Back to Journals » OncoTargets and Therapy » Volume 12
The role of PEG3 in the occurrence and prognosis of colon cancer
Authors Zhou T, Lin W, Zhu Q, Renaud H, Liu X, Li R, Tang C, Ma C, Rao T, Tan Z , Guo Y
Received 9 March 2019
Accepted for publication 16 July 2019
Published 25 July 2019 Volume 2019:12 Pages 6001—6012
DOI https://doi.org/10.2147/OTT.S208060
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Geoffrey Pietersz
Ting Zhou,1–4 Wei Lin,5 Qiongni Zhu,6 Helen Renaud,7 Xiaowei Liu,8 Ruidong Li,9 Cui Tang,1–4 Chong Ma,1–4 Tai Rao,1–4 Zhirong Tan,1–4 Ying Guo1–4
1Department of Clinical Pharmacology, Xiangya Hospital, Central South University, Changsha 410008, People’s Republic of China; 2Human Key Laboratory of Pharmacogenetics, Institute of Clinical Pharmacology, Central South University, Changsha 410078, People’s Republic of China; 3Engineering Research Center of Applied Technology of Pharmacogenomics, Ministry of Education, Changsha 410078, People’s Republic of China; 4National Clinical Research Center for Geriatric Disorders, Changsha, Hunan 410008, People’s Republic of China; 5Department of Pathology, Xiangya Hospital, Central South University, Changsha 410008, People’s Republic of China; 6Department of Pharmacy, Ruijin Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai 200025, People’s Republic of China; 7Department of Pharmacology, Toxicology and Therapeutics, University of Kansas Medical Center, Kansas City, KS 66160, USA; 8Department of Gastroenterology, Xiangya Hospital of Central South University, Changsha, Hunan 410008, People’s Republic of China; 9Graduate Program in Genetics, Genomics & Bioinformatics, University of California, Riverside, CA 92507, USA
Purpose: Imprinted genes are often identified as key players in the etiology and prognosis of many tumors; however, the role they play in colon cancer remains unclear. Along with the development of big data analysis came the discovery of a wealth of genetic prognostic factors, like microsatellite instability for colon cancer, which need to be taken into consideration when evaluating new biomarkers for the disease.
Methods: We systematically mined public databases to find recurrence free survival (RFS)-related imprinted genes for colon cancer patients on the mRNA level by univariate and multivariate survival analyses. We then investigated the association of methylation status and microRNA expression of the targeted imprinted genes with survival rate of colon cancer patients. Lastly, in a clinical study we used qRT-PCR and immunohistochemistry to quantify mRNA and protein expression of the imprinted genes that related to RFS in our bioinformatics screening, respectively, in 20 tumor tissues compared to paired adjacent tissues.
Results: The results show that paternally expressed gene 3 (PEG3) is the only imprinted gene related to colon cancer patient prognosis on the mRNA level in our datasets, and high mRNA expression of PEG3 is associated with a poor prognosis. Furthermore, the methylation beta value of cg13960339, as well as the expression of 4 microRNAs, negatively correlated with PEG3 mRNA level and were correlated with the prognosis of colon cancer patients. Moreover, the expression of PEG3 mRNA in colon cancer is significantly lower, but PEG3 protein expression is significantly higher compared to that in normal tissues.
Conclusion: PEG3 is likely associated with the progression and prognosis of colon cancer.
Keywords: colon cancer, prognosis, imprinted gene, PEG3
Introduction
Colon cancer is a public health problem and is a major cause of morbidity and mortality worldwide. Colon cancer is the third most common cancer diagnosis among men and women in the United States.1,2 Although surgical resection is a relatively effective method for the treatment of colon cancer, metastasis and recurrence often occur after surgery, which greatly shortens the survival time of patients. Along with the development of big data analysis came the discovery of a wealth of genetic prognostic factors, like microsatellite instability for colon cancer, which need to be taken into consideration when evaluating new biomarkers for this disease.
