Back to Journals » Drug Design, Development and Therapy » Volume 17
Therapeutic Mechanism of Baicalin in Experimental Colitis Analyzed Using Network Pharmacology and Metabolomics
Authors Wu Q , Wu X, Wang M , Liu K, Li Y, Ruan X, Qian L, Meng L, Sun Z , Zhu L, Wu J , Mu G
Received 18 December 2022
Accepted for publication 13 March 2023
Published 31 March 2023 Volume 2023:17 Pages 1007—1024
DOI https://doi.org/10.2147/DDDT.S399290
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Tin Wui Wong
Qi Wu,1,* Xingxing Wu,1,* Mao Wang,2,* Kexin Liu,1 Yuge Li,1 Xiaoyu Ruan,1 Lin Qian,1 Lingchang Meng,3 Zhiting Sun,3 Lei Zhu,1 Jing Wu,3 Genglin Mu3
1Gastroenterology Department, Affiliated Hospital of Nanjing University of Chinese Medicine, Nanjing, People’s Republic of China; 2Ethics Committee, Affiliated Hospital of Nanjing University of Chinese Medicine, Nanjing, People’s Republic of China; 3Institute of Chinese Medicine, Nanjing University, Nanjing Drum Tower Hospital, Drum Tower Clinical Medicine College of Nanjing University of Chinese Medicine, Nanjing, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Jing Wu; Genglin Mu, Nanjing Drum Tower Hospital, Drum Tower Clinical Medicine College of the Nanjing University of Chinese Medicine, 321, Zhongshan Road, Nanjing, Jiangsu, People’s Republic of China, Tel +86 13451825475, Email [email protected]; [email protected]
Background: Baicalin is an important active flavonoid isolated from the roots of Scutellaria baicalensis (S. baicalensis), a well-known traditional Chinese herb used in treating inflammatory bowel disease (IBD). The objectives of this study were to assess the potential benefit of baicalin in experimental colitis, as well as to investigate metabolic biomarkers of experimental colitis in conjunction with network pharmacology.
Methods: Using a widely utilized network pharmacology technique, baicalin’s targets and pathways were predicted. Simultaneously, experimental colitis was induced by intrarectal administration of TNBS. Histopathology examinations were performed to confirm pathological changes. Plasma samples were examined by using an untargeted metabolomics technique based on ultra-high performance liquid chromatography-high resolution mass spectrometry (UHPLC-HRMS) to screen differential metabolites and associated metabolic pathways. Additionally, network pharmacology and integrated analysis of metabolomics were used to identify the primary targets.
Results: Through network pharmacology research, tumor necrosis factor (TNF), interleukin 6 (IL6), serine/threonine-protein kinase (AKT1), and other 7 proteins were found to be the main targets of baicalin against IBD. The untargeted metabolomics results showed that 47 metabolites in glycerophospholipids and sphingolipid metabolism were involved as key pathways in the experimental colitis model group. 19 metabolites, including Sphingomyelin (SM d42:2, SM d42:1, SM d34:1), Lysophosphatidic acids (LPA 18:4), 1-Palmitoylglycerophosphocholine, and 17(18)-EpETE were demonstrated as key metabolites for baicalin to exert effects. Moreover, udp-glucose ceramide glucosyltransferase (UGCG), sphingomyelin synthase 1 (SGMS1), and sphingosine kinase (SPHK1) were predicted as sphingolipids-linked targets of baicalin against experimental colitis by integrative analysis.
Conclusion: Based on these results, it implies that sphingolipid metabolism and sphingolipid signaling pathway might be acted as therapeutic mechanism for baicalin against experimental colitis.
Keywords: baicalin, inflammatory bowel disease, sphingolipid metabolism, metabolomics, network pharmacology, inflammation
Introduction
Inflammatory bowel diseases (IBD) including Crohn’s disease (CD) and Ulcerative colitis (UC) are chronic, idiopathic gastrointestinal (GI) diseases. It is thought to be the result of a complex interaction between a genetic predisposition and an abnormal host immune response to environmental exposures. The study of IBD has made extensive use of metabolomic techniques, which have the potential to be applied to disease subtyping and the evaluation of therapy response as well as offering insights into the pathophysiology of the disease.1 Changes in the amino-acid and lipids metabolism have been observed in metabolic studies of IBD.2 A lipid profile study in patients with IBD showed that primary bile acid biosynthesis, arachidonic acid metabolism, sphingolipid metabolism, fatty acid elongation, and glycerophospholipid metabolism were all linked to IBD dysregulation.3,4
IBD treatment options currently available are relatively restricted, and they are mostly aimed at keeping the disease under control and maintaining remission.5 However, this has to be weighed against the side effect profiles and costs of different drugs. With the advancement of high-throughput screening and bioinformatics technologies, natural products with a variety of pharmacological targets in different diseases have turned into important sources of novel drug discovery. These compounds are obtained from a variety of plants, including the dried root of Scutellaria baicalensis, which is frequently called Huangqin, a well-known traditional Chinese herb. It has been used as medicine in China for a very long time. Baicalin (5, 6-dihydroxy-7-O-glucuronide) is a kind of flavonoid, which is isolated predominantly from the roots of Scutellaria baicalensis (S. baicalensis). In modern studies, baicalin is a typical multi-target, a multi-pathway agent with multiple pharmacological effects.6 By altering several signaling pathways, baicalin has been demonstrated to have a protective effect against disorders linked to the gut.7 Baicalin’s effect on experimental colitis was attributed to its ability to reduce oxidative stress, immunological control, and anti-inflammatory qualities by inhibiting the activation of the TLR4/NF-κB pathway, MAPK, and associated cytokines.8,9 Baicalin reduces the severity of UC induced by dextran sulfate sodium (DSS). It modifies the polarization of M1 macrophages toward the M2 phenotype.10 Nevertheless, the mechanisms underlying baicalin’s anti-UC actions were not quite clear.
