Back to Journals » OncoTargets and Therapy » Volume 10
The role of long noncoding RNA-LET in cell proliferation and invasion of nasopharyngeal carcinoma and its mechanism
Authors Chen L, Sun L, Dong L , Cui P, Xia Z, Li C, Zhu Y
Received 7 November 2016
Accepted for publication 26 January 2017
Published 30 May 2017 Volume 2017:10 Pages 2769—2778
DOI https://doi.org/10.2147/OTT.S126907
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr William C. Cho
Lei Chen,1,* Lingling Sun,1,* Lei Dong,2 Peng Cui,3 Ziwei Xia,4 Chao Li,1 Yu Zhu5
1Department of Otolaryngology, The Second Hospital of Tianjin Medical University, Tianjin, People’s Republic of China; 2Department of Pediatrics, Division of Hematology/Oncology, Aflac Cancer and Blood Disorders Center, Emory University School of Medicine, Atlanta, GA, USA; 3Department of Multidisciplinary Consultation Center of TCM and Western Medicine, The Affiliated Qingdao Hiser Hospital of Qingdao University, Qingdao, 4Department of Clinical Medicine, The Second Clinical Medical School of Tianjin Medical University, 5Department of Clinical Laboratory, Tianjin Huanhu Hospital, Tianjin Key Laboratory of Cerebral Vessels and Neural Degeneration, Tianjin, People’s Republic of China
*These authors contributed equally to this work
Abstract: LncRNA-LET, a recently identified long noncoding RNA, has been shown to act as a tumor suppressor; however, its biological function and mechanism have not been fully investigated. Our research found that there was less expression of LET in nasopharyngeal carcinoma (NPC) tissues than normal tissues and that LET might inhibit proliferation, adhesion and invasion of NPC in vitro by enhancing its expression. By contrast, decreased LET expression could promote the proliferation, adhesion and invasion of NPC. In addition, the expression profiles of related genes and MAPK/ERK pathway were also regulated effectively via overexpression or silencing of LET. This result provides comprehensive evidence of LET’s antitumor effect on NPC in vitro, which might provide a new approach for clinical treatment.
Keywords: LncRNA-LET, proliferation, invasion, nasopharyngeal carcinoma, MAPK/ERK pathway
Introduction
Nasopharyngeal carcinoma (NPC), which occurs in the epithelial lining of the nasopharynx, is one of the most common types of head and neck tumors.1,2 It has extremely skewed ethnic and geographic distributions, which is a rare malignant tumor in Western countries, but is one of the most prevalent cancers in the southern region of the People’s Republic of China and Southeast Asia.3–5 At present, radiotherapy and radiochemotherapy are the major means of treating NPC, but because of its high metastasis, a significant proportion of patients still develop neck and distant metastasis and the prognosis of these patients remains poor.6,7 Therefore, further explorations of the molecular mechanisms of NPC carcinogenesis and more effective therapeutic strategies are urgently required.
The long noncoding RNAs (lncRNAs) are non-protein-coding transcripts, longer than 200 nucleotides in length.8–10 Many studies have found that lncRNAs participate in many physiological processes by modulating gene expression at the epigenetic, transcriptional and posttranscriptional levels. Increasing evidence has indicated that several lncRNAs are linked to human disease, especially cancer,11,12 and their abnormal expression is closely related to tumor proliferation, differentiation and metastasis.
LncRNA-LET (with low expression in tumor) is a recently identified lncRNA, and recent studies have showed that LncRNA-LET plays key roles in diverse biological processes, such as cell cycle progression, differentiation, proliferation and so on, by regulating gene expression at the transcriptional level. Clinically, downregulation of LncRNA-LET has been detected in many human malignancies, such as esophageal cancer, osteosarcoma, pancreatic cancer and hepatocellular carcinoma. It has also been reported to exhibit tumor-suppressive roles in these tumors.13–17 However, its function in cell cycle and invasion of NPC, and related molecular mechanisms have not been fully discussed. Hence, this study investigates the biological function of LET in NPC. Our results show that LET exerts a significant antitumor effect via the regulation of the proliferation, adhesion, invasion and cell cycle of NPC. Our finding gives a deep insight into the antitumor mechanisms of LET and provides the theoretical basis for clinical treatment of NPC.
Materials and methods
Patients and tissue collection
A total of 95 NPC tissues and their pair-matched adjacent normal tissues were collected. The clinical stages of these tissues were determined according to the criteria of the American Joint Committee on Cancer. Written informed consent was obtained from all the patients including sample collection for the use of the NPC tissues and normal tissues in this study. This study was approved by the Ethics Committee of the Second Hospital of Tianjin Medical University, Tianjin, People’s Republic of China.