Imprinted genes usually express only one of the homologues from the parents, and are regarded to be important in mammalian growth, development, and cell differentiation.3,4 Monoallelic expression ensures that the levels of the proteins expressed by these genes are tightly regulated. Loss of imprinting is an epigenetic modification involving loss of parental allele-specific expression due to changes in the methylation status of imprinted genes, where the repressed allele becomes active and the tight regulation of protein expression from this gene is lost. Loss of imprinting is seen frequently and precociously in a large variety of human tumors.5 For example, loss of imprinting of insulin-like growth factor 2, leads to overexpression of the protein and is associated with a poor prognosis and resistance to chemotherapy in many cancers.6,7 Additionally the paternally imprinted gene, neuronatin, is a biomarker for breast cancer because high expression is associated with poor prognosis.8
CpG-rich regions, termed CpG islands, in the promoters of imprinted genes have allele-specific methylation patterns that govern monoallelic expression.9 Epigenetic instability of imprinted domains on these CpG islands is often regarded as a biomarker for human cancers.10,11 Additionally, the mRNA expression of most imprinted genes is downregulated in primary tumors compared to normal tissues and is typically accompanied by DNA methylation level changes in the imprinted domains; examples of genes that undergo these changes include PEG3, mesoderm-specific transcript, and stimulatory GTPase α.12 MicroRNA is another master epigenetic regulator that alters more than 30% of human gene expression. These molecules are a class of evolutionarily conserved, small non-coding RNAs, which exist widely in eukaryotes and do not encode proteins but function in RNA silencing and post-transciptional regulation of gene expression. By base-pairing with complementary sequences within mRNA molecules, miRNAs silence translation by either cleavage of the mRNA strand, or through binding to the 3ʹ untranslated region.13 MicroRNAs affect many physiological and pathological processes, such as tumor development.14 It is reported that microRNA expression is associated with survival prognosis of colon cancer patients.15 However, few studies have systematically investigated the independent role that microRNAs play in colon cancer survival prognosis together with multiple covariates.
In the current study, we first systematically mined public databases to find prognosis-related imprinted genes for colon cancer patients based on mRNA level by univariate and multivariate survival analysis. We then investigated whether an association exists between methylation status, predicted regulator microRNA level of the target imprinted gene, and prognosis of colon cancer patients. Our study describes the independent roles imprinted genes may play in colon cancer.
Materials and methods
Patients and samples
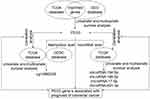
In this study, colon cancer datasets from TCGA (The Cancer Genome Atlas, https://cancergenome.nih.gov/) were downloaded and processed by GDCRNATools.16 An independent colon cancer dataset GSE39582 was downloaded from GEO (Gene Expression Omnibus). Clinical data including age, sex, histology, tumor stage, grade, tumor location, etc. are available in both datasets. IC50 value of 5-fluorouracil (5-fu) in different colon cancer cell lines was downloaded from the Genomics of Drug Sensitivity in Cancer (GDSC) database (www.cancerRxgene.org). The work flow of mining data is shown in Figure 1.
Twenty pairs of primary colon cancer and adjacent tissue were obtained from Xiangya Hospital, Central South University. The study was conducted in accordance with the Declaration of Helsinki and approved by the Ethics Committee of the Institute of Clinical Pharmacology of Central South University (registration no. CTXY-150001-2). In addition, prior to entering into the study, written informed consent was obtained from all the participants. Clinical data collected included gender, age, familial history, Histological type, lymphatic metastasis, stage, tumor location and differentiation. Staging was based on the TNM (tumor, node, and metastasis) classification system of the International Union against Cancer 2009 (UICC).17
Survival and statistical analysis
Recurrence free survival (RFS) is the period from resection to the first radiological evidence of tumor recurrence.18 The Kaplan-Meier method and multivariate Cox’s proportional were used to assess the prognosis of colon cancer patients for univariate and multivariate analyses, respectively, and the log-rank test was used to compare the curves among high-risk and low-risk groups. The above two analyses were done through the survival package in R language, covariates include age, stage, sex, tumor location, and MMR (mismatch repair) status. Hazard ratio (HR) and 95% confidence interval (CI) were used to describe the relative risk. Paired Student’s t-tests were used to compare the expression of PEG3 in primary tumor to that in normal tissues by GraphPad Prism 6.0 (Graphpad Inc., San Diego, CA, USA). Associations between PEG3 expression and immunostaining results were tested by chi-square or Fisher’s exact test by SPSS 23.0 (SPSS Inc., Chicago, IL, USA). Spearman’s correlation analysis was performed to calculate the coefficient of correlation between the levels of mRNA expression and methylation as well as microRNA, respectively, using R. All tests were two-tailed and P<0.05 was deemed as statistically significant.
