (Figure 3).
The results of virulence genes presence are shown in Figure 3. Several key virulence genes were screened, including siderophores-associated genes (yersiniabactin (encoding ybt, irp and fyu genes), aerobactin (encoding iut and iuc genes), enterobactin (encoding ent and fep genes) and salmochelin (encoding iro gene)), fimbriae-associated genes (fim, pil, chp and hif genes), capsule-associated genes (cps, ctr, wcb, wzt and ure genes), lipopolysaccharide-associated genes (rfb, waa, gul and gal genes). All the three isolates carried ybt, irp, fyuA, iut, ent, fep, iroN, pil, chp, cps, ctrD, wcb, wzt, ureA, waa, gluE and galE genes. Besides, strain 1 carried rfb gene, strain 2 and strain 3 harbored fimB genes, and strain 3 harbored iucA gene. And that, the serotypes were determined via the wzi locus, and the results showed that the serotypes of strain 1 and strain 2 manifested K35 and KL47, respectively.
Detailed results of resistance genes and virulence genes by WGS were shown in Supplementary Tables S2 and S3.
The Molecular Typing Results
With that, multi-locus sequence typing (MLST) was used to analyze the molecular epidemiology of the strains. Amplification by PCR, we finally discovered three novel ST types, ST5365, ST5587, ST5647. Through the software goeBURST analysis of MLST data, the results indicated that the novel ST5365 (gap-3, infB-3, mdh-1, pgi-1, phoE-1, rpoB-71, tonB-4) had similar structure to ST11 and belonged to CC258, while the novel ST5647 (gap-38, infB-19, mdh-39, pgi-20, phoE-417, rpoB-123, tonB-130), ST5587 (gap-2, infB-20, mdh-416, pgi-1, phoE-9, rpoB-146, tonB-93) both had rather distant relationship with clone lineage CC258 (Figure 4).
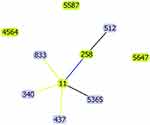 |
Figure 4 goeBurst- based genetic relationship of ST5365, ST5587, ST5647 CRKP and some common clone lineage CC258. |
Discussion
In recent years, K. pneumoniae is one of the multi-drug resistant microorganisms posing an urgent threat to public health. More importantly, the increase in antimicrobial resistance and the emergence of hypervirulent phenotype of this bacteria are of great concern to the scientific community. Whereas, the most optimal therapeutic strategy is hard to set out, especially with the novel sequences of the CRKP isolates appearing all the time.
In our study, three novel ST CRKP strains were isolated from two general tertiary hospitals in Shanxi Province, in North China, from November 2019 to April 2021. Of the three patients, two of them were from ICU, and the remaining one was from plastic burn surgery, all of whom were in serious condition, even with a variety of diseases, such as diabetes, electrolyte disturbance and cholecystitis. The patient who isolated strain 1 was in hospital for 164 days, and the patient who isolated strain 2 was in for 56 days. Thus, it can be seen that the hospitalization time of the two patients was much longer than the average length of stay (LOS) of the Shanxi Province in 2019 (10.3 days) and 2020 (10.15 days). But anyway, both patients had satisfying outcomes. As for the third patient who was already 81 years old, in spite of the shorter hospitalization, she was discharged with a still serious condition, making all of us depressed. Of note, CRKP accounts for over 60% of clinical CRE infections in China, resulting in mortality rate nearly 40% in nosocomial settings.20,33 Moreover, in a multicenter study in China, Zhen et al highlighted that CRKP were related to significantly increased economic costs, excess LOS, and in-hospital mortality rate.34 That being so, CRKP infection can not only prolong the LOS of patients but also increase their financial burden.