In order to investigate its mechanisms, approaches like metabolomics techniques and network pharmacology could be used. These approaches are based on the fact that traditional Chinese medicine (TCM) chemicals have a multitarget effect and the ability to change the entire body. Metabolomics is part of system biology. It is useful to identify as many metabolites related to diseases or biological processes as possible in the discovery phase. Metabolomics is effective for tracking the changes of metabolites in complicated diseases, like IBD. However, metabolomics provided too broad information and could not direct specific endogenous processes of disease, such as the proteins that make up their upstream pathways, and the proteins that a drug may affect. In this situation, network pharmacology is a good supplementary method and is helpful to build the entire drug-disease network.11 Network pharmacology is a methodology based on system biology. It evaluates drug polypharmacological effects at the molecular level to predict the interactions of natural products and proteins as well as to pinpoint the key mechanisms. Network pharmacology aims to provide a fresh perspective on how medications interact with diseases and to direct the creation of novel therapies.12
Therefore, the objective of this study is to link network pharmacology and metabolomics to help identify additional disease-related biological pathways.13 First, network pharmacology was used to anticipate the possible targets of baicalin for the treatment of IBD. Second, untargeted metabolomics and multivariate data analysis using UHPLC-HRMS were used to examine biomarkers and pathways. Finally, an analysis of the shared objectives of network pharmacology and untargeted metabolomics was performed to learn more about the therapeutic mechanisms of baicalin. This study offers a method for accurate biomarker screening while establishing the link between biomarkers and hub genes for defining the baicalin intervention mechanism.
Materials and Methods
Chemicals and Reagents
Baicalin (A0016, purity ≥98%, Chengdu Manstead Biotechnology Co. Ltd, China); 5% TNBS (2,4,6-Trinitrobenzenesulfonic acid) (P2297-5X,10ML, Sigma Corporation, USA); When utilized, baicalin was made into a 0.5% solution of carboxymethylcellulose sodium (CMC-Na). Methanol, acetonitrile, and formic acid were provided by Fisher Scientific(USA). Using a Milli-Q Integral Water Purification System from Merck Millipore, ultra-pure water was produced (Merck Millipore, USA).
Animal Experiments
30 male SD rats weighing 180–220g were purchased from SPF (Beijing) Biotechnology Co., Ltd. The animals were kept in an area with consistent humidity (45%± 5%), temperature (22~26 °C), and access to free food and water for at least a week prior to the experiment. Then rats were randomly assigned to each of the three groups after one week of adaptive feeding: control (DNC), model (DAT), and baicalin administration (DHG). All experiments and animal care were conducted in accordance with the Guide for the Care and Use of Laboratory Animals by the National Institutes of Health, and the animal study was approved by the IRB of the Affiliated Hospital of Nanjing University of Chinese Medicine (AEWC-20210515-182). Baicalin was administered orally to rats daily for 10 days prior to TNBS induction and daily thereafter. The dosage of baicalin in the DHG group was 100mg/kg.14–16 Other groups were given an equal volume of normal saline by gavage. Rats were starved for 24 hours prior to modeling, however, they were allowed to sip water. On the 11th day, rats were anesthetized with isoflurane. The DNC group received 1 mL of 0.9% saline through the rectum. The DAT group and 100mg/ (kg·d) baicalin group received 1mL TNBS solution (100mg/kg 5% TNBS dissolved in 50% ethanol) via the rectum. The rats were held in the vertical position for 60 seconds after the mixed solution was slowly injected into the proximal end of the descending colon, which was 4–6 cm long. The severity of colitis was recorded daily, including weight changes, diarrhea, and bloody stools. All rats were sacrificed on day 14 and the colon tissues and plasma were removed for further analysis. Body weight, stool character, and fecal occult blood were measured throughout the experiment. The scoring system was used to determine the disease activity index (DAI), as previously explained, DAI scores were assessed on the day of TNBS-induced colitis and three days after, recorded as DAI1-4.17
Segments of colonic tissues were fixed in 10% formalin for histology and demonstration of the established gross morphological changes associated with TNBS-induced colitis. Based on the tissue damage seen under the optical microscope, hematoxylin and eosin (H&E) staining was done to evaluate histopathological alterations. The damage score was assigned using the assessment scale.18
Network Pharmacology
Structural Information and ADME Criteria
Baicalin’s structural details were found on the NCBI PubChem (https://pubchem.ncbi.nlm.nih.gov/) database. A public database on the chemical compositions and biological characteristics of tiny molecules is called NCBI PubChem. Then, the screening criteria for absorption, distribution, metabolism, and excretion (ADME) from the Traditional Chinese Medicine Systems Pharmacology (TCMSP) database were used to evaluate baicalin (http://lsp.nwu.edu.cn/tcmsp.php). Two indexes, oral bioavailability (OB) and drug-likeness (DL) were used. The threshold values of the two indexes were OB ≥ 30% and DL ≥ 0.18.
Target Acquisition
In order to estimate the potential targets of baicalin, we first used the phrase “baicalin” to search literature from PubMed Central of the NCBI database (http://www.ncbi.nlm.nih.gov/pubmed/). We next filtered out the known targets of baicalin. We applied the pharmmapper Server (http://www.lilab-ecust.cn/pharmmapper/).19–21 The baicalin-associated targets were then created by combining the targets found in literature and databases. Additionally, the keywords “inflammatory bowel disease” was entered into GeneCards Database (http://www.genecards.org/),22 OMIM Database (http://omim.org/)23 and DisGeNET database (https://www.disgenet.org/)24 to search for and assess associated disease targets. The redundant targets were then eliminated and incorporated. In order to determine the prospective targets for baicalin’s treatment of IBD, the targets of baicalin and the targets of IBD were finally intersected.
Network Construction
Target proteins were uploaded as gene symbols to the STRING(https://string-db.org/)25 database for protein-protein (PPI) interaction analysis. Only output files with the suffix “TSV” and input proteins were included in the analysis range. Cytoscape 3.9.1 was utilized to display the files. Cytoscape 3.9.1’s NetworkAnalyzer was used to evaluate the degree and total scores. Data were imported into Cytoscape 3.9.1, using the cytohubba tool to calculate the scores of the nodes. Then, TOP10 nodes ranked by Maximal Clique Centrality (MCC)were selected. The top 10 high-node degree targets were chosen as the primary targets. Through the use of Metascape26 (https://metascape.org/), Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses, and Gene Ontology (GO) enrichment analyses, the potential functional roles of targets that appear on both the baicalin and IBD target lists were assessed.
Metabolomic
Plasma Sample Handling
Plasma samples (50 µL) were homogenized in a 200µL cold methanol, including internal standard substances (Table 1). The mixture was homogenized, vortexed for two minutes, allowed to stand at low temperature for 10 min, and then was centrifuged at 14,000 rpm for fifteen minutes at 4 °C. 200 µL of supernatant were absorbed and evaporated, then stored at −20°C. For LC-MS analysis, the extract samples were redissolved in 100 µL of methanol/water 1:5 (v:v). Additionally, to ensure quality control, equal portions of the supernatant from each sample were combined to create the quality control (QC) samples. Methanol/water in a blank extract was diluted 1:5(v:v). QC samples and blank samples underwent the same preparation as the analyzed samples.