Quantitative real-time reverse transcription PCR
For the quantitative real-time RT-PCR assay, 20 paired primary colon cancer and non-tumorous tissues were obtained and ground into powder in liquid nitrogen. Total RNA was extracted from the powder and cDNA synthesized using PrimeScript™ RT reagent Kit with gDNA Eraser (Perfect Real Time) (Takara, Shiga, Japan) according to the manufacturer’s protocol. Expression of mRNA was quantified by qRT-PCR with a LightCycler 480 (Roche Diagnostics, Basel, Switzerland) using SYBR® Premix DimerEraser™ (Takara). The primers were synthesized by Sangon Biotech Co., Ltd. (Shanghai, China) and the sequences of the primers were as follows: PEG3 forward: 5ʹ‑CCTACCCAAGCACCAGTCG‑3ʹ, and PEG3 reverse: 5ʹ‑GGAACTGCGTGACACATCCT‑3ʹ; PPIA forward: 5ʹ‑GTGGTATAAAAGGGGCGGGAG‑3ʹ, and PPIA reverse: 5ʹ‑GTGGGGTTGACCATGGCTAATAGTA‑3ʹ; B2M forward: 5ʹ‑ATGCCTGCCGTGTGAACCATGTGA‑3ʹ, and B2M reverse: 5ʹ‑TCCAAATGCGGCATCTTCAAACCTC-3ʹ. Amplification conditions were denaturation for 30 sec at 95°C and then 40 cycles of 92°C for 5 sec, 55°C for 30 sec, and 72°C for 30 sec. The mRNA expression of PEG3 was normalized to the average of the expressions of B2M and PPIA.19
Immunohistochemistry
Immunohistochemistry was performed as previously described.20 Paraffin embedded tissue sections were cut to 4 μm thick and blocked with endogenous peroxidase. The autoclave technique was used to achieve heat-induced antigen retrieval. Sections were then incubated overnight at 4°C with rabbit polyclonal antibody anti-PEG3 (PA5-51298; Thermo Fisher Scientific, Inc.) at a 1:40 dilution. Subsequently, the sections were washed three times with phosphate buffer saline, and then incubated with a biotin-labeled goat anti-rabbit antibody. Sections were then stained with peroxidase-conjugated streptavidin (Boster, Wuhan, China) with 3, 3ʹ-diaminobenzidine tetrahydrochloride as the chromogen. Finally, the sections were dyed with hematoxylin. Each slide was evaluated using light microscopy and the staining was scored semiquantitatively by assessing the intensity (on a 0–3 scale: 0, no staining of tumor cells; 1, weak; 2, moderate; 3, strong staining) and by estimating the percentage of positive cytoplasmic or membranous staining cells (on a 1–4 scale: 1,1–25% staining; 2, 26–50% staining; 3, 51–75% staining; or 4, >75% staining). Intensity score ≥2 and ≥25% cells were defined as being positive for levels of protein expression in this study.21
Results
PEG3 is the only imprinted gene related to colon cancer prognosis
Clinical information and mRNA expression data were downloaded from the TCGA database (280 colon cancer patients) and GEO database (GSE39582, 566 colon cancer patients). The characteristics of patients including sex, age, tumor location, stage, distant metastasis and adjuvant chemotherapy are presented in Table 1. We included 100 imprinted genes found in the human genome in our analyses.12 Data obtained from TCGA indicate that higher mRNA expression of PEG3, PEG10, SGCE and TFPI2 were significantly associated with shorter RFS in univariate and multivariate analyses.
 |
Table 1 Patient demographics and clinical characteristics |
Significance of these four genes in colon cancer prognosis was further validated in an independent cohort from GEO. The results showed that only PEG3 mRNA expression was correlated with colon cancer prognosis in the two databases (Table 2). Due to these findings, the imprinted gene PEG3 was chosen for further study. In both TCGA and GEO databases, univariate and multivariate survival analysis showed that the expression of PEG3 is a risk factor in colon cancer prognosis (HR>1), and the high-expression group had a shorter RFS compared to the low-expression group (Table 2). Combining the above results from the TCGA and GEO databases, we conclude that, of the imprinted genes we evaluated, PEG3 is the only one that affects the prognosis of patients with colon cancer, and it is a disadvantageous factor for prognosis on mRNA level.
 |
Table 2 Four imprinted genes associated with RFS in TCGA and validated in GEO by univariate and multivariate survival analysis |
Methylation of cg13960339 is an advantageous prognostic factor and negatively correlates with PEG3 mRNA level
In the TCGA database, 292 cases of colon cancer data containing both clinical prognostic information and genome-wide methylation data based on Illumina Methylation 450K chip were evaluated. Univariate and multivariate analyses were performed to identify methylation sites that affect prognosis. Among all the methylation sites associated with PEG3, cg13960339 was the only one associated with RFS of colon cancer in the TCGA dataset, the results of the univariate and multivariate survival analyses showed that high cg13960339 methylation levels significantly increased the RFS of colon cancer patients (Figure 2A).
