Back to Journals » International Journal of Nanomedicine » Volume 18
Therapeutic Effects of Mechanical Stress-Induced C2C12-Derived Exosomes on Glucocorticoid-Induced Osteoporosis Through miR-92a-3p/PTEN/AKT Signaling Pathway
Authors Xu N , Cui G, Zhao S , Li Y, Liu Q, Liu X, Zhao C , Feng R , Kuang M, Han S
Received 8 September 2023
Accepted for publication 1 December 2023
Published 12 December 2023 Volume 2023:18 Pages 7583—7603
DOI https://doi.org/10.2147/IJN.S435301
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Yan Shen
Ning Xu,1,* Guanzheng Cui,1,* Shengyin Zhao,1 Yu Li,1 Qian Liu,2 Xuchang Liu,1 Chuanliang Zhao,3 Rongjie Feng,1 Mingjie Kuang,1 Shijie Han1
1Department of Orthopedic Surgery, Shandong Provincial Hospital Affiliated to Shandong First Medical University, Shandong, 250014, People’s Republic of China; 2Department of Pain, Qilu Hospital of Shandong University, Jinan, 250012, People’s Republic of China; 3Department of Orthopedic Surgery, The Affiliated Taian City Central Hospital of Qingdao University, Taian, 271000, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Shijie Han; Mingjie Kuang, Department of Orthopedic Surgery, Shandong Provincial Hospital Affiliated to Shandong First Medical University, Jinan, Shandong, 250014, People’s Republic of China, Email [email protected]; [email protected]
Introduction: Osteoporosis is a common bone disease in which the bone loses density and strength and is prone to fracture. Bone marrow mesenchymal stem cells (BMSCs) are important in bone-related diseases. Exosomes, as mediators of cell communication, have potential in cell processes. Previous studies have focused on muscle factors’ regulation of bone remodeling, but research on exosomes is lacking.
Methods: In order to confirm the therapeutic effect of mechanically stimulated myocytes (C2C12) derived exosomes (Exosome-MS) on the Glucocorticoid-induced osteoporosis(GIOP) compared with unmechanically stimulated myocytes (C2C12) derived exosomes (Exosomes), we established a dexamethasone-induced osteoporosis model in vivo and in vitro. Cell viability and proliferation were assessed using CCK8 and EDU assays. Osteogenic potential was evaluated through Western blotting, real-time PCR, alkaline phosphatase activity assay, and alizarin red staining. Differential expression of miRNAs was determined by high-throughput sequencing. The regulatory mechanism of miR-92a-3p on cell proliferation and osteogenic differentiation via the PTEN/AKT pathway was investigated using real-time PCR, luciferase reporter gene assay, Western blotting, and immunofluorescence. The therapeutic effects of exosomes were evaluated in vivo using microCT, HE staining, Masson staining, and immunohistochemistry.
Results: In this study, we found that exosomes derived from mechanical stress had a positive impact on the proliferation and differentiation of bone marrow mesenchymal stem cells (BMSCs). Importantly, we demonstrated that miR-92a-3p mimics could reverse dexamethasone-induced osteoporosis in vitro and in vivo, indicating that mechanical stress-induced mouse myoblast-derived exosomes could promote osteogenesis and prevent the occurrence and progression of osteoporosis in mice through miR-92a-3p/PTEN/AKT signaling pathway.
Conclusion: Exosomes derived from mechanical stress-induced myoblasts can promote the proliferation and osteogenic differentiation of bone marrow mesenchymal stem cells through miR-92a-3p/PTEN/AKT signaling pathway, and can have a therapeutic effect on glucocorticoid-induced osteoporosis in mice in vivo.
Keywords: Human bone marrow mesenchymal stem cells, Mechanical stimulation, Exosome, miR-92a-3p, AKT signaling
Graphical Abstract:
Introduction
Osteoporosis is a prevalent bone metabolic disorder characterized by reduced bone mineral density, thinning bones, and an increased susceptibility to fractures and other bone injuries.1 Since the discovery of coronavirus disease-19 (COVID-19) in 2020, dexamethasone has been used in most parts of the world as an effective treatment for respiratory complications caused by COVID-19.2 Glucocorticoids accelerate bone loss in the elderly, making them more prone to Glucocorticoid-induced osteoporosis(GIOP).3 The clinical treatment of osteoporosis has limited efficacy, huge cost, and different degrees of adverse drug reactions.4,5 Therefore, extensive search for the pathogenesis of osteoporosis and search for accurate, economical, practical, cutting-edge and precise treatment methods play an extremely important role in the early detection and treatment of GIOP.
Bone marrow mesenchymal stem cells (BMSCs) are multipotent cells found in the bone marrow with the ability to differentiate into various cell types, including osteoblasts. Their role as precursor cells of osteoblasts is essential for bone development, repair, and overall bone health.6 Osteoblasts are responsible for bone formation and mineralization, and also play an important role in bone tissue regeneration and remodeling.7,8 Mechanical stimulation can promote the regeneration and remodeling of bone tissue, and activate osteoblasts through direct and indirect stimulation.9 Direct stimulation refers to the mechanism by which mechanical stimulation produces force that can directly act on osteoblasts, thereby activating them. For example, mechanical stimulation resulting from muscle movement, such as compression, stretching, bending, torsion, and shear, can directly affect the regeneration of bone tissue.10 Indirect stimulation refers to the mechanism by which mechanical stimulation activates other cells, such as immune cells, endothelial cells, and muscle cells, which then activate osteoblasts. Mechanical stimulation can cause muscle cells to secrete growth factors, such as insulin-like growth factor-1 (IGF-1), basic fibroblast growth factor (FGF-2), interleukin-7 (IL-7), osteonecin and matrix metalloproteinase-2 (MMP-2), which can then activate osteoblasts and promote bone tissue regeneration and remodeling.11 Research has shown that physical activity can directly regulate muscle cells through mechanical force or indirectly regulate bone metabolism through endocrine factors.12