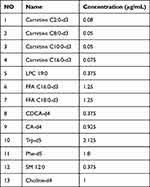 |
Table 1 Internal Standard Substances |
Chromatography-Mass Spectrometry Acquisition Conditions
Chromatography Analysis Conditions
Under positive-ion mode, the chromatography was performed on the BEH C8 column (2.1×100 mm×1.7 μm, Waters, Milford, MA, United States). The flow rate was 0.35 mL/min. The column temperature was 50°C. The injection volume was 5 μL. The mobile phase was composed of solvent A (water with 0.1% formic acid) and solvent B (acetonitrile with 0.1% formic acid). Gradient elution procedure: 0–1 min, 5%B; 1.1–11 min, 5–100%B; 11–13 min,100%B; 13.1–15 min, 5%B(Table S1-1). Under negative-ion mode, a 5µl reconstitution sample was injected for chromatography separation in the HSS T3 column (2.1×100 mm×1.8μm, Waters, Milford, MA, United States). The other chromatographic conditions Under negative-ion mode are the same as positive-ion mode (Table S1-2).
Mass Spectrometry Conditions
The data acquisition instrument system mainly includes Q-Exactive (Thermo Fisher, USA). Mass spectrometry main complete scan + data-dependent acquisition (DDA) secondary ion scan was employed in positive-ion mode (Table S2-1). The spray voltage was 3.8 kV in the positive ionization mode and −3.0 kV in the negative ionization mode when the electrospray ionization sources (ESI) of both positive and negative modes were used. The other mass spectrometry conditions under negative-ion mode are the same as positive-ion mode (Table S2-2). The Capillary temperature was maintained at 320 °C and the Sheath gas flow rate was 35 Arb. The Aux gas heater temperature was stipulated at 350 °C as well as the Aux gas flow rate was 8 Arb. The S-lens RF level was 50. The Mass range (m/z) was stipulated from 100 from 1200Da. The Full ms resolution was stipulated to be 70,000. The MS/MS resolution was 17,500. NCE/stepped NCE was 20, 40. During data acquisition, we developed DRMS / HRMS data quality online real-time monitoring software.
Data Processing and Statistical Analysis
All measurement data are collected by Xcalibur data acquisition software. The “80% rule” was used as a pretreatment on the data to lessen the entry of missing values. For multi-dimensional statistical analysis using principal component analysis, partial least squares discriminant analysis (PLS-DA), and orthogonal partial least squares discriminant analysis, the raw data were imported into the One-MAP/PTO software (OPLS-DA). With the variable importance in the projection VIP>1.0 as the standard to identify potential difference variables. Two sample unpaired t-tests are used in metabolomics research to demonstrate which metabolites have the ability to distinguish between the various groups in the data set, p<0.05 was used as a threshold. The FC was calculated relative to a given reference sample based on averaged raw signal intensities and serves as a metric for the relative change in a given concentration of the metabolite in the various settings under examination. A criterion of FC >1.5 or <2/3 was established. The possible biomarkers were screened using the two-sample unpaired t-test, fold change (FC) analysis, and variable importance projection (VIP) from the peak intensity. Finally, it was decided that data with VIP >1 and p< 0.05, FC > 1.5, or <2/3 were differential metabolites. The One-MAP platform was used to map the above already obtained metabolites, to metabolic pathways. One-step Metabolomics was used to analyze all the data.
Integrating Analysis
Metabolite targets derived from the KEGG website for major metabolic pathways enriched by differential metabolites which were regulated by baicalin. The combined projected targets of baicalin and metabolite targets were uploaded to STRING for protein-protein interaction (PPI) analysis. To obtain the protein-protein interaction (PPI) network, “Highest confidence>0.9” was chosen as the minimal interaction criterion. Based on the PPI data, we concentrated on the targets that were associated with them in order to identify plausible processes.
Statistical Methods
All data were expressed as mean ± standard deviation (X±SD). The software for graphing was GraphPad Prism 8.0, while SPSS 26.0 was utilized for statistical analysis. One-way ANOVA was used to compare group differences, and the threshold for statistical significance of p <0.05 was used.
Results
Baicalin Has a Therapeutic Effect on Experimental Colitis
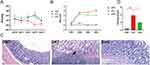
Rats in the Model group displayed sleepiness, sluggishness, loss of appetite, rough hair, diarrhea, bloody stool or fecal occult blood, and weight loss after modeling. We measured the weight loss and found that TNBS decreased the weight of DAT, whereas baicalin increased the body weight of DHG rats compared with the DAT group (Figure 1A). In comparison to the DNC group, the DAT group’s DAI was much higher (Figure 1B). The DAI3 and DAI4 were significantly lower in the DHG group compared to the Model group, which significantly alleviated the symptoms of diarrhea and hematochezia in UC model rats. Rats exposed to TNBS (DAT) suffered damage to their intestinal mucosa, including ulcerations, edema, and tissue thickening, whereas the DNC group kept their usual structure. The colonic tissue underwent a considerable improvement following treatment with baicalin(DHG)(Figure S1), with the damage score and extent of the damaged areas both declining.
Histologically, the DNC group showed a normal structure, whereas the TNBS group(DAT) displayed a serious transmural disruption, widespread ulceration, inflammation, and edema, mainly in the mucosa. The animals treated with baicalin (DHG) in the preventive strategy displayed decreased edema and ulceration as well as a repair of the mucosa, submucosa, and muscle layers (Figure 1C).
Compared with the control group, the histology score was significantly increased in the TNBS group (P<0.001). Compared with the TNBS group, the histology score of the baicalin group decreased significantly (P<0.05) (Figure 1D). In conclusion, baicalin ameliorates the severity of TNBS-induced colitis and suppresses colonic tissue damage.
Baicalin’s Potential Herb-Target Pathway Interaction on IBD Was Anticipated by Network Pharmacology in the Section
Structural Information and ADME-Related Properties of Baicalin
Baicalin’s structural details (Figure 2A) and information pertaining to its ADME were obtained from TCMSP. Baicalin’s OB and DL have been found to fit the cutoffs, which were determined to be 40.12% and 0.75, respectively. This demonstrated baicalin’s superior oral bioavailability and its similarity to well-known medications.
Target Prediction
Given that baicalin has various pharmacological effects, it is essential to precisely explore these targets. From the NCBI database’s PubMed Central, we were able to obtain targets for baicalin that had been mentioned in the literature. Then, PhamaMapper was used to predict the compound targets. In addition, we deleted the overlap targets and blended the objectives from the two approaches. Finally, 287 baicalin-associated targets were discovered and displayed. Additionally, the GeneCards, DisGeNET, and OMIM databases’ scores are used to screen possible disease-causing genes. The objectives from these three datasets were integrated, and the overlapped ones were eliminated. Finally, 1833 targets related to IBD were chosen and displayed in Figure 2B.