We used spearman’s correlation analysis to identify the relationship between methylation and mRNA expression. Degree of methylation was estimated using beta values (between 0 and 1, with 0 being unmethylated and 1 fully methylated). In the TCGA-Colon Adenocarcinoma (COAD) dataset, a lower methylation beta value of cg13960339 was negatively correlated with PEG3 mRNA level (Figure 2B) and was significantly associated with poor prognosis (Figure 2A). The results of this multivariate analysis are consistent with the above results (P=0.027, HR=0.481, 95% CI=0.251–0.919). These results suggest that methylation of cg13960339 may be a biomarker of patient prognosis and regulate the expression of PEG3.
In order to confirm whether the cg13960339 methylation level is indeed an indicator of prognosis, IC50 values in colon cancer cells to 5-fu therapeutics in the GDSC database were evaluated. IC50 values were divided into two groups by median: high or low. The beta value in the low IC50 group was 0.7387, and in the high IC50 group was 0.5989 (P=0.0068), which suggests that higher cg13960339 methylation could increase drug sensitivity and inhibit the survival of cancer cells. The correlation analysis between the beta value of cg13960339 and IC50 data showed that they are significantly correlated (Spearman r=−0.298, P=0.0468). The negative correlation indicates that methylation of PEG3 reduces the IC50 of 5-fu, which indicates that methylation at this methylation site may be beneficial to the therapeutic effect of 5-fu on patients with colon cancer, and suggests it is beneficial to the prognosis of patients.
Expression of 4 microRNAs associate with the prognosis of colon cancer patients and negatively correlate with PEG3 mRNA level
Prognostic data and microRNA expression data downloaded from the TCGA website were matched by patient ID. Clinical prognostic information from 441 cases of colon cancer and microRNA expression data were included. The results of the log-rank and Cox regression analyses are shown in Table 3. Patient prognosis was affected by the expression of 8 microRNAs.
 |
Table 3 Univariate and multivariate analysis of microRNA expression and RFS in TCGA |
Spearman’s correlation analysis was conducted to compare microRNA expression data with PEG3 mRNA expression data from TCGA. In 280 COAD patients, only 5 of the 8 microRNAs were significantly associated with PEG3 mRNA level (P<0.05). However, for one of these microRNAs (miR-181b-5p) the correlation coefficient is very small and the correlation is weak. The other 4 microRNAs showed higher and negative correlation with PEG3 expression (Table 4). Additionally, higher expression of these 4 microRNAs in colon cancer patients was associated with longer RFS (Table 3).
 |
Table 4 Correlation between selected microRNAs and the expression of PEG3 mRNA |
Expression of PEG3 mRNA is significantly lower but protein is significantly higher in colon cancer compared to adjacent tissue
To investigate the potential role of PEG3 in the etiology of colon cancer, the mRNA expression level of PEG3 in tumors and adjacent tissues of patients was explored in the TCGA dataset using R language, and validated in samples from Xiangya Hospital. In 33 types of different cancers from the TCGA database, the expression level of PEG3 was compared between the primary tumor and adjacent tissue. As shown in Figure 3A, in colon cancer patients, lower PEG3 mRNA expression was observed in tumors compared to adjacent tissue (P<0.05), which suggests that PEG3 may act as a tumor suppressor rather than act as an oncogene.
Subsequently, mRNA expression of PEG3 in 20 paired tissues were tested by qRT-PCR that confirmed our findings from the TCGA database. The patient information is shown in Table 5. PEG3 mRNA expression in colon cancer tissue was significantly lower compared to the adjacent tissue (P<0.01). The mean ± SD of colon cancer and para-cancer tissue of PEG3 is 0.025±0.0230 and 0.045±0.022, respectively (Figure 3B).
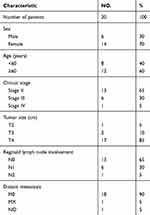 |
Table 5 Summary of 20 patients’ cohort information |
We performed immunohistochemical staining on the 20 pairs of colon cancer tissues and adjacent tissues, which were also used in our mRNA expression studies. Our immunohistochemical results showed that PEG3 protein expression is higher in cancer tissues than in adjacent tissues (P=0.016). The PEG3 protein is mainly distributed in the cytoplasm in colon cancer tissues, whereas very little protein was detected in the para-cancer tissue (Figure 4).
Discussion
This is the first study to identify PEG3 as the only imprinted gene that affects the prognosis of colon cancer. Additionally, we discovered that lower methylation levels of cg13960339 within the PEG3 promoter is associated with the prognosis of colon cancer. And lastly, several microRNAs that regulate PEG3 expression, are also associated with the outcome of colon cancer.