Cell lines and cell culture
The normal immortalized human nasopharyngeal epithelial cell line NP69, and the human NPC cell lines CNE1, CNE2, SUNE1, HNE1, HONE1 and C666-1 were purchased from the Cell Bank of Shanghai Institutes for Biological Sciences of Chinese Academy of Sciences, and maintained at 37°C in RPMI 1640 medium (Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS; Thermo Fisher Scientific) in a humidified incubator containing 5% CO2. The medium was routinely changed every 2 days. The cells in logarithmic growth phase were used for experiments.
Total RNA extraction and quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was extracted from normal tissues and NPC cells (seeded at a density of 5×104 cells/mL in a 24-well plate and cultured for 24 h) using TRIzol (Thermo Fisher Scientific) reagent according to the manufacturer’s instructions. RNA purity and integrity were analyzed using an Agilent Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA).
The cDNA was synthesized using an iScript cDNA Synthesis Kit (Bio-Rad Laboratories, Hercules, CA, USA) following the manufacturer’s instructions. qRT-PCR was performed using SYBR Premix ExTaq™ II kit (TaKaRa, Kusatsu, Japan). The conditions of qRT-PCR were as follows: 94°C for 10 s, 94°C for 5 s, 52°C for 30 s to anneal, and 72°C for 15 s followed by 40 cycles. The detection was set at 62°C. PCR amplifications were performed in three duplicates for each sample. The relative RNA expression was calculated using the 2−ΔΔCt method. The specific primer sequences are listed in Table 1.
  | Table 1 List of primers and their applications |
Construction of recombinant LET vector and transfection
The cDNA encoding LET was PCR-amplified and cloned into the pcDNA3.1 vector. The specific primer sequences are listed in Table 1. Transfection was performed using Lipofectamine2000 (Thermo Fisher Scietific) according to the manufacturer’s instructions. The transfection efficiency was monitored using qRT-PCR to quantify the post-transfection changes in LET expression level, and the data were analyzed using the same 2−ΔΔCt method.
Small interfering RNAs and transfection
Small interfering RNAs (siRNAs) targeting lncRNA-LET (si-LET) and negative control siRNA (si-Ctrl) were synthesized by GenePharma (Shanghai, People’s Republic of China) (Table 1). Transfection was performed using Lipofectamine2000 (Thermo Fisher Scietific) according to the manufacturer’s instructions. The transfection efficiency was monitored using qRT-PCR to quantify the post-transfection changes in LET expression level with the above condition, and the data were analyzed using the same 2−ΔΔCt method.
MTT assay
CNE2 cells were seeded at a density of 3×104 cells/mL in a 24-well plate, cultured for 24 h and then transfected with pcDNA3.1-LET or si-LET as described above. Cell viability was analyzed using MTT (Sigma-Aldrich, St Louise, MO, USA) assay for various periods of time (24, 48 and 72 h) as described previously. In brief, 1 μL/well of MTT was added and incubated at 37°C for an additional 4 h. Then, the medium was removed, and formazan was solubilized in isopropanol and measured spectrophotometrically at 563 nm. Each experiment was performed in triplicate and repeated three times.
Cell cycle assay
To determine the cell cycle distribution, appropriate cells were seeded at a density of 5×104 cells/mL in a 24-well plate and cultured for 24 h, and then they were collected, washed and suspended in phosphate-buffered saline (PBS) containing 1 mg/mL RNaseA and fixed in 75% ethanol. The fixed cells were stained with propidium iodide supplemented with RNaseA (Sigma-Aldrich) and analyzed with an FACScan flow cytometer (BD Biosciences, San Jose, CA, USA). Data were collected and analyzed with the ModFit software (BD Biosciences).
Cell–matrix adhesion assay
For measurement of cell adhesion, appropriate cells were seeded at a density of 5×104 cells/mL in a 24-well plate, and coated with 150 μL of type I collagen (10 μg/mL) and cultured for 30 min. Afterward, nonadherent cells were removed by washing with PBS, and adherent cells were fixed in ethanol. After staining with 0.1% crystal violet, fixed cells were lysed with 0.2% Triton-100, and measured spectrophotometrically at 550 nm.