Construction of PPI Networks and Core PPI Network Evaluation
PPI (Protein-Protein Interaction Network) networks can be employed to understand the function of diverse targets in IBD. The 287 baicalin-associated targets and 1833 IBD targets were inputted into Draw Venn Diagram (http://bioinformatics.psb.ugent.be/) to plot Wayne diagrams (Figure 2C). Then, The identified compound target overlapped with the IBD disease target.115 predicted targets were obtained for the treatment of IBD with baicalin.
The PPI network of intersection targets was analyzed by the STRING database (https://string-db.org/) and the targets with medium confidence scores>0.4 were screened for network construction. As a result, 115 nodes and 1476 edges were involved in this network. The average node degree is 25.7 and the PPI enrichment p < 1.0e-16. The lines represent protein interactions. More crucial targets were obtained based on the betweenness centrality of network topology analysis. Since STRING was unable to identify any of the targets acquired from the databases, they were removed. We utilized the merge function offered by Cytoscape to link medication targets with disease targets and disclose the pharmacological mechanisms of baicalin to treat IBD. The STRING results were exported in text and opened with Cytoscape 3.9.1 software to visualize protein interactions (Figure 2D). The crucial targets were the top 10 targets, which were positioned in the middle. The top 10 targets of baicalin against IBD were Albumin (ALB), tumor necrosis factor (TNF), interleukin 6 (IL6), RAC-alpha serine/threonine-protein kinase (AKT1), Proto-oncogene tyrosine-protein kinase Src (SRC), caspase 3 (CASP3), Matrix metalloproteinase-9 (MMP9), Insulin-like growth factor I (IGF1), mitogen-activated protein kinases (MAPK14), prostaglandin-endoperoxide synthase 2(PTGS2)(Figure 2E).
GO Enrichment Analyses and KEGG Pathway Enrichment Analysis
The 115 overlappings of IBD and Baicalin targets were subjected to KEGG pathway and GO enrichment studies using the Metascape platform. Three layers make up the GO enrichment analysis of 115 targets: molecular functions (MFs), biological processes (BPs), and cellular components (CCs). The top 10 of these enriched pathways were shown, and results were ordered based on increasing -lgP, which corresponds to the degree of connection with these targets (Figure 2F).
115 potential targets were chosen for KEGG pathway enrichment analysis in order to shed light on the crucial function that baicalin plays in the treatment of IBD. Through the Metascape platform, we learned more about the signaling pathways connected to these potential targets, and we imported this knowledge to create a pathway enrichment map (Figure 2G). Based on the P-value, the top 25 KEGG signaling pathways were found and created.
Metabolomics Analysis Identified Differential Plasma Metabolites in Groups
Metabolomics Data Analysis
Different metabolites in rat plasma samples were found and examined in order to comprehend the metabolic effects of baicalin on IBD. First and foremost, basic data analysis revealed that quality control samples from chromatography had satisfactory peak shapes and very uniform distributions under test conditions (Figure S2). The quality control data was then analyzed using PCA, and the score demonstrated the system’s repeatability and stability.
Metabolomics Profile of DAT versus DNC
To demonstrate that the DAT group was significantly separated from the control group, PLS-DA was more effectively applied (Figure 3A and B). From the plasma PLS-DA score plots, we found that the metabolic characteristics of the DAT group and the DNC group were significantly different. The validation parameters of two multivariate PLS-DA models included: Positive-ion mode R2Y=0.97, Q2Y=0.94, Q2X0=−1.17; Negative-ion mode R2Y=0.91, Q2Y=0.84, Q2X0=−0.95. The model has good predictive power and explanatory power for Y categorical variables. Q2X0 <0, indicating that none of the models are over-fitted. Furthermore, based on variable importance VIP > 1, P< 0.05, and FC> 1.5 or < 2/3 in PLS-DA, 47 differentiated metabolite ions were identified. In the DAT group, 24 ions were elevated and 23 ions were downregulated, showing that TNBS-induced experimental colitis causes metabolic abnormalities (Table S3-1). Through cluster analysis (heatmap) (Figure 3C and D), we can observe a distinct metabolic profile between DAT and DNC in plasma samples. As demonstrated in the visible volcano plot (Figure 3E and F), lysophosphatidic acids (LPA 18:4), sphingomyelin (SM d42:2, SM d42:1), ceramides (Cer-NP t33:2; Cer-NP t19:1/14:1), phosphatidylcholine (PC 34:4, PC 36:6) and 11β-Prostaglandin F2α were upregulated. Glycocholic acid, SM d34:1, PC 36:2, threoninyl-tryptophan, deoxyadenosine, Gly-Tyr-Ala, and fatty acids and conjugates (5,6-Epoxy-18R-HEPE, 17(18)-EpETE) and so on were downregulated.
Metabolomics Profile of DHG versus DAT
Moreover, PLS-DA shows that the baicalin group was significantly separated from the DAT group (Figure 4A and B). Each PLS-DA model displayed good separation with high R2Y and Q2, demonstrating the sample classification data’s strong explanatory power and cross-validated prediction power.
67 differentiated metabolite ions were found based on variable importance FC> 1.5 or <2/3, VIP > 1, and P<0.05 in PLS-DA. The DHG group had 40 ions that were upregulated (Fold change > 1) and 27 ions that were downregulated (Fold change < 1). (Table S3-2). In the advanced data analysis, differential metabolites in plasma were vividly displayed through cluster analysis (heatmap). The potential biomarkers were demonstrated using a volcano plot analysis (Figure 4C–F). Compared with the DAT group, the abundance of 17(18)-EpETE, 5.6-Epoxy-18R-HEPE, 15-Oxoete, ceramide (Cer 42:1, Cer(t18:0/24:0)), tetrahydrocorticosterone, PC 34:0, PC36:4, Prostaglandin D2, Prostaglandin E2, Prostaglandin I2, and SM d34:1 were upregulated in the DHG group. The abundance of lysophosphatidylcholine (LPC20:1,16:1), sphingomyelin (SM d42:2, SM d42:1), lysophosphatidic acids (LPA 18:4), 11β-Prostaglandin F2γ and PC 35:1 were downregulated in the DHG group. In the comparison of the three groups DNC group, DAT group, and DHG group, two sphingomyelins (SM d42:2, SM d42:1), LPA 18:4, 1-Methylxanthine, 1-Heptanesulfonic Acid and diethyl phenyl phosphate, in the DAT group, were significantly enhanced compared to the DNC group, while these metabolites were significantly downregulated in the DHG group. 17(18)-EpETE, 1-Palmitoylglycerophosphocholine, 3-hydroxytridecanoic acid, 5.6-Epoxy-18R-HEPE, tetrahydrocorticosterone, SM d34:1, and all-trans-4-Oxoretinoic acid, in the DAT group were significantly decreased compared to the DNC group, they significantly increased in baicalin group (Table 2).