Focusing on the resistance mechanism of CRKP isolates, the WGS results revealed that all the three CRKP isolates carried carbapenemase genes. What is noteworthy is that strain 1 co-existed blaNDM-1 and blaKPC-2, and strain 2 co-harbored blaKPC-2 and blaIMI, while strain 3 contained a variety of blaOXA enzymes. Hitherto, although KPC-producing and NDM-producing CRKP isolates have been widely distributed throughout the world, co-occurrence of blaKPC-2 and blaNDM-1 has rarely been reported yet. The conjugation assay in Gao et al's study reflected that blaNDM-1 plasmid is more prone to transfer than blaKPC-2 plasmid; therefore, it is possible that the emergence of KPC-2-NDM-1-CRKP was owing to blaNDM-1 plasmid transfered into blaKPC-2 plasmid, which can increase resistance of this type of bacteria. Gao et al also raised an alarm on the increased prevalence of co-existing blaKPC-2 and blaNDM-1 CRKP isolates with characteristics of high incidence, high stability and non-inferior fitness,35 which may increase the difficulty of treating and prolonging infections, thus leading to longer LOS, like the first patient in our study. In the meantime, this patient was also infected by other bacteria like Acinetobacter baumannii, P. aeruginosa and Enterococcus faecium, which visibly caused the therapy much harder as well. According to studies described previously, blaIMI-2 was often detected in Enterobacter cloacae, and regarded as the first inducible carbapenemase gene encoded by plasmids, conferring high resistance to imipenem and meropenem.36 Besides, blaIMI-2 is located in a plasmid that can transfer itself, whose upstream is identified as a LysR-type regulator gene that explains the inducibility of blaIMI-2 expression. Of note, in past studies, there were few reports about co-harbored blaKPC-2 and blaIMI, which needs our attention as well. To date, Evans et al summarized that the OXA- lactamases are originally rare and frequently plasmid mediated. K. pneumoniae strains usually exists carbapenemase blaOXA-48, however, in our study, no such enzyme was found. Instead, we detected a large number of other OXA enzymes, mainly dividing into four subgroups (blaOXA-23-like, blaOXA-24-like, blaOXA-51-like, blaOXA-58-like), which are scarcely found in K. pneumoniae but more commonly in Acinetobacter baumannii.37 Combined with the case, the third patient in our study came from the ICU of the First Hospital of Shanxi Medical University, where Acinetobacter baumannii were the main isolated strains in it. According to the data of this hospital, the isolation rate of carbapenem-resistant Acinetobacter baumannii in ICU is up to 82.6% in 2020. Based on the carbapenem genes detected in these three strains, due to the horizontal transfer of mobile genetic elements, we have sufficient reasons to suspect that there may have been emerged horizontal transfer of genes between the three CRKP isolates and other strains. Additionally, multiple other resistance genes, such as β-lactams genes, sulfonamides, fluoroquinolones, aminoglycosides and tetracyclines, were also detected, which are often in transmissible plasmids and thus spread readily among bacteria. In Zhu et al study, in a Chinese hospital from 2017 to 2019, the first CRKP outbreak which presumably put down to cross-transmitted among patients was successfully controlled via the multimodal and multidisciplinary infection control and effective surveillance.38 Thus, only strict prevention and effective control can be taken to commendably interdict the occurrence and rapid spread of nosocomial cross-infections.
Although the results of the string test and the sedimentation assay in our study both showed negative, the three CRKP strains still carried abundant virulent genes. Regarding virulence genes, they play an important role in hypervirulence of the strains mainly via encoding siderophores, fimbriae, capsular polysaccharides (CPS) and lipopolysaccharides (LPS). To our knowledge, iron obtaining is essential for bacterial replication and growth, and there are four major siderophores of K. pneumoniae, which are enterobactin (encoded by ent genes), yersiniabactin (encoded by ybt genes, regulated by irp genes, and the receptor is encoded by the fyuA gene), salmochelin (encoded by iro genes) and aerobactin (encoded by iuc genes). Enterobactin is produced by all K. pneumoniae, as reported by previous studies. Remarkably, lipocalin-2 (Lcn2) is a multifunctional protein that plays a role in its affinity for enterobacterin and gives rise to an inflammatory response. On the contrary, yersiniabactin avoids binding to Lcn2, thereby promoting bacterial growth.9 Based on the results of WGS analysis, all the three isolates harbored ybt, irp, fyuA, iut, ent, fep, and iroN genes; furthermore, strain 3 also detected iucA gene, makingstrain 3 more virulent.
The fim and mrk are two major virulence gene clusters that, respectively, encode bacterial type I and type III fimbriae to mediate bacterial adhesion, and further leading to bacterial colonization and pathogenicity. In our study, fimbriae (pilT, chpD), CPS (cps, ctrD, wcb, wzt, ureA) associated genes were detected in the three strains, furthermore,strain 3 also detected fim, hif genes. Studies have suggested that biofilm formation of K. pneumoniae is mainly influenced by fimbriae, CPS, efflux pump and other factors. The biofilm formation results showed that strain 1 and strain 2 were classified as moderate-producers, while strain 3 as weak-producer. Although all three isolates carried genes associated with factors mentioned above, strain 1 and strain 2 still detected capsule serotypes, which have an inseparable relationship to CPS, and this may explain why all three strains form biofilms, but strain 1 and strain 2 are more capable of biofilm formation than strain 3. Furthermore, compared to planktonic bacteria, biofilms exhibit significantly higher resistance to phagocytosis and antimicrobial resistance,9 which should also raise our grave concerns.