Transwell invasion assay
Transwell invasion assay was performed using the BioCoat Matrigel Invasion Chamber (BD Biosciences) according to the manufacturer’s instructions. In brief, 4×104 appropriate cells were plated in the upper chamber consisting of 8-mm membrane filter inserts coated with Matrigel (BD Biosciences). The bottom chamber contained Dulbecco’s Modified Eagle’s Medium with 10% FBS as an inducer of invasion. After 24 h, cells on the upper surface were removed, and those attached on the lower side of the membrane were fixed and stained with crystal violet before counting under a microscope in five randomly selected fields.
Western blot assay
Cells were seeded at a density of 5×104 cells/mL in a 24-well plate for 24 h, and then they were collected and lysed completely with cell lysis buffer (Beyotime) and protease inhibitors cocktail (Roche). Identical quantities of proteins were loaded and separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis, transferred to polyvinylidene fluoride membranes (Millipore), and then incubated with appropriate antibodies, which were against c-myc (Cell Signaling Technology), cyclin D1 (Cell Signaling Technology), COX-2 (Cell Signaling Technology), p-ERK1/2 (Cell Signaling Technology), p-MEK (Cell Signaling Technology), total-ERK1/2 (Cell Signaling Technology), total-MEK (Cell Signaling Technology) and β-actin (Abcam). The immuno-reactive bands were visualized using an enhanced chemiluminescence system (Pierce, Rockford, IL, USA) and obtained using a CCD camera system (Tanon), and the density of the bands was measured by ImageJ.
Statistical analysis
All experiments were performed independently for at least three times. All data are presented as the mean values ± standard deviations of each group. Statistically significant differences were calculated by two-tailed Student’s t-test using SPSS software (version 19.0). The graphs were generated with GraphPad Prism 5.0.
Results
Correlation of LET expression with clinicopathological features in NPC
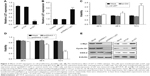
Previous research has shown that LET expression was decreased in NPC tissues. To further validate this result, we firstly examined the LET expression level and its correlation with clinicopathological characteristics in NPC. The expression level of LET was detected in 95 pairs of NPC tissues and pair-matched adjacent normal tissues using qRT-PCR. As shown in Figure 1A, LET transcript expression in NPC tissues was significantly downregulated compared with their corresponding adjacent normal tissues (P<0.01).
Furthermore, we also examined the LET expression levels in the NPC cell line CNE2, compared with immortalized normal nasopharyngeal epithelial cell line NP69 used as control groups. The results also showed that LET expression was greatly suppressed in CNE2 cells, compared with normal nasopharyngeal epithelial cell line (Figure 1B).
Effect of LET on NPC cell proliferation
To fully investigate the biological function of LET in NPC, we first studied the effect of downregulated LET on cell proliferation. For this purpose, we inhibited LET expression by transfecting LET-siRNA (si-LET) into CNE2 cells (named as si-LET group), using original cells and control siRNA (si-Ctrl) as control and the negative control, respectively. After 24 h of RNA interference, we examined the interference effect by qRT-PCR, and the result showed that LET expression level was decreased to 17.4%. This means that LET expression could be efficiently inhibited in CNE2 cells by transfecting with si-LET (Figure 2A). Then, we measured the proliferation of CNE2 cells. The MTT assays indicated that silencing of LET significantly promoted cell proliferation of CNE2 cells (Figure 2C).
To further investigate its effect on cell proliferation, we enhanced LET expression in CNE2 cells by transfecting an LET expression vector (pcDNA3.1-LET) (named as LET group) (Figure 2B). As expected, the MTT results revealed that overexpressing LET resulted in a significant decrease of cell viability in 72 h (Figure 2D).
Moreover, the expression levels of three proliferation-related genes, cyclin D1, c-myc and COX-2, were all upregulated in si-LET group and downregulated in LET group (Figure 2E).
Effect of LET on NPC cell cycle
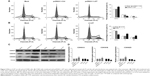
The finding of the antiproliferation effect of LET on NPC cells prompted us to investigate whether growth-inhibitory effect of LET was mediated through cell cycle arrest. For this purpose, we measured the cell cycle alteration in NPC cells by flow cytometry. As depicted in Figure 3A and B, in si-LET groups, the percentage of G0/G1 cells decreased from around 65.3% to around 34.2%, and the percentage of S cells increased from 21.7% to 40.5%, while in LET groups, there was a G0/G1 accumulation from around 64.6% to 80.4%, and a decrease of S phase from 21.5% to 10.8%.