TNM staging is a traditionally used prognostic classification system in clinical practice to select patients with colon cancer for adjuvant chemotherapy. However, this system often fails to accurately predict recurrence in many colon cancer patients who undergo surgical therapy. Approximately 10–20% stage II and 30–40% stage III colorectal cancer patients develop recurrence.22 With the wide spread use of modern biotechnology in research, several somatic mutations, including MMR and KRAS, were proved to influence the outcome and therapeutic effect of colon cancer patients, and thus applied as a basis of medical treatment decisions in routine clinical practices. Therefore, we chose the dataset GSE39582, which has the most comprehensive clinical information, as the validation data set for TCGA. Additionally, the current study includes MMR status as a covariant in the multivariate survival analysis to mine more reliable prognostic signatures for further study; however, the KRAS status was excluded because very few patients had this information in the discovery and validation datasets.
The work flow applied in this study (Figure 1) is effective to identify prognostic biomarkers for colon cancer. Data of PEG3 mRNA expression and promoter region methylation levels correlated with RFS, and the correlation of mRNA, microRNAs, and methylation changes, will be helpful to determine the mechanism behind the loss of imprinting of PEG3 in colon cancer. Although we weren’t able to validate the relationship of microRNA expressions with RFS in TCGA using another method, the overlapping of microRNA expression with solid PEG3 mRNA signatures, as well as the correlation analysis between these two variables, suggest these microRNAs as very promising candidates for further investigation. In addition, we evaluated PEG3 with two bioinformatics websites and forecast the log-rank analysis of prognosis as well as mRNA expression patterns between tumor and para-tumor tissue, and the results were very similar to ours even though different databases and algorithms were used (UCSC Xena and GEPIA, data not shown). Therefore, we provide the first evidence of a prognostic role for PEG3, its methylation sites, and targeting microRNAs in colon cancer.
The function of PEG3 is not completely understood and tends to vary between tissues. The role of PEG3 in survival rates of cancer patients is not fully understood; however, in ovarian cancer, down-regulation of PEG3 mRNA expression was included in a multi-gene prognostic signature for poor outcome of the disease,23 Additionally, upregulation of PEG3 was reported to be associated with a modest but significant reduction in distant metastases-free survival in breast cancer patients.24 A bioinformatic-screening study by Kaplan-Meier analyzing survival curves of all cancer patients in TCGA showed that, on the mRNA level, highly expressed PEG3 was related to better clinical outcomes in kidney renal clear cell carcinoma, brain lower-grade glioma, lung adenocarcinoma, pancreatic adeno-carcinoma, pheochromocytoma and paraganglioma, uveal melanoma, and thymoma.25 Although colon cancer was included in this study no statistically significant association was found between PEG3 expression and prognosis, and the discrepancy of this study and ours might be due to the different long-term efficacy evaluation index chosen (OS and DFS verse RFS) that altered the patients included in each study. Besides direct investigation of survival rates and PEG3 expression, PEG3-antisense RNA has also been reported to be associated with cancer prognosis. Down-regulation of PEG3-antisense RNA was revealed to be related to poor survival in squamous cell carcinoma of the head and neck, but related to a better prognosis for colorectal cancer patients based on bioinformatic screening of long noncoding RNA expression signatures.26,27 However, the function of PEG3-antisense RNA is poorly understood, and the idea that it might negatively regulate mRNA and protein levels of PEG3 was only a prediction according to neuron cell line-based transfection experiments.28 Therefore, the present study only evaluated and validated the independent prognostic role that PEG3 plays in colon cancer, and the mechanism of this needs further investigation.
Deregulation of imprinted genes can happen in somatic cells of adulthood, thus increasing cancer risk.29 Bioinformatics investigations by others, as well as our observations in this study show decreased mRNA expression of PEG3 in colon cancer.12,30 However, opposite PEG3 protein to mRNA expression patterns were observed in this study: decreased mRNA but increased protein levels of PEG3 in the cancer tissue. The discrepancy of PEG3 mRNA and protein expression was also found in testicular germ cell tumors.31 Thus, aberrant overexpression of PEG3 might contribute to the carcinogenesis of colon tissue. PEG3 protein was thought to be located mainly in nuclei,31 however, the immunohistochemical staining in the current study indicates that almost all the PEG3 protein is in the cytosol with little in the nuclei, which might be due to decreased mRNA expression and lack of protein degradation. PEG3 is quite a unique protein with opposite biochemical properties: it is a growth suppressor at the cellular level but a growth stimulator at the organismal level.28 Intriguingly, PEG3 might function as a tumor suppressor in the nucleus; however, accumulation of this protein in the cytosol might inhibit the expression of growth suppressors.28 Furthermore, it is reported that PEG3 plays an important role in autophagy and conveys the external signal to induce the formation of autophagosomes in the cell, which might be crucial for protein degradation.28 Detailed characterization of the role PEG3 plays in colon carcinogenesis with transcriptome, proteomics, and localization studies will be included in our follow-up investigations.