Exosome is a vesicle with a diameter of 30–130nm that can be transferred between various tissues and cells. It has been confirmed that multiple types of cells from different cell lineages can release exosomes.13 By releasing active molecules, such as proteins, lipids, and RNAs, exosomes can affect the function of recipient cells and generally have a similar function to the source cell. MicroRNA (miRNA) in exosomes have received extensive research attention, especially exosomes from various types of cells, such as mesenchymal stem cells (MSCs).14,15 The structure of the membrane of exosomes can protect miRNAs from degradation by RNAases, resulting in the regulation of multiple targets by many functional miRNAs, which are important factors in the research of exosomes miRNA. For example, miR-150-3p in exosomes from bone marrow mesenchymal stem cells (BMSCs) can promote the proliferation and differentiation of osteoblasts, and has important significance in the study of osteoporosis.16
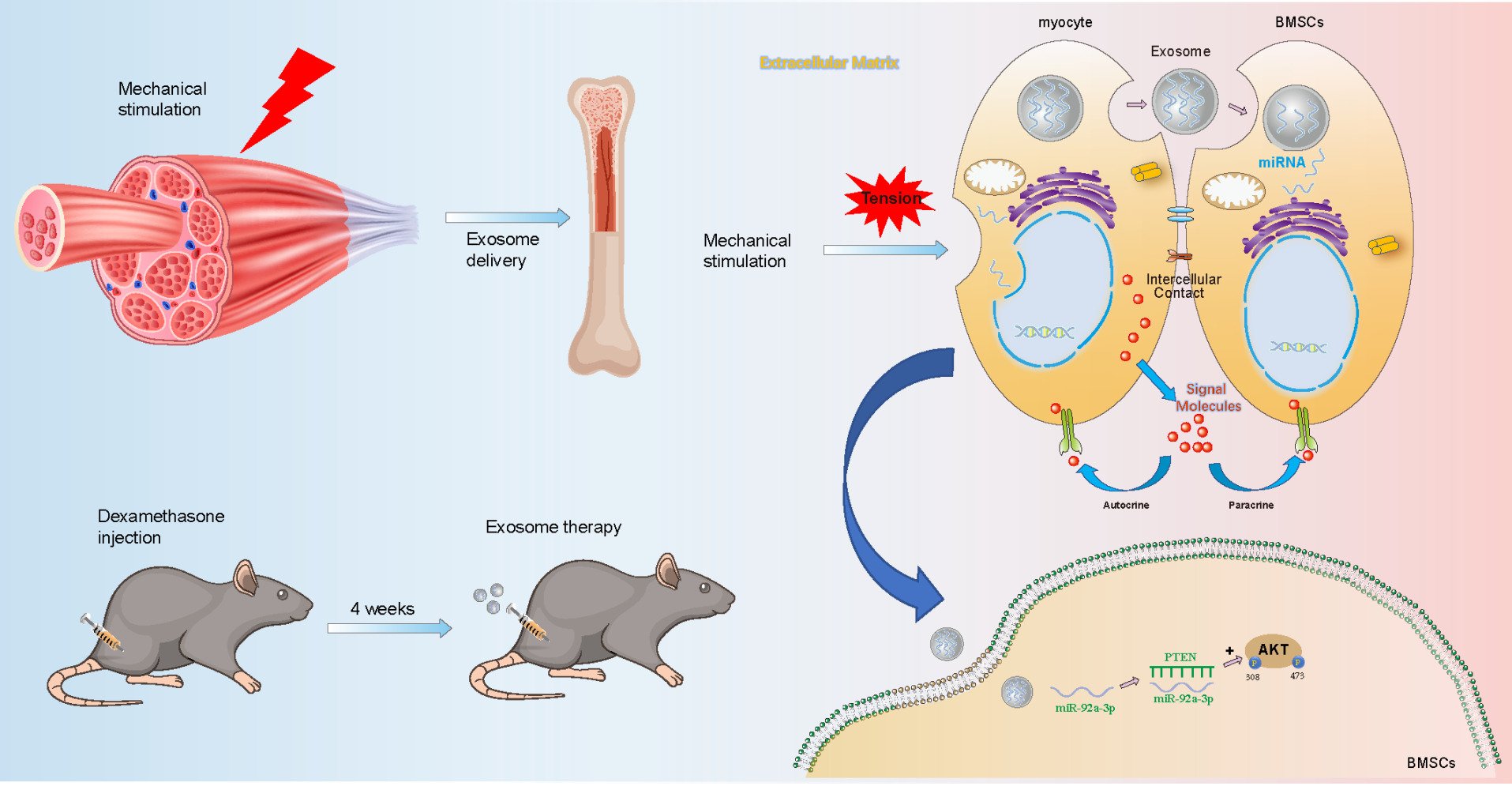
Considering the important role of exosomes in cell crosstalk and the positive effect of mechanical stimulation on osteogenesis, we speculated that mechanical stimulation such as stretching and compression between muscles can promote the secretion of exosomes by muscle cells and regulate bone metabolism. Further analysis was made to determine whether exosome microRNAs produced after mechanical application to muscle cells could also play a role in osteogenesis as special factors regulating bone metabolism. In this study, we provide the first in vitro and in vivo evidence that mechanically strain-induced myoblast-derived exosomes can be transferred to bone marrow mesenchymal stem cells, promote osteogenic differentiation and have a therapeutic effect on mice with hormone-induced osteoporosis. In addition, we found that miR-92a-3p is a key exosome component responsible for this effect.
Materials and Methods
Cell Culture
Human bone marrow mesenchymal stem cells (hBMSCs) and mouse myoblasts (C2C12) were obtained from Wuhan Bost Biotechnology Co., LTD. BMSCs cells were cultured in α-MEM containing 10% fetal bovine serum (FBS, Gibco, California, USA) and 1% penicillin-streptomycin (Gibco, California, USA).17 C2C12 cells were cultured in DMEM, FBS and the ratio of penicillin-streptomycin were the same as BMSCs.18
Osteogenic Differentiation Assay
BMSCs were cultured in osteogenic differentiation medium (Oricell, Guangzhou, China) until day 21.19 The old medium was discarded, and 1 mL Phosphate Buffered Saline (PBS) (Gibco, California, USA) was added to each well for 2–3 rounds of cleaning. Subsequently, 1 mL of 4% paraformaldehyde (Biosharp, Anhui, China) was added to each well for 20 minutes of fixation. The paraformaldehyde was then discarded, and the wells were rinsed with PBS three times. After absorption, 1 mL of 40 mM alizarin red solution (Aladdin, Shanghai, China) was added, and incubation took place at room temperature for 20–30 minutes away from light. Alkaline phosphatase (ALP) activity was detected using the BCIP/NBT alkaline phosphatase color development kit (Beyotime, Shanghai, China).20 Once the color reached the expected shade, the dye solution was aspirated. Excess dye solution was rinsed with PBS, and the culture plates were placed under a microscope to observe the osteogenic staining effect.
Oil Red O Staining Assay
BMSCs were cultured in adipogenic differentiation medium (Oricell, Guangzhou, China) until day 21.21 Aspirate the adipocyte-inducing differentiation complete media from the wells and wash three times with PBS (Gibco, California, USA) to remove residuals. Add 2 mL of 4% paraformaldehyde (Biosharp, Anhui, China) to each well. Then, carefully aspirate the fixative solution and wash the wells with PBS three times to ensure complete removal of the fixative solution. Followed by adding 2 mL of Oil red O staining solution (Cyagen, Suzhou, China) and staining for 30 minutes to show the effect of Oil Red O staining and the location and morphology of adipocyte droplets in the cells. Examine the dried samples under a microscope. Yellow or red colors will appear on substances of lipid nature, which is the result of binding of the Oil Red O dye to lipid substances.
Chondrogenic Differentiation Assay
BMSCs were cultured in chondrogenic differentiation medium (Oricell, Guangzhou, China) until day 21. Upon completion of the incubation period, the old medium was discarded, washed 2–3 times by adding 1mL PBS (Gibco, California, USA), and then fixed by adding 1mL 4% paraformaldehyde (Biosharp, Anhui, China). The cells were washed 2 to 3 times with PBS and then incubated with alcian blue staining solution (Cyagen, Suzhou, China).22 After aspiration of the dye solution, the excess dye solution was rinsed with PBS. Finally, the cartilage staining effect in the culture plates was observed using a microscope.
Mechanical Strain Loading on C2C12 Cells
An appropriate amount of FBS was first selected and then centrifuged in an ultracentrifuge at 120,000×g/min for 12 hours. The supernatant was taken and then filtered in a sterile ultra-clean table using a 0.22μm filter membrane to obtain sterile and exosomes-free FBS. C2C12 cells grown in the logarithmic phase were selected, the density was adjusted to 1×105 / mL, and seeded on Bioflex 6-well culture plates (Flexcell International Corporation, Burlington, NC). Cells were placed in a Flexcell Tension system (FX-5000 T, Flexcell International Corporation, Burlington, NC) to apply traction. The parameters of the FX-5000T application system were set as 10% variable, 0.25Hz frequency and 60 minutes. The supernatant was collected after continued incubation for 24h following the completion of the force application. C2C12 cells without mechanical stimulation were seeded in ordinary 6-well culture plates at the same density of 1×105 / mL. After the cells adhered to the wall, they were cultured for 24 hours, and the supernatant was collected.
