Back to Journals » Infection and Drug Resistance » Volume 16
The Resistance and Virulence Characteristics of Salmonella Enteritidis Strain Isolated from Patients with Food Poisoning Based on the Whole-Genome Sequencing and Quantitative Proteomic Analysis
Authors Xu B , Hou Z , Liu L, Yan R , Zhang J , Wei J, Du M, Xuan Y, Fan L, Li Z
Received 29 June 2023
Accepted for publication 27 September 2023
Published 6 October 2023 Volume 2023:16 Pages 6567—6586
DOI https://doi.org/10.2147/IDR.S411125
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Benjin Xu,1– 3,* Zhuru Hou,2,4,* Ling Liu,1– 3,* Rongrong Yan,3 Jinjing Zhang,3 Jianhong Wei,4 Miao Du,1,2 Yan Xuan,1,2 Lei Fan,2,4 Zhuoxi Li2,4
1Department of Medical Laboratory Science, Fenyang College of Shanxi Medical University, Fenyang, People’s Republic of China; 2Key Laboratory of Lvliang for Clinical Molecular Diagnostics, Fenyang, People’s Republic of China; 3Department of Clinical Laboratory, Fenyang Hospital of Shanxi Province, Fenyang, People’s Republic of China; 4Department of Basic Medicine, Fenyang College of Shanxi Medical University, Fenyang, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Ling Liu; Benjin Xu, Department of Medical Laboratory Science, Fenyang College of Shanxi Medical University, Fenyang, 032200, People’s Republic of China, Email [email protected]; [email protected]
Objective: This paper explores the drug resistance, genome and proteome expression characteristics of Salmonella from a food poisoning event.
Methods: A multidrug-resistant Salmonella Enteritidis strain, labeled as 27A, was isolated and identified from a food poisoning patient. Antimicrobial susceptibility testing determined the resistance of 27A strain to 14 antibiotics. Then, WGS analysis and comparative genomics analysis were performed on 27A, and the functional annotation of resistance genes, virulence genes were performed based on VFDB, ARDB, COG, CARD, GO, KEGG, and CAZY databases. Meanwhile, based on iTRAQ technology, quantitative proteomic analysis was conducted on 27A to analyze the functions and interactions of differentially expressed proteins related to bacterial resistance and pathogenicity.
Results: Strain 27A belonged to ST11 S. Enteritidis and was resistant to levofloxacin, ciprofloxacin, ampicillin, piperacillin, and ampicillin/sulbactam. There were 33 drug resistance genes, 384 virulence genes and 2 plasmid replicon, IncFIB(S) and IncFII(S), annotated by WGS. Proteomic analysis revealed significant changes in virulence and drug proteins, which were mainly involved in bacterial pathogenicity and metabolic processes. PPI prediction showed the relationship between virulence proteins and T3SS proteins, and PagN cooperated with proteins related to T3SS to jointly mediate the invasion of 27A strain on the human body. Phylogenetic analysis indicated that S. Enteritidis has potential transmission in humans, food, and animals.
Conclusion: This study comprehensively analyzed the drug resistance and virulence phenotypes of S. Enteritidis 27A using genomic and proteomic approaches. These helps reveal the drug resistance and virulence mechanisms of S. Enteritidis, and provides important information for the source tracing and the prevention of related diseases, which lays a foundation for research on food safety, public health monitoring, and the drug resistance and pathogenicity of S. Enteritidis.
Keywords: Salmonella, WGS, quantitative proteomics, resistance, virulence, evolution
Introduction
As an important zoonosis pathogen, Salmonella is widely distributed in nature and one of the four major pathogens causing global diarrhea diseases.1 Salmonella has six subspecies and 2659 serotypes, among which Salmonella Enteritidis (S. Enteritidis) are the most common serotypes causing Salmonella outbreaks.1 S. Enteritidis has a wide variety of host species and strong pathogenicity, which can be transmitted to humans through undercooked or raw infected foods (especially meat and eggs), causing fever, abdominal pain, diarrhea, as well as urinary tract infections, arthritis, meningitis, and even death.2 According to the reports, S. Enteritidis infections cause approximately 93.8 million illnesses and 155,000 deaths each year in the world, with about approximately 80.3 million cases being foodborne.3 Due to the lack of effective immune prevention strategies, the analysis of S. Enteritidis infections has attracted much attention.