Methylation is essential for genes to remain imprinted, and abnormal methylation levels of the differentially methylated regions are associated with many types of diseases,9,32 thus we did the preliminary data mining to analyze the relationship between PEG3 methylation and RFS in colon cancer. Interestingly, the current study revealed that cg13960339 methylation level is positively associated with survival probability. This methylation site exists in the CpG island of chromosome19 and the distance to the transcription start site of PEG3 is 200 bp, suggesting this site might be an important transcriptional regulation position for gene expression. Methylation is strongly associated with negative regulation of mRNA expression, which was also shown in our analysis for the association between cg13960339 and PEG3 mRNA level. Additionally, the influence of these factors on RFS is also consistent with this phenomenon. The impact of PEG3 whole methylation density on the etiology and prognosis of colon cancer will be systematically analyzed in our future clinical studies.
Four microRNAs in the current study are reported to be diagnostic or prognostic biomarkers for colon cancer: hsa-miR-194-5p, hsa-miR-186-5p, hsa-miR-17-5p, hsa-miR-491-3p. Past studies have shown that miR-17- suppresses cell proliferation and invasion by increasing p53, decreasing p62 expression, or affecting cell cycle machinery, and might be a potential treatment target for adjuvant therapy in several cancers.16,30,33–36 Additionally, a Spanish study of 110 patients undergoing surgery for colorectal cancer revealed that miR-17-5p had a trend to correlate with disease-free survival at early stages,37 A meta-analysis revealed that the increased expression of miR-17 is associated with unfavorable prognoses for colon cancer. MiR-194-5p, miR-181a-5p, miR-494-3p and miR-491-3p are also reported as prognosis biomarkers in clear cell kidney carcinoma, ovarian cancer, lung (breast) cancer, and tongue cancer patients, respectively.33,38–41 Tumor suppressive miR-296-3p and miR-186-5p also affect tumorigenesis of non-small cell lung cancer and colorectal cancer cells.42,43 Therefore, these microRNAs might be potential prognostic biomarkers of colon cancer and should be further investigated.
Conclusions
The current study revealed that the imprinted gene, PEG3, is a prognostic biomarker for colon cancer, and the potential mechanism might involve altered epigenetic regulation on different levels including microRNA and methylation of a CpG island for PEG3. Additionally, we speculate that the degradation and ectopic expression of PEG3 might be critical to the function it plays in the tumorigenesis of colon cancer.
Abbreviations
CI, Confidence interval; COAD, Colon Adenocarcinoma; GEO, Gene Expression Omnibus; GDSC, Genomics of Drug Sensitivity in Cancer; HR, Hazard ratio; MMR, Mismatch repair; PEG, Paternally expressed gene; RFS, Recurrence free survival; TCGA, The Cancer Genome Atlas; TNM, Tumor, node, metastasis; 5-fu, 5-fluorouracil.
Acknowledgments
This study was funded by the National Natured Science Foundation of China (Grant No 81503563). The authors thank Dr. Miao Liu for technical support.
Disclosure
The authors declare no conflicts of interest in this work.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. doi:10.3322/caac.21442
2. Xie XN, Yu J, Zhang LH, et al. Relationship between polymorphisms of the lipid metabolism-related gene PLA2G16 and risk of colorectal cancer in the Chinese population. Funct Integr Genomics. 2018;19:227–236.
3. Barton SC, Surani MA, Norris ML. Role of paternal and maternal genomes in mouse development. Nature. 1984;311(5984):374–376. doi:10.1038/311374a0
4. Surani MA, Barton SC, Norris ML. Nuclear transplantation in the mouse: heritable differences between parental genomes after activation of the embryonic genome. Cell. 1986;45(1):127–136. doi:10.1016/0092-8674(86)90544-1
5. Uribe-Lewis S, Woodfine K, Stojic L, Murrell A. Molecular mechanisms of genomic imprinting and clinical implications for cancer. Expert Rev Mol Med. 2011;13:e2. doi:10.1017/S1462399410001717
6. Livingstone C. IGF2 and cancer. Endocr Relat Cancer. 2013;20(6):R321–R339. doi:10.1530/ERC-12-0368
7. Brouwer-Visser J, Huang GS. IGF2 signaling and regulation in cancer. Cytokine Growth Factor Rev. 2015;26(3):371–377.
8. Nass N, Walter S, Jechorek D, et al. High neuronatin (NNAT) expression is associated with poor outcome in breast cancer. Virchows Archiv. 2017;471(1):23–30.