 |
Table 2 Comprehensive Analysis of the Differential Metabolites of Three Groups |
Pathway Analysis
As shown in Figure 5A and B, the rats in the DAT group damaged Sphingolipid metabolism and Glycerophospholipid metabolism compared with the DNC group. Compared with the DAT group, the baicalin group significantly regulated sphingolipid metabolism.
 |
Figure 5 Metabolic pathway (A) metabolic pathway of the DAT group compared to the DNC group. (B) metabolic pathway of the DHG group compared to the DAT group. |
Integrating Analysis
Differential metabolites engaged in several metabolic and signal transduction pathways, which pathway enrichment analysis can help to identify. Compared with the DAT group, two SMs(SM d42:2, SM d42:1) were significantly downregulated, four Cers (Cer 42:1, Cer(t18:0/24:0), Cer-NS d41:1, Cer-NS d43:1) and SM d34:1 were significantly upregulated in the DHG group. These metabolites are mainly enriched in the sphingolipid metabolic pathway in the DHG group, as shown in Figure 5B. Moreover, network pharmacology revealed that baicalin can regulate the hsa04071:Sphingolipid signaling pathway. Results from network pharmacology and untargeted metabolomics suggested that baicalin’s ability to treat IBD entailed controlling sphingolipid metabolism (Figure S3-1) and sphingolipid signaling pathway (Figure S3-2 and S3-3).
Additionally, we used STRING to link the 115 anticipated targets of baicalin against IBD with the 55 metabolite targets associated with sphingolipid metabolism, creating the PPI network (Figure 6). The associated targets were constructed to understand the underlying mechanism of baicalin’s treatment of IBD in accordance with the findings of the PPI network analysis. Based on the above screening, the targets with the top 8 betweenness values were selected as the key targets (Table 3). Of the top 8 targets, UGCG, SGMS1, and SPHK1 are important targets for sphingolipid metabolism.
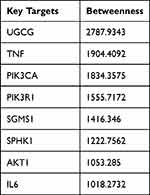 |
Table 3 Top 8 of Betweenness Values as the Key Targets |
 |
Figure 6 The PPI network of sphingolipid metabolism-related targets with predicted targets of baicalin against IBD. |
Discussion
In this study, we combined untargeted metabolomics with network pharmacology to explore new molecular targets of baicalin in the treatment of IBD. From our data, we found abnormalities in glycerophospholipid metabolism and sphingolipid metabolism in the TNBS-induced experimental colitis rats. Baicalin can correct sphingolipid metabolism. Baicalin’s ability to treat IBD might be associated with controlling sphingolipid metabolism and 8 related targets (UGCG, TNF, PIK3CA, PIK3R1, SGMS1, SPHK1, AKT1, IL6), according to both untargeted metabolomics and network pharmacology studies. Notably, baicalin’s expected sphingolipids-linked targets against IBD included UGCG, SGMS1, and SPHK1. In conclusion, baicalin might improve experimental colitis in model rats by targeting several proteins on the sphingolipid metabolic pathways.
Our metabolomics study showed considerable changes in the lipid profiles of the DAT group when compared with the DNC group. Sphingolipids, glycerophospholipids, fatty acyls, and sterol lipids are primarily affected. We will therefore focus on the changes in bioactive lipids in experimental colitis, the modulatory effect of baicalin on lipid profiles, exploring the causes of metabolite changes, and the mechanism of action of baicalin. In our study, the pharmacological effects of baicalin involve the sphingolipid signaling pathway and can regulate sphingolipid metabolism. An increasing number of studies indicate that IBD is associated with several sphingolipid species, including SM, ceramide (Cer), sphingosine (Sph), as well as their 1-phosphorylated derivatives ceramide-1-phosphate (C1P) and sphingosine-1-phosphate (S1P).27 Ceramide is the primary component of all complex sphingolipids, such as glycosphingolipids and sphingomyelin, which is created in the cytosolic leaflet of the endoplasmic reticulum (ER), is hydrolyzed to form sphingosine. Sphingosine kinases (SphK1 and SphK2) swiftly phosphorylate sphingosine to form sphingosine-1-phosphate (S1P).28 Ceramide (Cer) and sphingosine-1-phosphate (S1P), two specific sphingolipid metabolites, have emerged as novel classes of potent bioactive chemicals.29 Furthermore, Cer is transferred to the Golgi either by the ceramide transfer protein CERT or at ER-trans-Golgi interaction sites to generate sphingomyelin(SM).30 In a previous study of a mouse model of colitis, SM and Cer are dramatically elevated.31
These sphingolipids changes are consistent with the trend of SM and Cer in the DAT group of our study. Based on the predictions of network pharmacology and the results of metabolomics, we hypothesize that the increased SM and Cer in IBD appear to be caused by many enzymatic changes. N-SMase is downregulated in colon samples from mice with DSS-induced IBD and human chronic colitis was found to have a decrease in Alk-SMase activity, which may be a crucial mechanism causing SM accumulation.31,32 Both an increase in glucocerebrosidase (which converts GlcCer to Cer) and CerS activation were observed in IBD, and these were proposed as the causes of Cer elevation.33 Udp-glucose ceramide glucosyltransferase (UGCG) is the first rate-limiting step in the glucosylceramide biosynthetic pathway which is essential for intestinal endocytic function. After UGCG deletion, structural alterations in the intestine appeared.34 In the studies, data showed that in IBD patients, the expression of UGCG decreased. In line with this, the UGCG level in mesenteric lymph nodes and T cells from the spleen was decreased with Dextran sulfate sodium (DSS). Furthermore, T cells exacerbated the pathogenesis of colitis, and the knockdown of UGCG was accompanied by a decrease in Treg cell levels.35,36 Based on ceramide, more than 300 species of gangliosides are synthesized by the stepwise addition of carbohydrates. As we knew that UGCG plays an important role in ganglioside synthesis. Recently, several studies have been conducted on the therapeutic effects of gangliosides in models of inflammatory diseases.37 Ganglioside content and enzymatic regulation of ganglioside metabolism differ between IBD and healthy intestines. These differences may impact the intestinal barrier and immunity functions altered in IBD.38 Based on the targets derived from the combined analysis, in addition to SMase and UGCG, there is SPHK1. SphK1/S1P/S1PRs axis has recently been defined as a target for the treatment of GI diseases including IBD/colitis.39 Enhanced SphK1 protein expression exacerbated IBD conditions in a colitis mouse model and patients with UC.40,41 Investigations into the targeting of sphingolipids as a possible area of therapy for regulating the course and morbidity of this illness have been driven by the discovery that various sphingolipid changes are related with IBD.