Consistent with these results, the protein and mRNA expression levels of cell cycle-related genes, CDKN1A, CDKN1B and CDKN2D, were significantly upregulated in LET groups, and were remarkably downregulated in si-LET group (Figure 3C and D). This finding indicated that repression of LET could promote cell proliferation via inducing cell cycle into S phase by regulation of the expression of multiple cell cycle-related genes.
Effect of LET on NPC cell adhesion and invasion
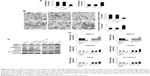
In addition, we further investigated the potential anti- metastatic activity of LET in CNE2 cells. CNE2 cells were transfected with si-LET or pcDNA3.1-LET and then analyzed for cell adhesion by cell–matrix adhesion assay, and cell invasion by transwell assay. In the cell–matrix adhesion assay, the cell adhesion of si-LET group was significantly increased, while that of the LET group was reduced (Figure 4A). The transwell assay showed that the invasiveness of CNE2 cells transfected with si-LET was promoted, while that of the LET group was remarkably suppressed compared with negative control cells (Figure 4B).
Besides that, we further examined the protein and mRNA expression level of invasion-related genes. The result was also consistent with the effect of LET on CNE2 cells. It suggested that LET might inhibit the invasion of NPC cells via regulating the expression of related genes, such as N-cadherin, CD44, CXCR4, HNRNPU, MMP-2 and MMP-9 (Figure 4C and D).
Effect of LET on ERK/MAPK signaling pathway
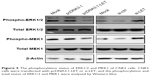
Since ERK1/2 pathway plays a crucial role in cancer biology and constitutively active ERK1/2 signaling has been shown to lead to increased tumor invasion,18–22 we next studied whether LET plays a role in activation of ERK signaling pathway. First, we analyzed the phosphorylation status of ERK1/2 by Western blot assays. As shown in Figure 5, p-ERK1/2 was remarkably downregulated and upregulated in LET group and si-LET group, respectively; however, the total-ERK1/2 was not changed. Since activation of the ERK pathway is regulated by MEK1, we then examined the phosphorylation status of MEK1, and found that the total MEK1 was also not changed. The results were similar to that of ERK; the phosphorylation of MEK1 in LET group was decreased, but was increased in si-LET group, which means that LET might suppress the cell growth and invasion of NPC cells through inhibiting the activation of ERK signaling pathway.
Discussion
LncRNAs are evolutionarily conserved noncoding RNAs that are more than 200 nucleotides in length. In recent years, an increasing number of research studies have demonstrated that lncRNAs are not the “dark matter” of the genome, but play integral and essential roles in various biological processes such as transcriptional and translational regulation of protein-coding sequences, cell proliferation and differentiation.23 In addition, accumulating evidence indicates that many classical lncRNAs participate in tumorigenesis by modulating the expression of many oncogenes or tumor suppressor genes.24–26 However, there are only several studies of lncRNAs in human NPC. HOTAIR, MALAT1 and ANRIL, three types of lncRNAs, have been found to be dysregulated in NPC, and recent research has demonstrated that lncRNA-LET might be involved in cell proliferation and cell apoptosis of NPC. However, the functions and mechanisms of lncRNAs in NPC remain largely unknown.27–31
In the present study, we fully investigated the biological functions and related mechanisms of lncRNA-LET in NPC. We knocked down or overexpressed lncRNA-LET to alter its expression level in NPC cells, and then examined its effect on cell growth and invasion. The results showed that the expression of LET was suppressed in NPC tissues, compared to their adjacent normal tissues. Thus, LET was found to exhibit various antitumor effects in NPC.
Matrix metalloproteinases, such as MMP-2 and MMP-9, could digest tissues and cells basement membrane, and adhesion molecules such as N-cadherin and CD44 could induce heterogeneous adhesion of tumor cells; therefore, the potential of invasion of the tumor cells is increased by the gene expression of the above molecules. HNRNPU is a cell cycle-related gene, and it could regulate the expression of CDKN1A, CDKN1B and CDKN2D which are anti-oncogenes and could suppress the activity of cyclin and CDKs, to influence cell cycle and proliferation. In this study, high expression of LET was found to inhibit cell proliferation, induce cell cycle arrest in G0/G1 phase and suppress cell invasion by regulating the expression of the related genes, while low expression of LET exhibited the opposite effects.