9. Huang JM, Kim J. DNA methylation analysis of the mammalian PEG3 imprinted domain. Gene. 2009;442(1–2):18–25.
10. Barrow TM, Barault L, Ellsworth RE, et al. Aberrant methylation of imprinted genes is associated with negative hormone receptor status in invasive breast cancer. Int J Cancer. 2015;137(3):537–547. doi:10.1002/ijc.29419
11. Ribarska T, Bastian KM, Koch A, Schulz WA. Specific changes in the expression of imprinted genes in prostate cancer–implications for cancer progression and epigenetic regulation. Asian J Androl. 2012;14(3):436–450. doi:10.1038/aja.2011.160
12. Kim J, Bretz CL, Lee S. Epigenetic instability of imprinted genes in human cancers. Nucleic Acids Res. 2015;43(22):10689–10699. doi:10.1093/nar/gkv867
13. Long LM, He BF, Huang GQ, Guo YH, Liu YS, Huo JR. microRNA-214 functions as a tumor suppressor in human colon cancer via the suppression of ADP-ribosylation factor-like protein 2. Oncol Lett. 2015;9(2):645–650. doi:10.3892/ol.2014.2746
14. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi:10.1016/s0092-8674(04)00045-5
15. Xu J, Zhao J, Zhang R. Four microRNAs signature for survival prognosis in colon cancer using TCGA data. Sci Rep. 2016;6:38306. doi:10.1038/srep38306
16. Li R, Qu H, Wang S, et al. GDCRNATools: an R/Bioconductor package for integrative analysis of lncRNA, miRNA and mRNA data in GDC. Bioinformatics. 2018;34(14):2515–2517. doi:10.1093/bioinformatics/bty124
17. Liu S, Chen X, Chen R, et al. Diagnostic role of Wnt pathway gene promoter methylation in non small cell lung cancer. Oncotarget. 2017;8(22):36354–36367. doi:10.18632/oncotarget.16754
18. Zeng WJ, Yang YL, Liu ZZ, et al. Integrative analysis of DNA methylation and gene expression identify a three-gene signature for predicting prognosis in lower-grade gliomas. Cell Physiol Biochem. 2018;47(1):428–439. doi:10.1159/000489954
19. Kheirelseid EA, Chang KH, Newell J, Kerin MJ, Miller N. Identification of endogenous control genes for normalisation of real-time quantitative PCR data in colorectal cancer. BMC Mol Biol. 2010;11:12. doi:10.1186/1471-2199-11-12
20. Li YQ, Chen J, Yin JY, Liu ZQ, Li XP. Gene expression and single nucleotide polymorphism of ATP7B are associated with platinum-based chemotherapy response in non-small cell lung cancer patients. J Cancer. 2018;9(19):3532–3539. doi:10.7150/jca.26286
21. Chen HH, Yan JJ, Chen WC, et al. Predictive and prognostic value of human copper transporter 1 (hCtr1) in patients with stage III non-small-cell lung cancer receiving first-line platinum-based doublet chemotherapy. Lung Cancer. 2012;75(2):228–234. doi:10.1016/j.lungcan.2011.06.011
22. Liu R, Zhang W, Liu ZQ, Zhou HH. Associating transcriptional modules with colon cancer survival through weighted gene co-expression network analysis. BMC Genomics. 2017;18(1):361. doi:10.1186/s12864-016-3396-5
23. Wang R, Ye XH, Zhao XL, Liu JL, Zhang CY. Development of a five-gene signature as a novel prognostic marker in ovarian cancer. Neoplasma. 2018;66:343–349.
24. Giuliano M, Herrera S, Christiny P, et al. Circulating and disseminated tumor cells from breast cancer patient-derived xenograft-bearing mice as a novel model to study metastasis. Breast Cancer Res. 2015;17:3. doi:10.1186/s13058-014-0508-5
25. Li M, Sun Q, Wang X. Transcriptional landscape of human cancers. Oncotarget. 2017;8(21):34534–34551. doi:10.18632/oncotarget.15837
26. Hsu CM, Lin PM, Lin HC, et al. Altered expression of imprinted genes in squamous cell carcinoma of the head and neck. Anticancer Res. 2016;36(5):2251–2258.