In addition, the capsule-associated genes wabG (encoding capsule), uge (encoding capsule lipoprotein), ycfM (promoting external membrane protein), ureA (urease) are highly involved in virulence on account of their ability to evade phagocytosis and complement-mediated killing and further inhibit complement activation of the host.39,40 CPS-associated genes (cps, ctrD, wcb, wzt, ureA) were detected in three strains. Moreover, the results of the serum resistance assay in our study revealed thatstrain 2 showed resistance, while strains 1 and 3 showed sensitivity. Su et al mentioned that K. pneumoniae can produce CPS that may make this species of bacteria resistant to serum, thus promoting their survival.41 However, Tomás et al proved that the CPS did not seem to play any important role in serum for survival of the bacterium, which may explain our findings why all three isolates carried virulence genes associated with CPS but varied in serum resistance.42 Thus, further investigations are needed to interpret the inconsistencies among these studies.
LPS, also known as endotoxin, is associated with O-antigen to protect the pathogens against complement-mediated killing. Specifically, O-antigen binds to the C3b, a complement component that is both an opsonin and part of the pore-forming process, allowing it to form pores that prevent bacterial cell membranes from perforating.43 There are four distinct gene clusters, lpx, waa, rfb and lpt, involved in LPS biosynthesis and output of K. pneumoniae. The WGS results showed that LPS-associated genes (waa, gluE, galE) were detected in all three isolates, and the rfb gene was extra detected in strain 1. Presently, it is unclear whether LPS produced has a unique role in hypervirulence of K. pneumoniae,29 however, we still need to place our attention on virulence of these three CRKP isolates with new sequence types.
MLST analysis uncovers the molecular epidemiological characteristics of K. pneumoniae, with three novel STs identified in our study: ST5647 (strain 1), ST5365 (strain 2), ST5587 (strain 3). Besides, it turned out that the serotypes of strains 1 and 2 were K35 and KL47, respectively, while the serotype of strain 3 was not detected yet. In line with a retrospective study of ST11-CRKP strains in China, there is no doubt that ST11 is the most predominant ST of high-risk CRKP strain, furthermore, KL47 and KL64 are two most common serotypes of ST11 isolates,44 which is also highly relevant to our findings. Another study by Zhao et al discovered a novel ST4496-KL47, and compared it with ST11-KL64 and ST11-KL47, which are two main the predominant groups of China, further proving that the novel ST4496-KL47 is less virulence than both of them.21 In our study, the ST of strain 2 is ST5365, and it is deemed as a part of the clonal complex CC258 like ST11. Hence, it is reasonable to presume that the novel ST5365-KL47 strain in our study is less virulent as well. With regard to the other two strains of the new STs (ST5587 and ST5647), they have a relatively further distant relationship with ST11, so we would prefer to concentrate more on their resistance. In addition, new ST discovery can not only help us understand the latest epidemic situation of local cloned strains but also help prevent the potential danger caused by such strains over time.
There are also some limitations in our study. Above all, the number of the strains collected in our study is small, even so, we evaluated the resistance and virulence of the three new STs more roundly via various aspects through our studies. Second, these three novel STs of the CRKP strains collected from two hospitals of Shanxi Province, which cannot well represent the situation of the whole North China, but it also points out that the three STs urgently need to enhance clinical awareness.
Conclusion
In summary, we comprehensively analyzed three strains in many aspects through molecular epidemiology, resistance pattern and virulence presence. Deserved to be mentioned, three novel sequence types (ST5365, ST5587, ST5647) were first identified in the course of our study, indicating that the brand-new types have spread in North China. Moreover, three novel STs manifested strong resistance, of which two of isolates co-harbored different carbapenemases (strain 1 co-harbored blaKPC-2 and blaNDM-1, and strain 2 co-harbored blaKPC-2 and blaIMI), while strain 3 carried a large number of other OXA enzymes, like blaOXA-23-like, blaOXA-24-like, blaOXA-51-like, blaOXA-58-like, which was differed from common carbapenemases in K. pneumoniae. For all this, while focusing on the virulence of the three strains with new sequence types, we should also pay more attention to their resistance. It also emphasizes that effective prevention and further control of a wide distribution of these three new sequence types are highly needed.