Isolation, Purification, and Characterization of Exosomes from the Supernatants of C2C12 Cell Cultures
Differential centrifugation is frequently employed as a prominent method for extracting exosomes, which typically involves three centrifugations: first, 300g ×10 min to remove cell debris, followed by 10000g ×30 min to remove macromolecular impurities, and then the supernatant was collected and centrifuged again at 120000g ×70 min to collect exosomes.23 Finally, the exosomes were stored at −80°C.
Nanoparticle tracking analysis (NTA) was utilized to determine the size distribution of exosomes. The extracted exosome solution was injected into the sample pool of Zetasizer Nano (Malvern, UK), and PBS was added to 2–3mL, then inserted into the sample chamber and left for 5 minutes until the sample stabilized. Using the accompanying operation procedure, parameter is set to aqueous solvent, default solute parameter. The results were determined automatically using Zeta view software and then exported.
The exosomes were characterized for their morphology and size through the utilization of transmission electron microscopy (TEM) (Hitachi HT7700 TEM, Tokyo, Japan). The exosomes were immobilized with glutaraldehyde and then added to the copper mesh. Next, a 2% phosphotungstic acid solution was added to the copper grid to stain the exosomes. After removal of excess solution, the mesh was dried and then imaged using an imaging system.
To detect the expression of exosome surface-specific proteins CD9 and TSG101, as well as the exosome-negative protein calnexin, Western blotting analysis was employed.
PKH26-Stained Exosome Tracing Assay
This experiment employed the PKH26 cell membrane staining kit (Solarbio, Beijing, China). Exosomes were initially incubated in PKH26 staining solution at room temperature in the dark for 5 minutes. Following termination with complete medium, the mixture was ultracentrifuged at 120,000g for 70 minutes to obtain a red precipitate. After resuspension in PBS and filtration through a 0.22μm membrane, the labeled exosomes were added to pre-cultured BMSCs and incubated for 24 hours. Finally, cells were fixed with 4% paraformaldehyde, stained with DAPI for 10 minutes in the dark, and observed under an inverted fluorescence microscope after gentle PBS washing.
Cell Counting Kit-8 Assay
Cell counting kit 8 (MedChemExpress, New Jersey, USA) was used to verify cell proliferation. Firstly, BMSCs were seeded in 96-well plates and cultured. Subsequently, we added treatment reagents to the different test Wells. On each of days 0–3, we removed the cultured cells from one 96-well plate and replaced them with 100 μL complete growth medium and 10 μL CCK-8 solution. The well plates were returned to the incubation box for another 1.5 hours. CCK-8 was reduced to a water-soluble purple product, and we measured the absorbance at 450nm using a microplate reader (Thermo Fisher Scientific, USA) and recorded the data.
5-Ethynyl-2′-Deoxyuridine (EdU) Staining Assay
This method involves the use of the EdU Apollo 567 in vitro kit (Ribobio, Guangzhou, China) to detect the DNA replication activity of BMSCs, further confirming their proliferation rate. This technique seeds 3000 BMSCs/well in a 96-well plate and allows them to attach to the bottom after 24 hours. Cells were treated according to the required conditions, and subsequent related experiments were carried out according to the corresponding instructions of the kit. Images are acquired using an inverted fluorescence microscope.
Western Blotting Assay and Antibodies
Cell and exosome samples were subjected to ice cracking by adding a mixture of Cell lysate (RIPA Lysis buffer, Solarbio, Beijing, China) and Protease inhibitors (Phenylmethyl sulfonyl fuoride, Solarbio, Beijing, China) to extract protein samples. The protein concentration of the samples was determined using the BCA Protein Assay Kit (Solarbio, Beijing, China). The protein samples of different groups were electrophoresis on SDS-PAGE in equal amounts. Following electrophoresis, the PVDF membranes were transferred and subsequently blocked with 5% skim milk for 2 hours. The membranes were then incubated overnight at 4°C with primary antibodies. The PVDF membrane was placed in secondary antibody (Goat Anti-Rabbit IgG H&L, Abcam, ab6721, 1:5000) for 1h. TBST was washed again for three times, and Amersham Imager 680 ultra-sensitive multifunctional imager was used for visualization and detection of protein bands. Normalized with GAPDH. The following antibodies were used: anti-ALP (Abcam, ab307726, 1:1000), anti-BMP2 (Abcam, ab284387, 1:1000), anti-Runx2 (Abcam, ab23981, 1:1000), anti-Osteopontin (Abcam, ab214050, 1:1000), anti-GAPDH (Abcam, ab9485, 1:1000), anti-Pan-AKT (CST, 4691, 1:1000), anti-Phospho S473-AKT (CST, 4060, 1:1000), anti-PhosphoT308-AKT (CST, 13038, 1:1000), anti-PTEN (CST, 9559, 1:1000), anti-CD9 (CST, 98327, 1:1000), anti-TSG101 (Abcam, ab30871, 1:1000), anti-Calnexin (Abcam, ab22595, 1:1000). All experiments were conducted in triplicates.
Rt-Pcr
Trizol (Invitrogen, Carlsbad, CA, USA) was mixed with chloroform in a 1:5 ratio and added to the cell sample. The mixture was centrifuged, resulting in the separation of the precipitated RNA into three layers: upper, middle, and lower. The supernatant was aspirated, and isopropyl alcohol was added followed by inverted mixing on ice to remove the water phase. Finally, the RNA was rapidly dried using anhydrous ethanol. The mRNA reverse transcription was carried out using the EVO M-MLV RT Prime for qPCR kit (Accurate Biological, China), while the miRNA reverse transcription was conducted using the miRNA 1st strand cDNA synthesis kit (Accurate Biological, China). Subsequently, qRT-PCR was performed using SYBR PreMix Pro TaqHS qPCR kit (Accurate Biological, China). The Roche Photocycle 480 sequence detection system (Roche Diagnostics, Switzerland) was utilized for the analysis. GAPDH and U6 were employed as internal controls, and the specific primers used for the qRT-PCR can be found in Table 1.
 |
Table 1 mRNA and miRNAs Primer Sequences |
High-Throughput miRNA Sequencing
To determine miRNA expression levels in exosomes and exosome-MS, we employed high-throughput RNA sequencing. Fragments of total RNA ranging from 18 to 30nt were selected through electrophoretic cutting on agarose gels. After ligating small RNAs to both ends of the 3’ and 5’ splice regions, PCR amplification was performed to selectively amplify the ligated fragments. The resulting PCR products were then purified using agarose gel electrophoresis and utilized for subsequent library construction. Quality control of the constructed library was performed using Agilent 2100 and qPCR, and the final library was sequenced using Illumina Novaseq6000. We utilized edgeR software for differential miRNA analysis, with screening criteria including a more than 2-fold change in expression level and P < 0.05. Gidio Biotechnology Co., Ltd. (Guangzhou, China) provided the high-throughput sequencing services and performed bioinformatics analyses.
Transient Transfection
After BMSCs were inoculated for 24h, miR-92a-3p mimic, mimic NC, miR-92a-3p inhibitor, and inhibitor NC(Ribobio, China) were transiently transfected into cells using the riboFECTTMCP transfection kit (Ribobio, China). Briefly, miR-92a-3p mimic (50 nM), mimic NC (50 nM), inhibitor (100 nM), and inhibitor NC (100 nM) were mixed with riboFECTTMCP reagent and riboFECTTMCP buffer for 15 min, and the following sequences were used: miR-92a-3p mimic 5 “-UAUUGCACUUGUCCCGGCCUGU-3” and miR-92a-3p inhibitor 5 “-ACAGGCCGGGACAAGUGCAAUA-3” (RiboBio, Guangzhou, China).