Baicalin (5, 6-dihydroxy-7-O-glucuronide) is a kind of flavonoid. Previous studies have shown that flavonoid treatment improved animals with HFD-induced dyslipidemia by reducing the levels of Cer and SM in the liver and plasma by preventing the de novo synthesis of sphingolipids.42 Flavonoids affected the expression of desaturases involved in the omega-3 fatty acid synthesis and ceramide (Cer) metabolism.43 Previous research has found that flavonoids can disrupt the sphingolipid rheostat in colon cancer cells by preventing S1P production and Cer traffic. Thus, flavonoids have a regulatory effect on the sphingolipid pathway. Based on these findings we infer that baicalin may also act on sphingolipid-related enzymes to regulate sphingolipid metabolism. Our work predicted this possibility. However, there are few studies on the direct effect of baicalin on sphingolipid metabolizing enzymes. In the past, researchers may have overlooked the effect of baicalin on sphingolipids. In our present study, we are pointing to a promising pathway for future studies on the mechanism of baicalin action.
Since baicalin referred to the sphingolipid signaling route as part of the primary mechanism for treating IBD, integrated network pharmacology demonstrated that it also belonged to this pathway. Sphingolipid signaling was identified as one of the key mediators of pro-inflammatory conditions, and, specifically, TNF-α related signaling.44 Several immunological responses trigger the activation of the sphingolipid metabolism. The initiation and resolution of inflammation are linked to the stimulation of sphingolipid metabolism by pro-inflammatory cytokines and coagulation-related chemicals.45 Numerous studies suggest that cytokines and other sphingolipids form a close network that is essential for controlling inflammatory pathways and immunological responses in IBD. Both innate and adaptive immune cells are sent to the tissues during inflammatory processes, and cytokine networks are also activated. Such changes are regulated by sphingolipids, and sphingolipid metabolism is altered in tissues as a response to these alterations.46 We also found that cytokines are associated with enzymes related to sphingolipid metabolism. Research shows that SphK1 has emerged as a significant player, triggered by a number of cytokines, TNF-α being one of them.47 Cytokines produced by innate and adaptive immune cells have a crucial role in the pathogenesis of IBD, where they control multiple aspects of the inflammatory response.48 Baicalin may be an alternative medication in the treatment of UC because of its effects on the suppression of inflammation. Researchers discovered that baicalin prevented the activation of the TLR4/NF-κB signaling pathway to ameliorate UC in addition to lowering the levels of the inflammatory markers IL-1, TNF-α, and IL-6 in rat intestinal tissues. An HTHE-induced UC model confirmed similar findings. IL-1, IL-17, and IL-6 serum levels were significantly decreased by baicalin at 100 mg/kg.9 Feng et al additionally discovered that 100 mg/kg baicalin reduced the symptoms of DSS-induced UC by inhibiting the TLR4/NF-κB-p65/IL-6 signaling pathway and repressing the expression of TNF-α, IL-6, and IL-13 mRNA.49 Thus, we presume that cytokines such as TNF-α may be closely related to sphingolipid metabolism. Baicalin may affect sphingolipid metabolism by down-regulating abnormal cytokines.
We also found abnormalities in glycerophospholipids. Baicalin down-regulates the abundance of LPA18:4. We found that in the DAT group prostaglandins metabolism was disturbed. Abnormal fatty acid metabolism was noticed in patients with IBD in both the stationary period and the active period.4 Based on the above metabolism results, we found that disorders of lipid metabolism were predominantly seen in the DAT group, such as glycerophospholipid metabolism and sphingolipid metabolism. Baicalin might regulate them at the same time, even though sphingolipid metabolism might be in the dominant role.
The combined use of network pharmacology and metabolomics facilitate the unraveling of potentially complex relationships between multiple metabolites and multiple targets, providing strategies for the study of the mechanisms of therapeutic diseases in Chinese medicine. However, there are some limitations. It is currently not possible to remove interference with the endogenous metabolome by herbal components. Incomplete information on the various types of databases, and different algorithms between databases without using them in conjunction, cause great problems in determining the final results. The gap between these predicted targets and the real situation is still large. Further experimental studies needed to be put forward to validate our findings.
Conclusion
Baicalin was found to regulate 19 different metabolites, including sphingomyelin, glycerophospholipids, and fatty acids, according to untargeted metabolomics data. The pathway study showed that baicalin’s protective effects might be influenced by controlling sphingolipid metabolism. After integrating the findings of metabolomics and network pharmacology analyses, sphingolipid-linked proteins UGCG, SGMS1, and SPHK1 were predicted as targets for baicalin against IBD. Taken altogether, this research provided a meaningful direction to future drug discovery and understanding of the pharmacological mechanism of baicalin to treat inflammatory bowel disease.
Abbreviations
TNBS, 2,4,6-trinitrobenzenesulfonic acid; UHPLC-HRMS, ultra-high performance liquid chromatography-high resolution mass spectrometry; IBD, inflammatory bowel disease; TNF, tumor necrosis factor; IL6, interleukin 6; AKT1, serine/threonine-protein kinase; SM, sphingomyelin; LPA, lysophosphatidic acids; UGCG, udp-glucose ceramide glucosyltransferase; SGMS1, sphingomyelin synthase 1; SPHK1, sphingosine kinase; CD, crohn’s disease; UC, ulcerative colitis; GI, gastrointestinal; TLR4, Toll Like Receptor 4; NF-κB, nuclear factor kappa-B; MAPK, mitogen-activated protein kinases; DSS, dextran sulfate sodium; TCM, traditional Chinese medicine; CMC-Na, carboxymethylcellulose sodium; DAI, disease activity index; H&E, hematoxylin and eosin; ADME, absorption, distribution, metabolism, and excretion; TCMSP, traditional Chinese medicine systems pharmacology; OB, oral bioavailability; DL, drug-likeness; OMIM, online Mendelian inheritance in man; PPI, protein-protein interaction; MCC, maximal Clique Centrality; KEGG, Kyoto Encyclopedia of genes and genomes; GO, gene ontology; QC, quality control; DDA, data-dependent acquisition; PLS-DA, partial least squares discriminant analysis; OPLS-DA, orthogonal partial least squares discriminant analysis; FC, fold change; VIP, variable importance projection; ALB, albumin; SRC, proto-oncogene tyrosine-protein kinase; CASP3, caspase 3; MMP9, matrix metalloproteinase-9; IGF1, insulin-like growth factor I; PTGS2, prostaglandin-endoperoxide synthase 2; MFs, molecular functions; BPs, biological processes; CCs, cellular components; LPC, lysophosphatidylcholine; Cer, ceramide; Sph, sphingosine, C1P, ceramide-1-phosphate; S1P, sphingosine-1-phosphate; ER, endoplasmic reticulum; 17(18)-EpETE, 17R,18S-Epoxy-5Z,8Z,11Z,14Z-icosatetraenoic acid.