27. Zhang L, Chen S, Wang B, et al. An eight-long noncoding RNA expression signature for colorectal cancer patients’ prognosis. J Cell Biochem. 2018;120:5636–5643.
28. He H, Kim J. Regulation and function of the peg3 imprinted domain. Genomics Inform. 2014;12(3):105–113. doi:10.5808/GI.2014.12.3.105
29. Jirtle RL, Skinner MK. Environmental epigenomics and disease susceptibility. Nature Rev Genet. 2007;8(4):253–262. doi:10.1038/nrg2045
30. Liao XH, Xiang Y, Yu CX, et al. STAT3 is required for MiR-17-5p-mediated sensitization to chemotherapy-induced apoptosis in breast cancer cells. Oncotarget. 2017;8(9):15763–15774. doi:10.18632/oncotarget.15000
31. Ozata DM, Li X, Lee L, et al. Loss of miR-514a-3p regulation of PEG3 activates the NF-kappa B pathway in human testicular germ cell tumors. Cell Death Dis. 2017;8(5):e2759. doi:10.1038/cddis.2016.464
32. Murphy SK, Wylie AA, Jirtle RL. Imprinting of PEG3, the human homologue of a mouse gene involved in nurturing behavior. Genomics. 2001;71(1):110–117. doi:10.1006/geno.2000.6419
33. Faversani A, Amatori S, Augello C, et al. miR-494-3p is a novel tumor driver of lung carcinogenesis. Oncotarget. 2017;8(5):7231–7247. doi:10.18632/oncotarget.13933
34. Li HH, Miao DY, Zhu Q, Huang JH, Lu GX, Xu WG. MicroRNA-17-5p contributes to osteoarthritis progression by binding p62/SQSTM1. Exp Ther Med. 2018;15(2):1789–1794. doi:10.3892/etm.2017.5622
35. Jia J, Zhan D, Li J, Li Z, Li H, Qian J. The contrary functions of lncRNA HOTAIR/miR-17-5p/PTEN axis and Shenqifuzheng injection on chemosensitivity of gastric cancer cells. J Cell Mol Med. 2019;23(1):656–669. doi:10.1111/jcmm.13970
36. Hong P, Jiang M, Li H. Functional requirement of dicer1 and miR-17-5p in reactive astrocyte proliferation after spinal cord injury in the mouse. Glia. 2014;62(12):2044–2060. doi:10.1002/glia.22725
37. Diaz R, Silva J, Garcia JM, et al. Deregulated expression of miR-106a predicts survival in human colon cancer patients. Gene Chromosome Canc. 2008;47(9):794–802. doi:10.1002/gcc.20580
38. Ye Y, Zhang F, Chen Q, Huang Z, Li M. LncRNA MALAT1 modified progression of clear cell kidney carcinoma (KIRC) by regulation of miR-194-5p/ACVR2B signaling. Mol Carcinog. 2019;58(2):279–292. doi:10.1002/mc.22926
39. Petrillo M, Zannoni GF, Beltrame L, et al. Identification of high-grade serous ovarian cancer miRNA species associated with survival and drug response in patients receiving neoadjuvant chemotherapy: a retrospective longitudinal analysis using matched tumor biopsies. Ann Oncol. 2016;27(4):625–634. doi:10.1093/annonc/mdw007
40. Chen SM, Wang BY, Lee CH, et al. Hinokitiol up-regulates miR-494-3p to suppress BMI1 expression and inhibits self-renewal of breast cancer stem/progenitor cells. Oncotarget. 2017;8(44):76057–76068. doi:10.18632/oncotarget.18648
41. Zheng G, Jia X, Peng C, et al. The miR-491-3p/mTORC2/FOXO1 regulatory loop modulates chemo-sensitivity in human tongue cancer. Oncotarget. 2015;6(9):6931–6943. doi:10.18632/oncotarget.3165
42. Luo W, Lin Y, Meng S, Guo Y, Zhang J, Zhang W. miRNA-296-3p modulates chemosensitivity of lung cancer cells by targeting CX3CR1. Am J Transl Res. 2016;8(4):1848–1856.
43. Li J, Xia L, Zhou Z, et al. MiR-186-5p upregulation inhibits proliferation, metastasis and epithelial-to-mesenchymal transition of colorectal cancer cell by targeting ZEB1. Arch Biochem Biophys. 2018;640:53–60. doi:10.1016/j.abb.2018.01.002
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.