Ethics Approval and Consent to Participate
This study was reviewed and approved by the research ethics committee of the Second Hospital of Shanxi Medical University (2019 YX-181). The data of patients’ clinical variables were collected from their medical records and did not contain name, address, or other personal information. The patients’ written informed consent was exempt. This study was also in line with the guidelines outlined in the Declaration of Helsinki.
Acknowledgments
The authors thank the second Hospital of Shanxi Medical University for supporting this study.
Funding
This work was supported by the Shanxi Province Natural Science Foundation (grant number 201803D31124). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Martin RM, Bachman MA. Colonization, infection, and the accessory genome of Klebsiella pneumoniae. Front Cell Infect Microbiol. 2018;8:4. doi:10.3389/fcimb.2018.00004
2. Hu Y, Liu C, Shen Z, et al. Prevalence, risk factors and molecular epidemiology of carbapenem-resistant Klebsiella pneumoniae in patients from Zhejiang, China, 2008–2018. Emerg Microbes Infect. 2020;9(1):1771–1779. doi:10.1080/22221751.2020.1799721
3. Hu F, Guo Y, Yang Y, et al. Resistance reported from China antimicrobial surveillance network (CHINET) in 2018. Eur J Clin Microbiol Infect Dis. 2019;38(12):2275–2281. doi:10.1007/s10096-019-03673-1
4. Lin D, Chen J, Yang Y, Cheng J, Sun C. Epidemiological Study of Carbapenem-resistant Klebsiella pneumoniae. Open Med. 2018;13:460–466. doi:10.1515/med-2018-0070
5. Zhao Y, Liao Y, Zhang N, et al. Four types of ST11 novel mutations from increasing Carbapenem-Resistant Klebsiella pneumoniae in Guangdong, 2016–2020. Front Microbiol. 2021;12:702941. doi:10.3389/fmicb.2021.702941
6. Zhang X, Li F, Cui S, et al. Prevalence and distribution characteristics of blaKPC-2 and blaNDM-1 genes in Klebsiella pneumoniae. Infect Drug Resist. 2020;13:2901–2910. doi:10.2147/IDR.S253631
7. Kong Y, Sun Q, Chen H, et al. Transmission dynamics of Carbapenem-Resistant Klebsiella pneumoniae sequence type 11 strains carrying capsular loci KL64 and rmpA/rmpA2 genes. Front Microbiol. 2021;12:736896. doi:10.3389/fmicb.2021.736896
8. Wang G, Zhao G, Chao X, Xie L, Wang H. The characteristic of virulence, biofilm and antibiotic resistance of Klebsiella pneumoniae. Int J Environ Res Public Health. 2020;17(17):6278. doi:10.3390/ijerph17176278
9. Ballen V, Gabasa Y, Ratia C, Ortega R, Tejero M, Soto S. Antibiotic resistance and virulence profiles of Klebsiella pneumoniae strains isolated from different clinical sources. Front Cell Infect Microbiol. 2021;11:738223. doi:10.3389/fcimb.2021.738223
10. Liu L, Feng Y, Long H, McNally A, Zong Z. Sequence Type 273 Carbapenem-Resistant Klebsiella pneumoniae carrying bla NDM-1 and bla IMP-4. Antimicrob Agents Chemother. 2018;62(6). doi:10.1128/AAC.00160-18
11. Qi Y, Wei Z, Ji S, Du X, Shen P, Yu Y. ST11, the dominant clone of KPC-producing Klebsiella pneumoniae in China. J Antimicrob Chemother. 2011;66(2):307–312. doi:10.1093/jac/dkq431
12. Choby JE, Howard Anderson J, Weiss DS. Hypervirulent Klebsiella pneumoniae – clinical and molecular perspectives. J Intern Med. 2020;287(3):283–300. doi:10.1111/joim.13007
13. Gato E, Vazquez-Ucha JC, Rumbo-Feal S, et al. Kpi, a chaperone-usher pili system associated with the worldwide-disseminated high-risk clone Klebsiella pneumoniae ST-15. Proc Natl Acad Sci U S A. 2020;117(29):17249–17259. doi:10.1073/pnas.1921393117