Institutional Review Board Statement
Animal care and experimentation were carried out strictly in accordance with the Guide for the Care and Use of Laboratory Animals by the National Institutes of Health, and the animal study was approved by the IRB of the Affiliated Hospital of Nanjing University of Chinese Medicine (AEWC-20210515-182). This paper is based on a study involving publicly available data, which is exempt from ethical review. It was approved by the IRB of the Affiliated Hospital of the Nanjing University of Chinese Medicine.
Acknowledgments
The authors appreciate Dalian Chem Data Solution Information Technology Co. Ltd. for the technical support provided in this study.
Funding
This research was funded by the National Natural Science Foundation of China (no.81873160).
Disclosure
Prof. Dr. Jing Wu reports grants from national natural science foundation of China, grants from Nanjing Drum Tower Hospital, during the conduct of the study. The authors report no conflicts of interest in this work.
References
1. Gallagher K, Catesson A, Griffin JL, Holmes E, Williams HRT. Metabolomic analysis in inflammatory bowel disease: a systematic review. J Crohns Colitis. 2021;15(5):813–826. doi:10.1093/ecco-jcc/jjaa227
2. Aldars-García L, Gisbert JP, Chaparro M. Metabolomics insights into inflammatory bowel disease: a comprehensive review. Pharmaceuticals. 2021;14:11. doi:10.3390/ph14111190
3. Santoru ML, Piras C, Murgia F, et al. Metabolic alteration in plasma and biopsies from patients with IBD. Inflamm Bowel Dis. 2021;27(8):1335–1345. doi:10.1093/ibd/izab012
4. Guan S, Jia B, Chao K, et al. UPLC-QTOF-MS-based plasma lipidomic profiling reveals biomarkers for inflammatory bowel disease diagnosis. J Proteome Res. 2020;19(2):600–609. doi:10.1021/acs.jproteome.9b00440
5. Nanda K, Moss AC. Update on the management of ulcerative colitis: treatment and maintenance approaches focused on MMX(®) mesalamine. Clin Pharmacol. 2012;4:41–50. doi:10.2147/CPAA.S26556
6. Ganguly R, Gupta A, Pandey AK. Role of baicalin as a potential therapeutic agent in hepatobiliary and gastrointestinal disorders: a review. World J Gastroenterol. 2022;28(26):3047–3062. doi:10.3748/wjg.v28.i26.3047
7. Hu Q, Zhang W, Wu Z, et al. Baicalin and the liver-gut system: pharmacological bases explaining its therapeutic effects. Pharmacol Res. 2021;165:105444. doi:10.1016/j.phrs.2021.105444
8. Shen J, Cheng J, Zhu S, et al. Regulating effect of baicalin on IKK/IKB/NF-kB signaling pathway and apoptosis-related proteins in rats with ulcerative colitis. Int Immunopharmacol. 2019;73:193–200. doi:10.1016/j.intimp.2019.04.052
9. Liang S, Deng X, Lei L, et al. The comparative study of the therapeutic effects and mechanism of baicalin, baicalein, and their combination on ulcerative colitis rat. Front Pharmacol. 2019;10:1466. doi:10.3389/fphar.2019.01466
10. Rizzo V, Ferlazzo N, Currò M, et al. Baicalin-induced autophagy preserved LPS-stimulated intestinal cells from inflammation and alterations of paracellular permeability. Int J Mol Sci. 2021;22:5. doi:10.3390/ijms22052315
11. Gertsch J. Botanical drugs, synergy, and network pharmacology: forth and back to intelligent mixtures. Planta Med. 2011;77(11):1086–1098. doi:10.1055/s-0030-1270904
12. Zhang W, Chen Y, Jiang H, et al. Integrated strategy for accurately screening biomarkers based on metabolomics coupled with network pharmacology. Talanta. 2020;211:120710. doi:10.1016/j.talanta.2020.120710
13. Wei S, Ma X, Niu M, et al. Mechanism of paeoniflorin in the treatment of bile duct ligation-induced cholestatic liver injury using integrated metabolomics and network pharmacology. Front Pharmacol. 2020;11:586806. doi:10.3389/fphar.2020.586806
14. Zhu L, Shen H, Gu P, et al. Effects of baicalin on the inflammation and apoptosis in ulcerative colitis rats relating to PI3K/AKT pathway. CJTCMP. 2017;32:9.
15. Zhu L, Shen H, Gu P, et al. Effect of baicalin on Expression of NF-κB in ulcerative colitis rats. J Nanjing univTraditChinMed. 2016;2016:1.
