Back to Journals » Journal of Multidisciplinary Healthcare » Volume 15
The Search for Cyclooxygenase-2 (COX-2) Inhibitors for the Treatment of Inflammation Disease: An in-silico Study
Authors Ruslin R , Yamin Y , Kasmawati H , Mangrura S, Kadidae L, Alroem A, Arba M
Received 25 January 2022
Accepted for publication 22 March 2022
Published 12 April 2022 Volume 2022:15 Pages 783—791
DOI https://doi.org/10.2147/JMDH.S359429
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Scott Fraser
Ruslin Ruslin,1 Yamin Yamin,1 Henny Kasmawati,1 Samuel Mangrura,2 Laode Kadidae,2 Armid Alroem,2 Muhammad Arba1
1Faculty of Pharmacy, Universitas Halu Oleo, Kendari, 93232, Indonesia; 2Department of Chemistry, Faculty of Mathematics and Natural; Sciences, Universitas Halu Oleo, Kendari, 93232, Indonesia
Correspondence: Ruslin Ruslin, Email [email protected]
Purpose: Cyclooxygenase (COX-2) has been validated as a molecular target for treating inflammatory diseases. The present work was performed to identify potential COX-2 inhibitors by employing pharmacophore modeling.
Methods: The pharmacophore features consisted of seven features, ie, three hydrophobic, one negative ion, and three hydrogen bond acceptors, which were developed based on the structure of COX-2 inhibitor, (R)-naproxen.
Results: The pharmacophore model was validated with a Goodness of Hit (GH score) of 0.754 and the values of AUC100% 0.51. Screening against the ZINC database retrieved 1675 hits, while the molecular docking procedure identified four best hit molecules in term of binding orientation and binding energies, ie, Lig_1805/ZINC103584272 (E = − 11.03 kcal/mol), Lig_553/ZINC408573132 (E = − 10.92 kcal/mol), Lig_680/ZINC103584263 (E = − 10.90 kcal/mol), and Lig_2006/ZINC19324645 (E = − 10.62 kcal/mol).
Conclusion: The interactions of the four hits occurred in the binding site as (R)-naproxen did, and interestingly, their binding affinities were stronger than (R)-naproxen, implying their potential as COX-2 inhibitors.
Keywords: COX-2 inhibitor, inflammation disease, molecular docking, pharmacophore modeling, ZINC database
Introduction
Inflammation is the body’s natural response associated with infection, irritation, or tissue damage. It is limited to the affected tissues under normal conditions; however, in several cases, it could become uncontrolled conditions such as those occurring in asthma, nephropathy, type-2 diabetes mellitus, cancers, and Alzheimer’s disease.1 Inflammation treatment involves narcotic (opioids) analgesic agents and Nonsteroidal Anti-inflammatory Drugs (NSAID). The cyclooxygenase 2 (COX-2) inhibitors have been in the main course for the treatment of inflammation. Inflammation treatment involves narcotic (opioids) analgesic agents and Nonsteroidal Anti-inflammatory Drugs (NSAID). The cyclooxygenase 2 (COX-2) inhibitors have been the main course for treating inflammation. Cyclooxygenase (COX-2) is widely known for its crucial role in converting arachidonic acid to prostaglandins (PG), which is necessary for the inflammation process.2
Several inhibitors of the COX enzyme have been used for decades to treat inflammation. They include celecoxib, etoricoxib, ketorolac, naproxen, acetylsalicylic acid, and indomethacin.3 However, during their use, several side effects such as bronchospasm, vasomotor and angioedema, renal dysfunction, meningeal syndrome, bone marrow depression, headache, and vertigo.4–6 Thus, there is increasing concern over the discovery of novel COX-2 inhibitors having more potent and selective COX-2 inhibition properties.
In view of the abovementioned issue, we used computational drug design to identify potential COX-2 inhibitors. The structure-based pharmacophore modeling was used to screen new molecular hits against many small molecules. In addition, the molecular docking protocol was employed to explore the binding orientation of ligands with certain protein.7
Materials and Methods
LigandScout Advanced 4.38 enabled current pharmacophore modeling based on (R)-naproxen, which is the COX-2 inhibitor, employing 426 actives and 13,159 decoys of a Directory of Useful Decoys: Enhanced (DUDE).9 The validated pharmacophore model was used for screening compounds from ZINC database10 by employing Pharmit (http://pharmit.csb.pitt.edu).
The COX-2 structure complexed with (R)-naproxen was taken from the Protein Data Bank (PDB code 3Q7D),11 which was prepared by using AutoDockTools 1.5.612,13 and submitted for molecular docking enabled with the help of iDock software.14 The ligand docking was centered by following the (R)-naproxen conformation (center x = −40.447, center y = 49.257, center z = 20.41) and arranging the grid width 40 x 40×40 and point spacing 0.375 Å.