14. Zou H, Shen Y, Li C, Li Q. Two phenotypes of Klebsiella pneumoniae ST147 outbreak from neonatal sepsis with a slight increase in virulence. Infect Drug Resist. 2022;15:1–12. doi:10.2147/IDR.S343292
15. David S, Reuter S, Harris SR, et al. Epidemic of carbapenem-resistant Klebsiella pneumoniae in Europe is driven by nosocomial spread. Nat Microbiol. 2019;4(11):1919–1929. doi:10.1038/s41564-019-0492-8
16. Lan P, Jiang Y, Zhou J, Yu Y. A global perspective on the convergence of hypervirulence and carbapenem resistance in Klebsiella pneumoniae. J Glob Antimicrob Resist. 2021;25:26–34. doi:10.1016/j.jgar.2021.02.020
17. Lu B, Lin C, Liu H, et al. Molecular characteristics of Klebsiella pneumoniae isolates from outpatients in Sentinel Hospitals, Beijing, China, 2010–2019. Front Cell Infect Microbiol. 2020;10:85. doi:10.3389/fcimb.2020.00085
18. Zhou C, Wu Q, He L, et al. Clinical and molecular characteristics of Carbapenem-Resistant Hypervirulent Klebsiella pneumoniae isolates in a Tertiary Hospital in Shanghai, China. Infect Drug Resist. 2021;14:2697–2706. doi:10.2147/IDR.S321704
19. Zhang S, Zhang X, Wu Q, et al. Clinical, microbiological, and molecular epidemiological characteristics of Klebsiella pneumoniae-induced pyogenic liver abscess in southeastern China. Antimicrob Resist Infect Control. 2019;8(1):166. doi:10.1186/s13756-019-0615-2
20. Zhou K, Xiao T, David S, et al. Novel subclone of Carbapenem-Resistant Klebsiella pneumoniae sequence type 11 with enhanced virulence and transmissibility, China. Emerg Infect Dis. 2020;26(2):289–297. doi:10.3201/eid2602.190594
21. Zhao D, Shi Q, Hu D, et al. The emergence of novel sequence type strains reveals an evolutionary process of intraspecies clone shifting in ICU-spreading Carbapenem-Resistant Klebsiella pneumoniae. Front Microbiol. 2021;12:691406. doi:10.3389/fmicb.2021.691406
22. Wang X, Li Q, Kang J, et al. Co-production of NDM-1, CTX-M-9 family and mcr-1 in a Klebsiella pneumoniae ST4564 Strain in China. Infect Drug Resist. 2021;14:449–457. doi:10.2147/IDR.S292820
23. Wang Q, Wang X, Wang J, et al. Phenotypic and genotypic characterization of Carbapenem-Resistant Enterobacteriaceae: data from a Longitudinal Large-scale CRE Study in China (2012–2016). Clin Infect Dis. 2018;67(suppl_2):S196–S205. doi:10.1093/cid/ciy660
24. Chen CM, Wang M, Li XP, et al. Homology analysis between clinically isolated extraintestinal and enteral Klebsiella pneumoniae among neonates. BMC Microbiol. 2021;21(1):25. doi:10.1186/s12866-020-02073-2
25. Jin C, Shi R, Jiang X, Zhou F, Qiang J, An C. Epidemic characteristics of Carbapenem-Resistant Klebsiella pneumoniae in the pediatric intensive care unit of Yanbian University Hospital, China. Infect Drug Resist. 2020;13:1439–1446. doi:10.2147/IDR.S245397
26. Li Z, Ding Z, Yang J, et al. Carbapenem-Resistant Klebsiella pneumoniae in Southwest China: molecular characteristics and risk factors caused by KPC and NDM producers. Infect Drug Resist. 2021;14:3145–3158. doi:10.2147/IDR.S324244
27. Wang TC, Lin JC, Chang JC, et al. Virulence among different types of hypervirulent Klebsiella pneumoniae with multi-locus sequence type (MLST)-11, Serotype K1 or K2 strains. Gut Pathog. 2021;13(1):40. doi:10.1186/s13099-021-00439-z
28. Walker KA, Miller VL. The intersection of capsule gene expression, hypermucoviscosity and hypervirulence in Klebsiella pneumoniae. Curr Opin Microbiol. 2020;54:95–102. doi:10.1016/j.mib.2020.01.006
29. Zhu J, Wang T, Chen L, Du H. Virulence Factors in Hypervirulent Klebsiella pneumoniae. Front Microbiol. 2021;12:642484. doi:10.3389/fmicb.2021.642484