Luciferase Reporter Assay
To identify miR-92a-3p binding sites in wild-type and mutant PTEN, TargetScan, RNAhybrid, mirTarBase, and Miranda were used. In a luciferase reporter assay, BMSCs were seeded into 12-well plates. The pmir-GLO dual luciferase miRNA target expression vector (containing wild-type or mutant PTEN 3’-UTR, Yibaike, Beijing, China) and related conditions were co-transfected, and the luciferase activity was detected using the dual luciferase reporter kit (Promega, USA) after transfection.
Animal Model
A cohort of sixty male C57BL/6J mice, aged 8 weeks, was procured from Beijing Weitonghe Laboratory Animal Technology Co., LTD. All animal experiments conducted in this study received approval from the Animal Ethics Committee of the Provincial Hospital Affiliated to Shandong First Medical University. The randomization is performed as follows. After the mice arrived from the company, a total of 60 animals were randomly assigned to 12 cages. The cages are given numerical names based on their position on the shelf. The mice were then randomly labeled from 1 to 60, and the cages were then randomly assigned between the exposed and control groups. Which were randomly divided into six groups: (1) control group (PBS-treated rats), (2) Dex group, (3) Dex+Exosome group, (4) Dex+Exosome+miR-92a-3p agomir group, (5) Dex+Exosome-MS group, and (6) Dex+Exosome-MS+miR-92a-3p antagomir group. Each group contained 10 mice (n=10). To induce GIOP in mice, dexamethasone (100mg/kg/day) was injected into both thigh muscles for 4 weeks, and miRNA was injected at the same site on days 1, 3, and 7 after modeling, with a single injection of 20μmol/L in 100 μL PBS. Exosomes and Exosome-MS 100μg/ mouse were also injected simultaneously at the same site. The team assessed health by taking weight measurements twice a week, food and water intake once a day, and general information about the animals’ activity and fur condition. Mice with body weight of 25±5 g were selected for data statistics. Animals are excluded if they die prematurely, preventing the collection of behavioral and histological data. After 4 weeks, the mice were sacrificed and the degree of tibial plateau osteoporosis was evaluated by micro-CT and histomorphological analysis. The analyst was unaware of the grouping of mice.
Biophotonic Imaging Analysis
Exo-MS was incubated with 1 mM fluorescent lipophilic tracer DiR (1, 1-octadecyl-3,3,3, 3-tetramethylindotricarbamine iodide) (Invitrogen, D12731, Life Technologies) at room temperature in the dark for 15 minutes. The exosomes were then separated at 120000g×70 min and the excess dye was washed. Exo-MS with a concentration of 100μg/100μL/mice was injected into the anesthetized mice via the tail vein. In vivo imaging was performed 24 h after injection, after which the mice were sacrificed and the relevant organs were removed. In the control group, 100ul PBS was injected into the tail vein and observed 24 hours later. DiR-EVs distribution was analyzed using an IVIS Spectrum system (Perkin Elmer).24
Micro-CT Analysis
Bone scanning was performed by using an in vivo micro-CT microscopic imaging system (Scanco, Switzerland). In brief, we used microCT to scan the tibial plateau to reconstruct its three-dimensional structure of the tibial plateau and performed calculations using the μCT Evaluation Program. Bone volume fraction (BV/TV, BV: Bone volume, TV: Total volume), trabecular number (Tb.N), trabecular thickness (Tb.Th) and trabecular spacing (Tb.Sp) were used to determine the bone quality.
Hematoxylin and Eosin (H&E) Staining
The mice were euthanized, and the muscles were dissected to extract the tibia. The extracted tibia was fixed in 4% paraformaldehyde and then subjected to decalcification using 10% EDTA. After decalcification, the samples were embedded in paraffin and sectioned. The sections underwent dealkylation in xylene for 10 minutes followed by dehydration in ethanol. Generally, the concentration gradually increased from low concentration to high concentration, and the control of the density and time of each ethanol had a great influence on the staining results. Each soaking time was 2min, and the sample tissue was cleaned with dimethyl tetraacetate and other media to make the sample tissue transparent. The tissue samples were immersed in hematoxylin solution and stained for 5 min to stain out the nuclear and cytoplasmic colors, followed by washing three times with PBS. Following a 1-minute treatment with 5% acetic acid, the cells were stained with eosin for an additional 1 minute. Subsequently, the stained cells were visualized and captured using a microscope.
IHC Staining
After fixation in 4% paraformaldehyde, tibial samples were embedded in paraffin and sectioned. The samples were subsequently dealkylated in xylene and rehydrated with ethanol in a graded fashion. Then, As a catalase quenching agent, 3% hydrogen peroxide was used, and the tissue sections were blocked with 10% goat serum (Sigma-Aldrich, St. Louis, Missouri, USA) for 1 hour at 4°C before being incubated with each antibody overnight. Samples were further stained with biotin-labeled secondary antibodies (Proteintech, SA00004-6, 1:100) and stained with diamine benzidine followed by counterstaining with hematoxylin. The antibodies used were as follows: anti-PTEN (CST, 9559T, 1:200) and anti-Osteopontin (Proteintech, 22953-1-AP, 1:200).
Masson Staining
Tibial specimens were fixed in 4% formaldehyde for 7 d before embedding in paraffin. After cutting the samples into 1-mm sections and de-alkanizing them in xylene for 10 minutes, they were rehydrated with ethanol and stained with Weigert’s iron hematoxylin. They were further soaked in 1% hydrochloric acid-ethanol and rinsed with water. The tissue sections were then stained with Ponceau solution in the Masson staining kit (Servicebio, G1006, China) for 5 minutes. After the cells were treated with phosphomolybdic acid, they were stained again with aniline blue solution.
Statistical Analysis
The data were expressed as mean ± standard deviation (SD). Statistical analyses were conducted using GraphPad Prism 8.0 software (La Jolla, CA, USA). One-way analysis of variance (ANOVA) combined with Tukey’s multiple comparison test was used for comparison between groups. The significance of the differences between the two groups was determined using a two-sided unpaired t-test. Statistical significance was indicated by p < 0.05. Symbols *p < 0.05, **p < 0.01, ***p < 0.001, and NS denoted no statistical significance.
Results
Characterization of BMSCs
When the cells were placed under the microscope, we found that the BMSCs were typical spindle-shaped and grew in a radial and whirlpool shape (Figure S1A). In osteogenic, chondrogenic, and adipogenic differentiation assays, induced BMSCs displayed mineralization nodules, cartilage tissue, and lipid droplets (Figure S1B–D). Furthermore, Mesenchymal stem cell markers CD105, CD90, CD29 and CD73 were positively expressed, but CD45 and CD34 were negatively expressed (Figure S1E). These results indicate that the cells we obtained are BMSCs.
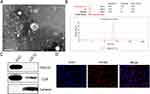
Identification of C2C12-Derived Exosomes
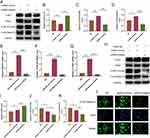
We used three different methods to identify the extracted exosomes. TEM analysis showed that the C2C12-derived exosome was round in appearance and about 100 nm in size (Figure 1A). According to NTA analysis, these particles are between 30 and 70 nm in size (Figure 1B). The Western blot analysis showed C2C12-Exos to be positive for exosome surface markers, CD9 and TSG101, and negatively for Calnexin, an endoplasmic membrane protein (Figure 1C). Additionally, exosomes labeled PKH26 were absorbed by BMSCs (Figure 1D). These data indicated that exosomes were successfully extracted and internalized by BMSCs, which laid the foundation for the following experiments.