Results
The (R)-naproxen consist of seven pharmacophores, ie, three hydrophobic, one negative ion, and three hydrogen bond acceptors (Figure 1). The 12 pharmacophore models were generated as listed in Table 1 with AUC-ROC values > 0.5. One model (NPX-8) with GH score 0.754 and AUC100% 0.51 was selected (Table 2), in which both GH score and AUC values indicated its ability to identify the actives from decoys (Figure 2).
 |  |  |
Table 1 The Pharmacophore Models |
 |
Table 2 The Parameter and GH Score Obtained for Pharmacophore Model NPX-8 |
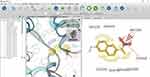 |
Figure 1 Display the pharmacophore features of (R)-naproxen, in which hydrophobic, negative ion, and hydrogen bond acceptors were assigned as yellow, brown, and red arrows, respectively. |
 |
Figure 2 The area under curve (AUC) of receiver operating characteristic plot (ROC) of NPX-8. |
The selected pharmacophore model was employed in a virtual screening campaign on the Pharmit webserver against small molecules of ZINC database, which further retrieved 1675 small molecules. All 1675 small molecules were docked to COX-2, while docking (R)-naproxen was performed to validate the docking protocol. The pose of the reference ligand after the docking process was similar to its experimental pose, as shown by the lower RMSD (0.71 Å).15,16 The conformation of (R)-naproxen contained one hydrogen bonding interaction with Tyr355 and hydrophobic interactions with Ala527, Gly526, Trp387, Tyr385, and Leu352 (Figure 3).11
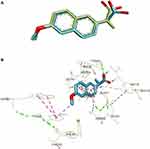 |
Figure 3 (A) Superimposed of (R)-naproxen between X-ray (green) and docked (blue) conformations with RMSD 0.71 Å. (B) The interaction of (R)-naproxen in the cavity of COX-2. |
Further, 1675 small molecules gave binding energies between −4.86 and −11.03 kcal/mol in which Lig_1805/ZINC103584272 (E=−11.03 kcal/mol), Lig_553/ZINC408573132 (E=−10.92 kcal/mol), Lig_680/ZINC103584263 (E=−10.90 kcal/mol), and Lig_2006/ZINC19324645 (E=−10.62 kcal/mol) had the binding energy lower than (R)-naproxen (Figure 4).
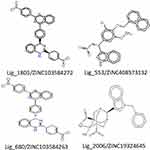 |
Figure 4 The molecular structures of best-docked hit molecules. |
Lig_2006/ZINC19324645 formed H-bond interaction with Arg120, Tyr115, and Ser119. In addition, hydrophobic interactions were observed with Val89, Leu93, and Val116. The H-bond with Arg120 was also noticed in the experimental structure of (R)-naproxen. It was also observed that H-bonds with Arg120 was established with Lig_1805/ZINC103584272 and Lig_680/ZINC103584263, while Lig_553/ZINC408573132 formed H-bond interactions with Gln454, His214, and His388. Figure 5 shows the interaction of Lig_1805/ZINC103584272, Lig_553/ZINC408573132, Lig_680/ZINC103584263, and Lig_2006/ZINC19324645, with COX-2.
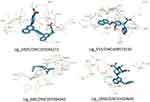 |
Figure 5 The interactions of Lig_1805/ZINC103584272, Lig_553/ZINC408573132, Lig_680/ZINC103584263, and Lig_2006/ZINC19324645, each with COX-2. |
Discussion
Nowadays, inflammatory diseases are the focus of research worldwide due to their effects on human health. One enzyme regulating inflammation in the human body is cyclooxygenase 2 (COX-2). The COX‐2 enzyme is known to be expressed when proinflammatory mediators are released. The use of COX-2 inhibitors such as celecoxib, rofecoxib, and valdecoxib has offered a better safety profile than classical NSAIDs, such as aspirin, ibuprofen, and indomethacin. However, several side effects were noticed, such as bleeding, gastrointestinal irritation, and perforation, which indicated the urgent need for more selective COX-2 inhibitors12,13. In the present study, a pharmacophore model was developed, validated, and utilized in the ZINC small molecules for the virtual screening process. The best pharmacophore features of naproxen were successfully validated. These include the hydrophobic feature from the naphthalene ring, the negative ion, and the hydrogen bond acceptors feature from the carboxyl group. This pharmacophore greatly affects the inhibition of cox-2 activity. Therefore, we screened the ZINC database to identify compounds with very similar pharmacophore features and better activity than naproxen. A total of 1675 molecules were docked to the COX-2 enzyme to predict their binding mode and affinity. The best four hits were evaluated in terms of their binding modes, which were revealed that all the top hits were able to bind to the active site of COX-2.
Heidarpoor et al14 employed docking-based high throughput virtual screening against 18 million compounds and recommended two top hits (ZINC16934653 and ZINC40484701) as a potential COX-2 inhibitor, which are different from our study. In other studies, Abdellatif et al15 The experimental and docking study explored new indomethacin analogs as selective COX-2 inhibitors. In another study, Szczukowski et al16 identify pyrrolo[3,4-d]pyridazinone-based 1,3,4-Oxadiazole as potential COX-2 inhibitors. The current study offers new hits different from those studies based on pharmacophore modeling and molecular docking study. However, experimental validation needs to be performed to evaluate those hits.