30. Chen L, Yu K, Chen L, et al. Synergistic activity and biofilm formation effect of colistin combined with PFK-158 against colistin-resistant gram-negative bacteria. Infect Drug Resist. 2021;14:2143–2154. doi:10.2147/IDR.S309912
31. Andrade NL, Da CCAC, Cabral AM, et al. Infective endocarditis caused by Enterobacteriaceae: phenotypic and molecular characterization of Escherichia coli and Klebsiella pneumoniae in Rio de Janeiro, Brazil. Braz J Microbiol. 2021;52(4):1887–1896. doi:10.1007/s42770-021-00528-w
32. Podschun R, Sievers D, Fischer A, Ullmann U. Serotypes, hemagglutinins, siderophore synthesis, and serum resistance of Klebsiella isolates causing human urinary tract infections. J Infect Dis. 1993;168(6):1415–1421. doi:10.1093/infdis/168.6.1415
33. Wang M, Earley M, Chen L, et al. Clinical outcomes and bacterial characteristics of carbapenem-resistant Klebsiella pneumoniae complex among patients from different global regions (CRACKLE-2): a prospective, multicentre, cohort study. Lancet Infect Dis. 2022;22(3):401–412. doi:10.1016/S1473-3099(21)00399-6
34. Zhen X, Stålsby LC, Sun X, Gu S, Dong H. Clinical and economic burden of Carbapenem-Resistant infection or colonization caused by Klebsiella pneumoniae, Pseudomonas aeruginosa, Acinetobacter baumannii: a Multicenter Study in China. Antibiotics. 2020;9(8). doi:10.3390/antibiotics9080514
35. Gao H, Liu Y, Wang R, Wang Q, Jin L, Wang H. The transferability and evolution of NDM-1 and KPC-2 co-producing Klebsiella pneumoniae from clinical settings. EBioMedicine. 2020;51:102599. doi:10.1016/j.ebiom.2019.102599
36. Rasmussen BA, Bush K, Keeney D, et al. Characterization of IMI-1 beta-lactamase, a class A carbapenem-hydrolyzing enzyme from Enterobacter cloacae. Antimicrob Agents Chemother. 1996;40(9):2080–2086. doi:10.1128/AAC.40.9.2080
37. Evans BA, Amyes SG. OXA β-lactamases. Clin Microbiol Rev. 2014;27(2):241–263. doi:10.1128/CMR.00117-13
38. Zhu J, Li Q, Li X, et al. Successful control of the first carbapenem-resistant Klebsiella pneumoniae outbreak in a Chinese hospital 2017–2019. Antimicrob Resist Infect Control. 2020;9(1):91. doi:10.1186/s13756-020-00757-y
39. Wu X, Shi Q, Shen S, Huang C, Wu H. Clinical and bacterial characteristics of Klebsiella pneumoniae affecting 30-day mortality in patients with bloodstream infection. Front Cell Infect Microbiol. 2021;11:688989. doi:10.3389/fcimb.2021.688989
40. Fang CT, Chuang YP, Shun CT, Chang SC, Wang JT. A novel virulence gene in Klebsiella pneumoniae strains causing primary liver abscess and septic metastatic complications. J Exp Med. 2004;199(5):697–705. doi:10.1084/jem.20030857
41. Su S, Zhang J, Zhao Y, et al. Outbreak of KPC-2 Carbapenem-Resistant Klebsiella pneumoniae ST76 and Carbapenem-Resistant K2 Hypervirulent Klebsiella pneumoniae ST375 strains in Northeast China: molecular and virulent characteristics. BMC Infect Dis. 2020;20(1):472. doi:10.1186/s12879-020-05143-y
42. Tomas JM, Benedi VJ, Ciurana B, Jofre J. Role of capsule and O antigen in resistance of Klebsiella pneumoniae to serum bactericidal activity. Infect Immun. 1986;54(1):85–89. doi:10.1128/iai.54.1.85-89.1986
43. Paczosa MK, Mecsas J. Klebsiella pneumoniae: going on the offense with a strong defense. Microbiol Mol Biol Rev. 2016;80(3):629–661. doi:10.1128/MMBR.00078-15
44. Liao W, Liu Y, Zhang W. Virulence evolution, molecular mechanisms of resistance and prevalence of ST11 carbapenem-resistant Klebsiella pneumoniae in China: a review over the last 10 years. J Glob Antimicrob Resist. 2020;23:174–180. doi:10.1016/j.jgar.2020.09.004
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.