Proliferation of BMSCs by Exosome-MS
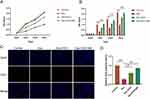
A study of the effects of exosomes originated from C2C12 cells under mechanical strain (Exosome-MS) on the proliferation of BMSCs treated with dexamethasone was conducted. In the CCK-8 experiment, the precultured cells were treated with 10−4 M dexamethasone for 0–3 days, and the EDU experiment was treated with the same concentration of dexamethasone for 24 hours. The above experiments were treated with 50μg/mL exosomes or exosome-MS for correlation analysis. CCK-8 results showed that dexamethasone treatment significantly decreased cell proliferation from day 1 onwards (P < 0.05). However, exosomes and exosomes-MS rescued the dexamethasone-induced downregulation of proliferation, but exosomes-MS was more effective than exosomes (Figure 2A and B). Similarly, EdU assay showed that the effect of exosome-MS on cell proliferation was similar to that detected by CCK-8 (Figure 2C and D). These results confirmed that mechanically stimulated exosomes could significantly promote the proliferation of BMSCs under dexamethasone inhibition compared with conventional exosomes.
Osteogenic Differentiation of BMSCs by Exosome-MS
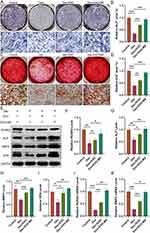
This part describes the osteogenic differentiation of BMSCs under dexamethason treatment using exosomes derived from C2C12 cells subjected to mechanical stimulation. In the following experiments, dexamethasone was used at a concentration of 10−4 M, and 50μg/mL was used for exosomes or exosome-MS. We cultivated BMSCs in osteogenic differentiation medium and evaluated their ability to form calcium nodules using alizarin red staining. According to ALP staining results, it was observed that the Dex group exhibited the smallest staining area, while the Dex+Exo-MS group showed a significantly larger staining area compared to both the Dex group and Dex+Exo group (Figure 3A and B). The findings from alizarin red staining were in agreement with the ALP staining results (Figure 3C and D). In dexamethasone-treated cells, the Western blot analysis revealed elevated protein expression levels in the Dex+Exosome-MS group and Dex+Exosome group compared to the Dex group. Furthermore, the Dex+Exosome-MS group exhibited higher expression levels than the Dex+Exosome group. This difference was statistically significant (Figure 3E–I). The results of RT-PCR were similar to those described above (Figure 3J and K). Even in a dexamethasone-induced environment, exosome-MS has been shown to promote osteogenic differentiation of BMSCs, as indicated by these findings.
Analysis of Functional Attributes Related to Gene Sets and Signaling Pathways in Exosome-MS
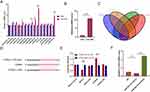
We utilized high-throughput sequencing to examine the molecular mechanisms implicated in exosomes-MS interactions. The heat map displays all significantly differentially expressed miRNAs (Figure 4A). In comparison to exosomes, Exosome-MS exhibited a total of 47 up-regulated targets and 44 down-regulated targets, which were represented in the heat map (Figure 4B and C). According to the GO enrichment analysis, miRNAs in Exosome-MS were found to be involved in crucial roles related to signal transduction, cell communication, multicellular biological processes, and cellular response to stimulus (Figure 4D). In addition, Gene Ontology analysis revealed that Exosome-MS primarily modulates gene networks associated with cellular processes, biological regulation, metabolic processes, and stimulus response within biological processes. At the same time, it mainly regulates catalytic activity, binding and molecular transducer activity in molecular function (Figure 4E). Further KEGG pathway enrichment analysis of Exosome MS-mediated gene expression showed that signaling pathways related to osteogenesis, including RAS, cAMP, and signaling pathways regulating stem cell pluripotency, were enriched (Figure 4F) In addition, Signaling pathway functions are mainly involved in environmental information processing and in human disease (Figure 4G). The chromosome mapping showed the mutation location and proportion of related genes on the chromosome (Figure 4H).
miR-92a-3p Targets to PTEN
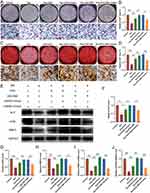
After co-culturing exosomes and exosome-MS with pre-cultured BMSCs for 48 hours, qRT-PCR analysis was performed on the top 10 upregulated and 5 downregulated miRNAs with the highest expression levels from high-throughput sequencing to detect differential expression in the cells. The results revealed an upregulation of miR-92a-3p expression specifically in the exosome-MS group (Figure 5A). RT-PCR analysis was carried out to authenticate the extracted exosomes and exosomes-MS samples, uncovering an elevation in miR-92a-3p expression specifically within the exosomes-MS samples (Figure 5B). By utilizing miRTarBase, RNAhybrid, TargetScan, and miRnada databases, we were able to predict the target genes of miR-92a-3p. This analysis yielded a total of 109 intersecting genes. Our research uncovered significant interactions between miR-92a-3p and both the wild-type and mutant forms of PTEN (Figure 5C and D). These interactions were subsequently validated through a luciferase assay, reinforcing our findings (Figure 5E). RT-PCR analysis demonstrated that the overexpression of miR-92a-3p inhibited the expression of PTEN. Conversely, the inhibition of miR-92a-3p resulted in an upregulation of PTEN expression (Figure 5F).
The PTEN/AKT Signaling Pathway is Involved in the Regulatory Role of Exosomal miR-92a-3p on the Proliferation of BMSCs
To elucidate the mechanism by which exosomal miR-92a-3p promotes the proliferation of BMSCs, we performed Western blot analysis to assess the phosphorylation status of AKT as well as the expression levels of PTEN (Figure 6A–D). Treatment with miR-92a-3p mimics led to a decrease in PTEN expression, indicating its downregulation. On the other hand, there was a noticeable increase in the phosphorylation levels of AKT at both Thr308 and Ser473 sites. On the contrary, when the group was treated with the miR-92a-3p inhibitor, the findings revealed a contrasting observation. Furthermore, we employed immunofluorescence technology to assess the expression of p-AKT (Ser473) (Figure 6L). According to the results, the fluorescence intensity of the miR-92a-3p mimic-treated group was found to be increased, while the fluorescence intensity of the miR-92a-3p inhibitor-treated group was observed to be decreased, when compared to the control group. Furthermore, RT-PCR was utilized to examine the expression levels of key genes involved in the PTEN/PI3K/AKT signaling pathway, namely CCDN1, PI3KCA, and PI3KCB. According to the findings, the miR-92a-3p mimics group exhibited an up-regulation of CCDN1, PI3KCA, and PIK3CB, whereas the miR-92a-3p inhibitor group displayed a down-regulation of these genes (Figure 6E–G). Furthermore, Western blot results revealed that the introduction of PTEN overexpressing lentivirus into the mimics group led to a significant decrease in P-AKT expression and a noticeable increase in PTEN expression in the miR-92a-3p mimics group, as compared to Figure A (Figure 6H–K). The results revealed that miR-92a-3p functions as a suppressor of PTEN, leading to AKT phosphorylation and subsequent modulation of downstream components in the signaling cascade.