Conclusion
The present work successfully identified four better hit molecules for COX-2 than (R)-naproxen in terms of their binding affinities employing the structure-based pharmacophore modeling. A pharmacophore model was developed based on the structure of (R)-naproxen complexed with COX-2. The validated model was utilized to screen against the ZINC database containing 13 million compounds. This hit molecule can be used as a reference for detailed analysis to be developed into new drugs. In addition, this hit molecule is able to be an alternative molecule for potential COX-2 inhibitors. The four molecules deserve further experimental verification to prove their potential as COX-2 inhibitors.
Acknowledgments
This research was funded by The Ministry of Education, Research, Research and Technology, Republic of Indonesia 2021.
Disclosure
The authors declare no conflicts of interest in this work.
References
1. Leláková V, Šmejkal K, Jakubczyk K, et al. Parallel in vitro and in silico investigations into anti-inflammatory effects of non-prenylated stilbenoids. Food Chem. 2019;285:431–440.
2. Ahmed EM, Kassab AE, El-Malah AA, Hassan MSA. Synthesis and biological evaluation of pyridazinone derivatives as selective COX-2 inhibitors and potential anti-inflammatory agents. Eur J Med Chem. 2019;171:25–37.
3. de Souza G, Silva RJ, Milián ICB, et al. Cyclooxygenase (COX)-2 modulates Toxoplasma gondii infection, immune response and lipid droplets formation in human trophoblast cells and villous explants. Scie Rep. 2021;11:12709.
4. El-Sayed NA, Nour MS, Salem MA, Arafa RK. New oxadiazoles with selective-COX-2 and EGFR dual inhibitory activity: design, synthesis, cytotoxicity evaluation and in silico studies. Eur J Med Chem. 2019;183:111693.
5. Kumar R, Saha N, Purohit P, et al. Cyclic enaminone as new chemotype for selective cyclooxygenase-2 inhibitory, anti-inflammatory, and analgesic activities. Eur J Med Chem. 2019;182:111601.
6. Atatreh N, Youssef AM, Ghattas MA, et al. Anti-inflammatory drug approach: synthesis and biological evaluation of novel pyrazolo[3,4-d]pyrimidine compounds. Bioorg Chem. 2019;86:393–400.
7. Arba M, Wahyudi ST, Brunt DJ, Paradis N, Wu C. Mechanistic insight on the remdesivir binding to RNA-Dependent RNA polymerase (RdRp) of SARS-cov-2. Comput Biol Med. 2021;129:104156.
8. Wolber G, Langer T. LigandScout: 3-D Pharmacophores Derived from Protein-Bound Ligands and Their Use as Virtual Screening Filters. J Chem Inf Model. 2005;45:160–169.
9. Mysinger MM, Carchia M, Irwin JJ, Shoichet BK. Directory of Useful Decoys, Enhanced (DUD-E): better Ligands and Decoys for Better Benchmarking. J Med Chem. 2012;55:6582–6594.
10. Irwin JJ, Sterling T, Mysinger MM, Bolstad ES, Coleman RG. ZINC: a Free Tool to Discover Chemistry for Biology. J Chem Inf Model. 2012;52:1757–1768.
11. Duggan KC, Hermanson DJ, Musee J, et al. (R)-Profens are substrate-selective inhibitors of endocannabinoid oxygenation by COX-2. Nat Chem Bio. 2011;7:803–809.
12. Sanad SMH, Mekky AEM. Synthesis, cytotoxicity and in vitro antibacterial screening of novel hydrazones bearing thienopyridine moiety as potent COX-2 inhibitors. J Iran Chem Soc. 2020;17:3299–3315.
13. Zhang B, Hu X-T, Gu J, Yang Y-S, Duan Y-T, Zhu H-L. Discovery of novel sulfonamide-containing aminophosphonate derivatives as selective COX-2 inhibitors and anti-tumor candidates. Bioorg Chem. 2020;105:104390.
14. Heidarpoor SL, Ebrahimi A, Lagzian M. Identification of new potential cyclooxygenase-2 inhibitors: insight from high throughput virtual screening of 18 million compounds combined with molecular dynamic simulation and quantum mechanics. J Biomol Struct Dyn. 2021;39:1717–1734.
15. Abdellatif KRA, Abdelall EKA, Elshemy HAH, El-Nahass E-S, Abdel-Fattah MM, Abdelgawad YYM. New indomethacin analogs as selective COX-2 inhibitors: synthesis, COX-1/2 inhibitory activity, anti-inflammatory, ulcerogenicity, histopathological, and docking studies. Arch Pharm. 2021;354:2000328.
16. Szczukowski Ł, Krzyżak E, Zborowska A, et al. Design, Synthesis and Comprehensive Investigations of Pyrrolo[3,4-d]pyridazinone-Based 1,3,4-Oxadiazole as New Class of Selective COX-2 Inhibitors. Int J Mol Sci. 2020;21(24):356.
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