The MAPK pathways serve to coordinate key cellular processes. The ERK1/2 pathway, activated by MEK1/2,32 plays key roles in the regulation of multiple biological activities, including cell proliferation, differentiation, cell cycle transition and survival, and constitutive activation of the ERK1/2 pathway contributes to tumorigenesis, or cancer growth, and increases the cell death threshold.33 The ERK1/2 pathway can regulate the expression of CXCR4 and cyclin D1, which is responsible for the G1/S transition.34 Recent research has demonstrated that in the human NPC cell line CNE1, cell growth and cell cycle were suppressed mainly by inhibiting the phosphorylation and kinase activity of ERK1/2.35 Our study also showed that overexpressing LET could inhibit the phosphorylation of ERK1/2, while silencing of LET could promote its activation.
Collectively, all these results suggest that lncRNA-LET could effectively regulate the cell proliferation, adhesion and invasion of NPC. This is the first report to fully demonstrate its potential anticancer effect in NPC cells, and the results may provide a theoretical support for its application in clinical treatment of NPC.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (grant no 31501159), Tianjin Public Health Key Research Project (grant no 15KG108) and Tianjin Research Program of Application Foundation and Advanced Technology (grant no 14JCQNJC12000).
Disclosure
The authors report no conflicts of interest in this work.
References
Hu C, Wei W, Chen X, et al. A global view of the oncogenic landscape in nasopharyngeal carcinoma: an integrated analysis at the genetic and expression levels. PLoS One. 2012;7(7):e41055. | ||
Burgos JS. Involvement of the Epstein-Barr virus in the nasopharyngeal carcinoma pathogenesis. Med Oncol. 2005;22(2):113–121. | ||
Jiang C, Zhou L, Wang H, Zhang Q, Xu Y. Axl is a potential cancer prognostic marker for the migration and invasion of nasopharyngeal carcinoma. Adv Clin Exp Med. 2016;25(3):531–537. | ||
Lee V, Lam KO, Kwong D, Leung TW. Radiation recall after capecitabine in a patient with recurrent nasopharyngeal carcinoma: a case report. J Med Case Rep. 2016;10(1):247. | ||
Li G, Liu Y, Liu C, et al. Genome-wide analyses of long noncoding RNA expression profiles correlated with radioresistance in nasopharyngeal carcinoma via next-generation deep sequencing. BMC Cancer. 2016;16:719. | ||
Jiang Q, Zhou Y, Yang H, et al. A directly negative interaction of miR-203 and ZEB2 modulates tumor stemness and chemotherapy resistance in nasopharyngeal carcinoma. Oncotarget. 2016;7(41):67288–67301. | ||
Wang HZ, Cao CN, Luo JW, et al. High-risk factors of parotid lymph node metastasis in nasopharyngeal carcinoma: a case-control study. Radiat Oncol. 2016;11(1):113. | ||
Feldman ER, Kara M, Oko LM, et al. A gammaherpesvirus noncoding RNA is essential for hematogenous dissemination and establishment of peripheral latency. mSphere. 2016;1(2):e00105–e00115. | ||
Fu WM, Lu YF, Hu BG, et al. Long noncoding RNA Hotair mediated angiogenesis in nasopharyngeal carcinoma by direct and indirect signaling pathways. Oncotarget. 2016;7(4):4712–4723. | ||
Zhang W, Wang L, Zheng F, et al. Long noncoding RNA expression signatures of metastatic nasopharyngeal carcinoma and their prognostic value. Biomed Res Int. 2015;2015:618924. | ||
Gong Z, Zhang S, Zeng Z, et al. LOC401317, a p53-regulated long non-coding RNA, inhibits cell proliferation and induces apoptosis in the nasopharyngeal carcinoma cell line HNE2. PLoS One. 2014; 9(11):e110674. | ||
Nie Y, Liu X, Qu S, Song E, Zou H, Gong C. Long non-coding RNA HOTAIR is an independent prognostic marker for nasopharyngeal carcinoma progression and survival. Cancer Sci. 2013;104(4):458–464. | ||
Jiang S, Wang HL, Yang J. Low expression of long non-coding RNA LET inhibits carcinogenesis of cervical cancer. Int J Clin Exp Pathol. 2015;8(1):806–811. | ||