Exosomal-MS Regulates BMSCs Osteogenic Differentiation via miR-92a-3p
To explore the underlying mechanism by which exosome-MS enhances the osteogenic differentiation of BMSCs in the presence of dexamethasone, we employed ALP staining (Figure 7A and B) and alizarin red staining as assays for verification (Figure 7C and D). Furthermore, to assess the expression levels of BMP2, ALP, and OPN, Western blotting and RT-PCR techniques were employed (Figure 7E–J). The concentration of dexamethasone was 10−4 M, and 50μg/mL was used for exosomes or exosomes-MS in related experiments. In the Dex+Exosome+miR-92a-3p mimics group, an increase in the expressions of BMP2, Runx2, ALP, and OPN was observed compared with the Dex+Exosome group. Likewise, Dex+Exosome-MS+inhibitor downregulated the expression of the same genes relative to Dex+Exosome-MS. These findings were further supported by the consistent results obtained from the ALP staining and alizarin red staining. Collectively, these findings strongly imply that exoses-MS is crucial in regulating bone formation and differentiation by regulating miR-92a-3p.
Exosome-MS Regulates Mouse GIOP Through miR-92a-3p
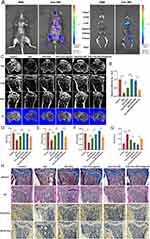
In order to assess the in vivo osteogenic potential of exosome-MS, a study was conducted to explore the interactions between exosome-MS and the GIOP model. To validate the functional effects of exosomes in mouse skeleton, in vivo imaging experiments were performed. The results showed that Exo-MS had specific dye enrichment in the bones of both lower limbs, indicating that Exo-MS could play a role in bone (Figure 8A). Subsequently, in order to verify the expression of miR-92a-3p in the tibia of mice, we performed qRT-PCR experiments on the tibia tissues of mice. The results showed that the expression of miR-92a-3p was the lowest in the Dex group, and the expression of miR-92a-3p in the Dex+Exo+miRNA agomir group was significantly higher than that in the Dex+Exo group. The expression of miR-92a-3p in Dex+Exo+miRNA antagomir group was significantly lower than that in Dex+Exo-MS group. These results indicate that miR-92a-3p can be expressed in vivo and plays an important role in osteogenesis (Figure 8B). MicroCT results showed that there was almost no trabecular formation in the Dex group and that the repair of trabeculae was facilitated by exosomes, which was further enhanced by the application of exosome-MS and miR-92a-3p agomir. The miR-92a-3p antagomir inhibited bone trabecular repair compared to the exosome-MS group (Figure 8C). Therefore, dexamethasone-induced bone loss was significantly reversed by the administration of the miR-92a-3p agomir and exosom-MS. In addition, the BV/TV, Tb.N and Tb.Th were significantly lower in the Dex group than in the control group, while the administration of miRNAagomir or exosome-MS significantly increased the above values (Figure 8D–G). The same trend was also observed in the changes of body weight from week 12 to week 16 (Figure S1F). Histological analysis using H&E and Masson staining revealed a notable occurrence of osteonecrosis in the Dex group. Trabecular bone was absent and bone formation was reduced in the tibial plateau. Conversely, the Dex+Exo-MS group and Dex+Exo+Agomir group exhibited a well-organized arrangement of bone trabeculae, and there was a higher amount of newly formed bone tissue. Compared to the two groups mentioned above, the Dex+Exo group and Dex+Exo-MS+Antagomir group exhibited sparse bone trabeculae and a lower amount of newly formed bone tissue, but the condition was better than that of the Dex group. Immunohistochemical staining (IHC) revealed a notable decrease in the expression of OPN in the tibial plateau of the Dex group. However, this decrease could be effectively reversed by exosomes. Notably, the combination of exosomes with miR-92a-3p agomir and exosome-MS demonstrated even more prominent restorative effects on OPN expression. In addition, when miR-92a-3p antagomir was transfected in the Dex+Exo-MS group, the expression of OPN decreased, and the expression of AKT pathway negative regulatory protein PTEN was opposite (Figure 8H). These data suggested that exosome-MS regulates GIOP in a mouse model via miR-92a-3p.
Discussion
The long-term use of glucocorticoids (GC) is associated with a myriad of detrimental side effects, with osteoporosis being one of the most significant among them. This condition dramatically heightens the vulnerability to fractures.25 In recent years, some studies have provided insights into the molecular mechanisms of GIOP occurrence and development, including increased apoptosis of mature osteoblasts and osteocytes, impairment of osteoblast differentiation, and prolonged lifespan of osteoclasts.26 BMSCs serve as pivotal cellular components in the process of bone regeneration. According to O’Brien et al, osteoblasts and osteocytes are direct targets of glucocorticoid action in vivo, and GCs can stimulate apoptosis, leading to decreased bone formation and bone strength.27 In our research, it was observed that glucocorticoids (GCs) exerted a substantial inhibitory impact on both the proliferation and osteogenic activity of BMSCs.
The mechanical coupling between bone and skeletal muscle is of great importance, with muscle serving as the primary load-bearing tissue and bone serving as the primary attachment site. In recent years, research has gradually emerged demonstrating that muscle and bone have important endocrine and paracrine functions, which provides a better direction for studying bone and muscle-related diseases and may lead to unprecedented treatments. Muscle load can be directly expressed on bone, which adjusts its shape and mass in response to the mechanical action of muscles. This principle encompasses both the mechanical effect of muscle on bone and the important role of muscle on the endocrine function of bone. Previous studies have found that muscle and bone have the role of endocrine organs, and muscle can secrete “muscle factor” to regulate bone metabolism when subjected to mechanical stimulation. However, all muscle-derived bone regulatory factors found to date are secreted proteins, and there are few studies on exosomes.28 In a recent study, it was demonstrated that C2C12 myoblast exosomes, which carry miR-27a-3p, have the capacity to enhance the osteogenic differentiation of MC3T3-E1 preosteoblasts.23 Furthermore, in a separate investigation, the exosome Prrx2 derived from C2C12 myoblasts was observed to offer potential osteoporosis mitigation by activating the Hippo pathway through the modulation of lncRNA-MIR22HG transcription.29 All the above studies have investigated exosomes as modulators of bone metabolism in the musculoskeletal system, but the effect of myocell-derived exosomes induced by mechanical stimulation on bone remodeling has not been investigated. Our findings revealed that C2C12-Exos-MS stimulated the proliferation and differentiation of BMSCs by activating the miR-92a-3p/PTEN/AKT signaling pathway. This pathway holds promising potential as a therapeutic target for addressing osteoporosis.