16. Zhu W, Jin Z, Yu J, et al. Baicalin ameliorates experimental inflammatory bowel disease through polarization of macrophages to an M2 phenotype. Int Immunopharmacol. 2016;35:119–126. doi:10.1016/j.intimp.2016.03.030
17. Saber S, El-Kader EMA. Novel complementary coloprotective effects of metformin and MCC950 by modulating HSP90/NLRP3 interaction and inducing autophagy in rats. Inflammopharmacology. 2021;29(1):237–251. doi:10.1007/s10787-020-00730-6
18. He W, Li Y, Liu M, et al. Citrus aurantium L. and Its Flavonoids Regulate TNBS-Induced Inflammatory bowel disease through anti-inflammation and suppressing isolated jejunum contraction. Int J Mol Sci. 2018;19(10):3057. doi:10.3390/ijms19103057
19. Liu X, Ouyang S, Yu B, et al. PharmMapper server: a web server for potential drug target identification using pharmacophore mapping approach. Nucleic Acids Res. 2010;38:W609–W614. doi:10.1093/nar/gkq300
20. Wang X, Pan C, Gong J, Liu X, Li H. Enhancing the enrichment of pharmacophore-based target prediction for the polypharmacological profiles of drugs. J Chem Inf Model. 2016;56(6):1175–1183. doi:10.1021/acs.jcim.5b00690
21. Wang X, Shen Y, Wang S, et al. PharmMapper 2017 update: a web server for potential drug target identification with a comprehensive target pharmacophore database. Nucleic Acids Res. 2017;45(W1):W356–W360. doi:10.1093/nar/gkx374
22. Safran M, Dalah I, Alexander J, et al. GeneCards Version 3: the human gene integrator. Database. 2010;2010:baq020. doi:10.1093/database/baq020
23. McKusick VA. Mendelian Inheritance in Man and its online version, OMIM. Am J Hum Genet. 2007;80(4):588–604. doi:10.1086/514346
24. Piñero J, À B, Queralt-Rosinach N, et al. DisGeNET: a comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. 2017;45(D1):D833–D839. doi:10.1093/nar/gkw943
25. Szklarczyk D, Gable AL, Nastou KC, et al. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021;49(D1):D605–D612. doi:10.1093/nar/gkaa1074
26. Zhou Y, Zhou B, Pache L, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10(1):1523. doi:10.1038/s41467-019-09234-6
27. Abdel Hadi L, Di Vito C, Riboni L. Fostering inflammatory bowel disease: sphingolipid strategies to join forces. Mediators Inflamm. 2016;2016:3827684. doi:10.1155/2016/3827684
28. Maceyka M, Spiegel S. Sphingolipid metabolites in inflammatory disease. Nature. 2014;510(7503):58–67. doi:10.1038/nature13475
29. Hannun YA, Obeid LM. Principles of bioactive lipid signalling: lessons from sphingolipids. Nat Rev Mol Cell Biol. 2008;9(2):139–150. doi:10.1038/nrm2329
30. Green CD, Maceyka M, Cowart LA, Spiegel S. Sphingolipids in metabolic disease: the good, the bad, and the unknown. Cell Metab. 2021;33(7):1293–1306. doi:10.1016/j.cmet.2021.06.006
31. Qi Y, Jiang C, Tanaka N, et al. PPARα-dependent exacerbation of experimental colitis by the hypolipidemic drug fenofibrate. Am J Physiol Gastrointest Liver Physiol. 2014;307(5):G564–G573. doi:10.1152/ajpgi.00153.2014
32. Sjöqvist U, Hertervig E, Nilsson A, et al. Chronic colitis is associated with a reduction of mucosal alkaline sphingomyelinase activity. Inflamm Bowel Dis. 2002;8(4):258–263. doi:10.1097/00054725-200207000-00004
33. Scharl M, Leucht K, Frey-Wagner I, et al. Knock-out of β-glucosidase 2 has no influence on dextran sulfate sodium-induced colitis. Digestion. 2011;84(2):156–167. doi:10.1159/000327380
34. Jennemann R, Kaden S, Sandhoff R, et al. Glycosphingolipids are essential for intestinal endocytic function. J Biol Chem. 2012;287(39):32598–32616. doi:10.1074/jbc.M112.371005
35. Komuro M, Nagane M, Endo R, et al. Glucosylceramide in T cells regulates the pathology of inflammatory bowel disease. Biochem Biophys Res Commun. 2022;599:24–30. doi:10.1016/j.bbrc.2022.02.004
36. Tanaka M, Saka-Tanaka M, Ochi K, et al. C-type lectin Mincle mediates cell death-triggered inflammation in acute kidney injury. J Exp Med. 2020;217:11. doi:10.1084/jem.20192230
37. Lalazar G, Acov A, Lador A, et al. Modulation of intracellular machinery by beta-glycolipids is associated with alteration of NKT lipid rafts and amelioration of concanavalin-induced hepatitis. Mol Immunol. 2008;45(13):3517–3525. doi:10.1016/j.molimm.2008.05.009
38. Miklavcic JJ, Hart TDL, Lees GM, et al. Increased catabolism and decreased unsaturation of ganglioside in patients with inflammatory bowel disease. World J Gastroenterol. 2015;21(35):10080–10090. doi:10.3748/wjg.v21.i35.10080
39. Mahajan-Thakur S, Sostmann BD, Fender AC, et al. Sphingosine-1-phosphate induces thrombin receptor PAR-4 expression to enhance cell migration and COX-2 formation in human monocytes. J Leukocyte Biol. 2014;96(4):611–618. doi:10.1189/jlb.3AB1013-567R
40. Crespo I, San-Miguel B, Mauriz JL, et al. Protective effect of protocatechuic acid on TNBS-induced colitis in mice is associated with modulation of the SphK/S1P signaling pathway. Nutrients. 2017;9:3. doi:10.3390/nu9030288
41. Karuppuchamy T, Behrens EH, González-Cabrera P, et al. Sphingosine-1-phosphate receptor-1 (S1P1) is expressed by lymphocytes, dendritic cells, and endothelium and modulated during inflammatory bowel disease. Mucosal Immunol. 2017;10(1):162–171. doi:10.1038/mi.2016.35
42. Shon JC, Kim WC, Ryu R, et al. Plasma lipidomics reveals insights into anti-obesity effect of chrysanthemum morifolium Ramat leaves and its constituent luteolin in high-fat diet-induced dyslipidemic mice. Nutrients. 2020;12:10. doi:10.3390/nu12102973
43. Kühn G, Pallauf K, Schulz C, Rimbach G. Flavonoids as putative modulators of Δ4-, Δ5-, and Δ6-desaturases: studies in cultured hepatocytes, myocytes, and adipocytes. Biofactors. 2018;44(5):485–495. doi:10.1002/biof.1443
44. Hait NC, Maiti A. The role of sphingosine-1-phosphate and ceramide-1-phosphate in inflammation and cancer. Mediators Inflamm. 2017;2017:4806541. doi:10.1155/2017/4806541
45. Proia RL, Hla T. Emerging biology of sphingosine-1-phosphate: its role in pathogenesis and therapy. J Clin Invest. 2015;125(4):1379–1387. doi:10.1172/JCI76369
46. Hla T, Dannenberg AJ. Sphingolipid signaling in metabolic disorders. Cell Metab. 2012;16(4):420–434. doi:10.1016/j.cmet.2012.06.017
47. De Palma C, Meacci E, Perrotta C, Bruni P, Clementi E. Endothelial nitric oxide synthase activation by tumor necrosis factor alpha through neutral sphingomyelinase 2, sphingosine kinase 1, and sphingosine 1 phosphate receptors: a novel pathway relevant to the pathophysiology of endothelium. Arterioscler Thromb Vasc Biol. 2006;26:1. doi:10.1161/01.ATV.0000194074.59584.42
48. Neurath MF. Cytokines in inflammatory bowel disease. Nat Rev Immunol. 2014;14(5):329–342. doi:10.1038/nri3661
49. Feng J, Guo C, Zhu Y, et al. Baicalin down regulates the expression of TLR4 and NFkB-p65 in colon tissue in mice with colitis induced by dextran sulfate sodium. Int J Clin Exp Med. 2014;7(11):4063–4072.
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.