Yang F, Huo XS, Yuan SX, et al. Repression of the long noncoding RNA-LET by histone deacetylase 3 contributes to hypoxia-mediated metastasis. Mol Cell. 2013;49(6):1083–1096. | ||
Ma MZ, Kong X, Weng MZ, et al. Long non-coding RNA-LET is a positive prognostic factor and exhibits tumor-suppressive activity in gallbladder cancer. Mol Carcinog. 2015;54(11):1397–1406. | ||
Sun Q, Liu H, Li L, et al. Long noncoding RNA-LET, which is repressed by EZH2, inhibits cell proliferation and induces apoptosis of nasopharyngeal carcinoma cell. Med Oncol. 2015;32(9):226. | ||
Liu FT, Zhu PQ, Ou YX, Lin QS, Qiu C, Luo HL. Long non-coding RNA-LET can indicate metastasis and a poor prognosis: a meta-analysis. Minerva Med. 2016;107(2):101–107. | ||
Roskoski R Jr. ERK1/2 MAP kinases: structure, function, and regulation. Pharmacol Res. 2012;66(2):105–143. | ||
Sulzmaier FJ, Ramos JW. RSK isoforms in cancer cell invasion and metastasis. Cancer Res. 2013;73(20):6099–6105. | ||
Whyte J, Bergin O, Bianchi A, McNally S, Martin F. Key signalling nodes in mammary gland development and cancer. Mitogen-activated protein kinase signalling in experimental models of breast cancer progression and in mammary gland development. Breast Cancer Res. 2009;11(5):209. | ||
Giannelli G, Napoli N, Antonaci S. Tyrosine kinase inhibitors: a potential approach to the treatment of hepatocellular carcinoma. Curr Pharm Des. 2007;13(32):3301–3304. | ||
Luo X, Yang L, Xiao L, et al. Grifolin directly targets ERK1/2 to epigenetically suppress cancer cell metastasis. Oncotarget. 2015;6(40): 42704–42716. | ||
Sun J, Bie B, Zhang S, Yang J, Li Z. Long non-coding RNAs: critical players in hepatocellular carcinoma. Int J Mol Sci. 2014;15(11): 20434–20448. | ||
Shi D, Zheng H, Zhuo C, et al. Low expression of novel lncRNA RP11-462C24.1 suggests a biomarker of poor prognosis in colorectal cancer. Med Oncol. 2014;31(7):31. | ||
Feng S, Yao J, Chen Y, et al. Expression and functional role of reprogramming-related long noncoding RNA (lincRNA-ROR) in glioma. J Mol Neurosci. 2015;56(3):623–630. | ||
Yin D, He X, Zhang E, Kong R, De W, Zhang Z. Long noncoding RNA GAS5 affects cell proliferation and predicts a poor prognosis in patients with colorectal cancer. Med Oncol. 2014;31(11):253. | ||
Zhang W, Huang C, Gong Z, et al. Expression of LINC00312, a long intergenic non-coding RNA, is negatively correlated with tumor size but positively correlated with lymph node metastasis in nasopharyngeal carcinoma. J Mol Histol. 2013;44(5):545–554. | ||
Zhang XM, Wang XY, Sheng SR, Wang JR, Li J. Expression of tumor related genes NGX6, NAG-7, BRD7 in gastric and colorectal cancer. World J Gastroenterol. 2003;9(8):1729–1933. | ||
Gao W, Chan JY, Wong TS. Differential expression of long noncoding RNA in primary and recurrent nasopharyngeal carcinoma. Biomed Res Int. 2014;2014:404567. | ||
Yang QQ, Deng YF. Genome-wide analysis of long non-coding RNA in primary nasopharyngeal carcinoma by microarray. Histopathology. 2015;66(7):1022–1030. | ||
Yang QQ, Deng YF. Long non-coding RNAs as novel biomarkers and therapeutic targets in head and neck cancers. Int J Clin Exp Pathol. 2014; 7(4):1286–1292. | ||
Ellinger-Ziegelbauer H, Brown K, Kelly K, Siebenlist U. Direct activation of the stress-activated protein kinase (SAPK) and extracellular signal-regulated protein kinase (ERK) pathways by an inducible mitogen-activated protein kinase/ERK kinase kinase 3 (MEKK) derivative.J Biol Chem. 1997;272(5):2668–2674. | ||
Ishikawa Y, Kitamura M. Dual potential of extracellular signal-regulated kinase for the control of cell survival. Biochem Biophys Res Commun. 1999;264(3):696–701. | ||
Zhai N, Xia Y, Yin R, Liu J, Gao F. A negative regulation loop of long noncoding RNA HOTAIR and p53 in non-small-cell lung cancer. Onco Targets Ther. 2016;9:5713–5720. | ||
Zhang SQ, Cai WJ, Huang JH, et al. Icariin, a natural flavonol glycoside, extends healthspan in mice. Exp Gerontol. 2015;69:226–235. |
 © 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.