Mechanical stimulation plays a crucial role in regulating bone mass and strength. During critical stages of growth and development, increasing the mechanical load can lead to bone mineral accumulation, increased bone mass, and strengthened bones. In contrast, reducing the mechanical load can result in loss of bone mass and strength.30 A previous study demonstrated that mechanical stimulation of myoblasts can modulate exosomes secretion. Mullen et al found that mechanical strain increased the total production of exosomes from C2C12 cells and could be regulated by different tensile loading methods. Additionally, their findings demonstrated that exosomes derived from cells exposed to mechanical stimulation enhanced both the proliferation and myogenic differentiation of naive C2C12 cells.31 Additionally, research has demonstrated the osteogenic potential of exosomes in facilitating bone regeneration and repair processes. For instance, Lu et al conducted a study showcasing the ability of exosomal miR-29a derived from BMSCs to enhance both angiogenesis and osteogenesis in experimental settings both in vitro and in vivo.32 Moreover, the study conducted by Lai et al provided evidence showing that the upregulation of miR-27a-3p and miR-196b-5p in exosomes derived from hBMSCs resulted in the downregulation of osteoclast markers, thereby exerting a beneficial effect on bone remodeling.33 Based on the above studies, we hypothesised that muscle under mechanical stimulation can promote exosome secretion and promote bone formation. Our relevant experiments also confirmed this hypothesis. Following that, our research proceeded with bioinformatics analysis and RNA-seq to explore the underlying mechanism behind the impact of Exosome-MS on cell proliferation and bone formation. Notably, among the differentially expressed miRNAs, miR-92a-3p showed the highest level of differential expression. Despite RT-PCR also showing an increase in miR-351-5p in BMSCs, contrary to sequencing results, we opted not to investigate miR-351-5p further and instead proceeded with miR-92a-3p. The results from both Figure 6A and B indicate that the expression of miR-92a-3p is not only high in exosome-MS but also increases in BMSCs after the addition of exosome-MS. Additionally, Figure 1D demonstrates that exosomes can be internalized by cells, indicating that miR-92a-3p can be transferred to and exert its function in mesenchymal stem cells. In a recent study conducted by Hu et al, it was observed that patients with both fracture and traumatic brain injury (TBI) exhibited heightened levels of miR-92a-3p expression. Notably, the healing process of fractures was found to be markedly accelerated in these individuals as compared to those with sole fractures. This study provides additional evidence supporting the significance of miR-92a-3p in the regulation of osteogenic differentiation.34 Nonetheless, up until now, there has been a dearth of studies investigating the impact of exosomal miR-92a-3p on osteoporosis. Considering the incomplete complementarity between miRNA and non-specific target genes or RNA sequences, leading to the regulation of these non-specific genes by miRNA and resulting in off-target effects.35 To avoid the off-target effects of miR-92a-3p, we conducted experiments using miRNA mimics and miRNA agomir. These mimics have a more stable and specific structure, which can reduce off-target effects. In order to fill this research gap, we conducted a thorough database search, eventually selecting PTEN as the target gene due to its strong correlation, to further validate its involvement. Previous research has substantiated that PTEN functions as a negative regulator within the PI3K/AKT signaling pathway. This pathway is primarily responsible for governing cell growth and survival, while also playing a significant role in cellular processes such as proliferation, differentiation, and osteogenesis.36,37 Our study elucidates the role of exosomal miRNA-92a-3p in osteoporosis and proposes a new mechanism by which miRNA-92a-3p promotes osteogenesis by targeting PTEN.
The PI3K/AKT signaling pathway serves as a crucial modulator of diverse cell phenotypes, spanning multiple diseases’ pathogenesis. Multiple studies have confirmed the intricate association between the PI3K/AKT signaling pathway and the intricate processes governing bone metabolism. Consequently, these findings indicate that promoting the activation of the PI3K/AKT pathway could hold therapeutic potential for mitigating osteoporosis and other disorders affecting bone health. Recent experimental research conducted by Zhao et al uncovered a significant role played by the macrophage scavenger receptor 1 (MSR1) in mediating the PI3K/AKT/GSK3β/β-catenin signaling pathway.38 Yang et al have unraveled the pivotal role of miRNA-21 in driving maxillofacial bone regeneration. Their investigations shed light on the PTEN/PI3K/Akt/HIF-1α signaling pathway, which serves as a key mediator of miRNA-21’s pro-regenerative effects.39 This crucial pathway has been found to enhance the osteogenic differentiation capacity of bone marrow mesenchymal stem cells, shedding light on the potential therapeutic implications for promoting bone formation. Our findings indicate that the upregulation of miRNA-92a-3p effectively suppresses PTEN activity, leading to the activation of the PI3K/AKT pathway and enhanced expression of target genes in osteoblasts. These insights deepen our understanding of the regulatory mechanisms involved in bone formation and present potential avenues for therapeutic intervention in promoting bone regeneration.
To summarize, our research focused on comparing the effects of C2C12-Exo-MS and C2C12-Exo in promoting the proliferation and osteogenesis of BMSCs. Furthermore, we examined the impact of C2C12-Exo-MS on miR-92a-3p/PTEN/AKT signaling in vivo and in vitro, shedding light on musculoskeletal crosstalk regulation by myofactors. This study offers a fresh perspective on the interplay between different cell types and provides insights for understanding and manipulating bone and muscle interactions. It also highlights the possibility of using muscle exosomes as indicators and interaction targets for bone remodeling, providing more directions and possibilities for osteoporosis treatment. Considering that mechanically induced exosomes derived from C2C12 cells can enhance bone density and improve trabecular bone structure, they are likely to have a positive therapeutic effect on other types of bone loss models, including ovariectomy-induced osteoporosis. However, this study was only conducted in mice, and it is hoped that large-scale animal experiments can be carried out in the future and gradually applied to the clinic to provide more options for the treatment of hormone-induced osteoporosis.
Conclusions
In conclusion, exosomes derived from myoblasts under appropriate mechanical stimulation can promote osteogenesis of BMSCs in vitro and treat GIOP in mice in vivo. The current findings not only implicate miR-92a-3p/PTEN/AKT signaling as a potential novel mechanism to promote bone metabolism and bone homeostasis through intercellular communication of muscle cell-derived exosomes, but also reveal the therapeutic effect of mechanical stress-induced muscle cell-derived exosomes as a treatment for GIOP.
Ethics Approval
The animal experiments involved in this experiment were approved by the Experimental Animal Ethics Committee of the Shandong Provincial Hospital Affiliated to Shandong First Medical University. All animal studies complied with the principles based on the International Guiding Principles for Biomedical Research Involving Animals.
Funding
This study received funding from the National Natural Science Foundation of China (NO. 82002302), the Natural Science Foundation of Shandong Province (NO. ZR2021MH040 and ZR2020QH074), the China Postdoctoral Science Foundation (2022M721986), the Clinical medicine technology innovation program of Jinan (NO. 202225048) and the Medical and health technology development program of Shandong Province (NO. 2019WS467).
Disclosure
All authors declared no competing of interests in this work.
References
1. Rachner TD, Khosla S, Hofbauer LC. Osteoporosis: now and the future. Lancet. 2011;377(9773):1276–1287. doi:10.1016/S0140-6736(10)62349-5
2. Vogel F, Reincke M. Endocrine risk factors for COVID-19: endogenous and exogenous glucocorticoid excess. Rev Endocr Metab Disord. 2022;23(2):233–250. doi:10.1007/s11154-021-09670-0
3. Tang J. COVID-19 pandemic and osteoporosis in elderly patients. Aging Dis. 2022;13(4):960–969. doi:10.14336/AD.2021.1201
4. Liel Y. Teriparatide vs risedronate for osteoporosis. Lancet. 2018;391(10133):1895. doi:10.1016/S0140-6736(18)30754-2
5. Reid IR. Denosumab for glucocorticoid-induced osteoporosis. Nat Rev Endocrinol. 2018;14(12):383–384. doi:10.1038/s41574-018-0035-z
6. Charbord P. Bone marrow mesenchymal stem cells: historical overview and concepts. Hum Gene Ther. 2010;21(9):1045–1056. doi:10.1089/hum.2010.115
7. Hu K, Olsen BR. Osteoblast-derived VEGF regulates osteoblast differentiation and bone formation during bone repair. J Clin Invest. 2016;126(2):509–526. doi:10.1172/JCI82585
8. Ponzetti M, Rucci N. Osteoblast differentiation and signaling: established concepts and emerging topics. Int J Mol Sci. 2021;22(13):6651. doi:10.3390/ijms22136651
9. Tagliaferri C, Wittrant Y, Davicco MJ, Walrand S, Coxam V. Muscle and bone, two interconnected tissues. Ageing Res Rev. 2015;21:55–70. doi:10.1016/j.arr.2015.03.002
10. Sharir A, Stern T, Rot C, Shahar R, Zelzer E. Muscle force regulates bone shaping for optimal load-bearing capacity during embryogenesis. Development. 2011;138(15):3247–3259. doi:10.1242/dev.063768
11. Kaji H. Effects of myokines on bone. BoneKEy Rep. 2016;5:826. doi:10.1038/bonekey.2016.48
12. Litmanovitz I, Dolfin T, Friedland O, et al. Early physical activity intervention prevents decrease of bone strength in very low birth weight infants. Pediatrics. 2003;112(1):15–19. doi:10.1542/peds.112.1.15
13. Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200(4):373–383. doi:10.1083/jcb.201211138
14. Koga Y, Yasunaga M, Moriya Y, et al. Exosome can prevent RNase from degrading microRNA in feces. J Gastrointest Oncol. 2011;2(4):215–222. doi:10.3978/j.issn.2078-6891.2011.015
15. Valadi H, Ekström K, Bossios A, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9(6):654–659. doi:10.1038/ncb1596
16. Qiu M, Zhai S, Fu Q, Liu D. Bone marrow mesenchymal stem cells-derived exosomal MicroRNA-150-3p promotes osteoblast proliferation and differentiation in osteoporosis. Hum Gene Ther. 2021;32(13–14):717–729. doi:10.1089/hum.2020.005
17. Chen W, Sun Y, Gu X, et al. Conditioned medium of human bone marrow-derived stem cells promotes tendon-bone healing of the rotator cuff in a rat model. Biomaterials. 2021;271:120714. doi:10.1016/j.biomaterials.2021.120714
18. Wang Y, Liu S, Yan Y, Li S, Tong H. SPARCL1 promotes C2C12 cell differentiation via BMP7-mediated BMP/TGF-β cell signaling pathway. Cell Death Dis. 2019;10(11):852. doi:10.1038/s41419-019-2049-4
19. Liu ZZ, Hong CG, Hu WB, et al. Autophagy receptor OPTN (optineurin) regulates mesenchymal stem cell fate and bone-fat balance during aging by clearing FABP3. Autophagy. 2021;17(10):2766–2782. doi:10.1080/15548627.2020.1839286
20. Sun Y, Zhu Y, Liu X, Chai Y, Xu J. Morroniside attenuates high glucose-induced BMSC dysfunction by regulating the Glo1/AGE/RAGE axis. Cell Prolif. 2020;53(8):e12866. doi:10.1111/cpr.12866
21. Luo H, Guo Y, Liu Y, et al. Growth differentiation factor 11 inhibits adipogenic differentiation by activating TGF-beta/Smad signalling pathway. Cell Prolif. 2019;52(4):e12631. doi:10.1111/cpr.12631
22. Prosser A, Scotchford C, Roberts G, Grant D, Sottile V. Integrated Multi-Assay Culture Model for Stem Cell Chondrogenic Differentiation. Int J Mol Sci. 2019;20(4):951. doi:10.3390/ijms20040951
23. Xu Q, Cui Y, Luan J, et al. Exosomes from C2C12 myoblasts enhance osteogenic differentiation of MC3T3-E1 pre-osteoblasts by delivering miR-27a-3p. Biochem Biophys Res Commun. 2018;498(1):32–37. doi:10.1016/j.bbrc.2018.02.144
24. Wiklander OP, Nordin JZ, O’Loughlin A, et al. Extracellular vesicle in vivo biodistribution is determined by cell source, route of administration and targeting. J Extracell Vesicles. 2015;4(1):26316. doi:10.3402/jev.v4.26316
25. Weinstein RS. Clinical practice. Glucocorticoid-induced bone disease. N Engl J Med. 2011;365(1):62–70. doi:10.1056/NEJMcp1012926
26. Raterman HG, Bultink IEM, Lems WF. Current treatments and new developments in the management of glucocorticoid-induced osteoporosis. Drugs. 2019;79(10):1065–1087. doi:10.1007/s40265-019-01145-6
27. O’Brien CA, Jia D, Plotkin LI, et al. Glucocorticoids act directly on osteoblasts and osteocytes to induce their apoptosis and reduce bone formation and strength. Endocrinology. 2004;145(4):1835–1841. doi:10.1210/en.2003-0990
28. Goodman CA, Hornberger TA, Robling AG. Bone and skeletal muscle: key players in mechanotransduction and potential overlapping mechanisms. Bone. 2015;80:24–36. doi:10.1016/j.bone.2015.04.014
29. Li Y, Wang X, Pan C, et al. Myoblast-derived exosomal Prrx2 attenuates osteoporosis via transcriptional regulation of lncRNA-MIR22HG to activate Hippo pathway. Mol Med. 2023;29(1):54. doi:10.1186/s10020-023-00649-y
30. Gomarasca M, Banfi G, Lombardi G. Myokines: the endocrine coupling of skeletal muscle and bone. Adv Clin Chem. 2020;94:155–218. doi:10.1016/bs.acc.2019.07.010
31. Mullen M, Williams K, LaRocca T, et al. Mechanical strain drives exosome production, function, and miRNA cargo in C2C12 muscle progenitor cells [published online ahead of print, 2022 Oct 17]. J Orthop Res. 2022;41(6):1186–1197. doi:10.1002/jor.25467
32. Lu GD, Cheng P, Liu T, Wang Z. BMSC-derived exosomal miR-29a promotes angiogenesis and osteogenesis. Front Cell Dev Biol. 2020;8:608521. doi:10.3389/fcell.2020.608521
33. Lai G, Zhao R, Zhuang W, et al. BMSC-derived exosomal miR-27a-3p and miR-196b-5p regulate bone remodeling in ovariectomized rats. PeerJ. 2022;10:e13744. doi:10.7717/peerj.13744
34. Hu L, Liu J, Xue H, et al. miRNA-92a-3p regulates osteoblast differentiation in patients with concomitant limb fractures and TBI via IBSP/PI3K-AKT inhibition. Mol Ther Nucleic Acids. 2021;23:1345–1359. doi:10.1016/j.omtn.2021.02.008
35. Singh S, Narang AS, Mahato RI. Subcellular fate and off-target effects of siRNA, shRNA, and miRNA. Pharm Res. 2011;28(12):2996–3015. doi:10.1007/s11095-011-0608-1
36. Downes CP, Ross S, Maccario H, et al. Stimulation of PI 3-kinase signaling via inhibition of the tumor suppressor phosphatase, PTEN. Adv Enzyme Regul. 2007;47(1):184–194. doi:10.1016/j.advenzreg.2006.12.018
37. Li H, Liu D, Li C, et al. Exosomes secreted from mutant-HIF-1α-modified bone-marrow-derived mesenchymal stem cells attenuate early steroid-induced avascular necrosis of femoral head in rabbit. Cell Biol Int. 2017;41(12):1379–1390. doi:10.1002/cbin.10869
38. Zhao SJ, Kong F-Q, Jie J, et al. Macrophage MSR1 promotes BMSC osteogenic differentiation and M2-like polarization by activating PI3K/AKT/GSK3β/β-catenin pathway. Theranostics. 2020;10(1):17–35. doi:10.7150/thno.36930
39. Yang C, Liu X, Zhao K, et al. miRNA-21 promotes osteogenesis via the PTEN/PI3K/Akt/HIF-1α pathway and enhances bone regeneration in critical size defects. Stem Cell Res Ther. 2019;10(1):65. doi:10.1186/s13287-019-1168-2
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.