Antibiotics are commonly used strategies for treating S. Enteritidis infections and are widely used in clinical, livestock, and food processing. However, antibiotic resistance has become one of the greatest public health threats of the 21st century in the past decades of development. Multidrug resistant S. Enteritidis appears and has a significant impact on public health. These bacteria have developed resistance to multiple antibiotics, making it difficult to treat infections caused by them and resulting in longer hospital stays, increased morbidity, and higher healthcare costs.4 Studies have shown that S. Enteritidis was resistant to nalidixic acid (94.5%), ampicillin (75%), streptomycin (67%), cefoperazone (52%), and sensitive to cephalosporin and ciprofloxacin.5 Cephalosporins and ciprofloxacin are the first-line drugs for treating Salmonella infections, with azithromycin as an adjuvant treatment. However, as the sensitivity of Salmonella to cephalosporins and ciprofloxacin gradually decreases, strains that are resistant to both cephalosporins, ciprofloxacin, and azithromycin have emerged.6 In addition to increasing bacterial resistance to antibiotics, resistance genes, such as quinolone resistance (qnrA, qnrB), β-lactam resistance (blaTEM, blaSHV, blaCTX-M, blaCMY-2), have also emerged. These genes are usually located on the plasmid and can transfer among multiple bacterial genera as the plasmid moves, thereby expressing resistance to kinds of antibiotics and increasing the difficulty of anti-infection treatment.7
The virulence mechanisms of S. Enteritidis are complex, as it contains the type III secretion system (T3SS) encoded by Salmonella pathogenic island-1 (SPI-1) and Salmonella pathogenic island-2 (SPI-2). T3SS is the core of S. Enteritidis pathogenicity. SPI-1 T3SS encodes Salmonella invasion proteins (Sips) and Salmonella outer proteins (Sops), which change the actin cytoskeleton of intestinal epithelial cells, leading to membrane folding and bacterial internalization. Among them, SopE can induce the production of nitrate by the host, promoting the growth of Salmonella within host cells.8 Besides, after engulfed by cells, the host cell membrane will rearrange to form a membrane-bound organelle, called Salmonella containing vacuole (SCV). SPI-2 T3SS genes are expressed within the SCV, and contribute to the survival and large-scale replication of Salmonella in host cells.9 In addition to SPIs, the plasmids of S. Enteritidis also carry virulence genes that play roles in infecting host cells, ensuring nutrient supply, competing with symbiotic bacteria, and evading the innate immune system.10
Recently, whole-genome sequencing (WGS) has developed rapidly, and genomic analysis can provide detailed data on pathogen genes and identify serotypes as well as virulence, drug-resistant determinants. WGS-based Salmonella serotyping can be obtained through open access tools, avoiding the costs associated with traditional methods and allowing for efficient and accurate serotyping of Salmonella. In addition, WGS can predict drug resistance by identifying resistance genes, which perfectly matches the resistance phenotype obtained through standard broth microdilution methods.11 The research has shown that the all-round informations provided by WGS enhances the monitoring of multidrug-resistant strains transmitted in types of hosts. Genomic data can be used to identify the source of the epidemic, describe its development, and understand the consequences of antibiotic use, greatly improving the speed of tracing research.12 WGS was used as a prospective monitoring tool for foodborne diseases as early as 2016. Compared to traditional microbial typing and characterization techniques, WGS offers faster and more accurate monitoring results. Proteins are the main functional performers in organisms, and the process of translating genetic information into functional proteins is complex and multi-step. Although the mechanisms of transcription and translation are highly refined processes, they still have a certain error rate. These transcription and translation errors are the main causes of diseases. Therefore, relevant information about proteins cannot be simply read from genes or transcripts.13 Proteomics quantitative analysis is an important approach for studying the complex proteome profiles of bacteria, post-translational modifications of proteins, interactions between pathogens and hosts, antimicrobial resistance, and the discovery of novel protein biomarkers. It serves as an effective complement to genomics and transcriptomics.14 Proteomics is used to annotate bacterial resistance proteins and virulence factors, monitor the molecular responses of bacteria to external stimuli, such as antibiotic damage, and capture changes in metabolic pathways that contribute to the development of antibiotic resistance, which will offer more important information on bacterial resistance mechanisms and the lifecycle of strains in food production.15
In this study, we performed whole-genome sequencing and quantitative proteomic analysis on a multidrug resistant S. Enteritidis strain isolated from a food poisoning incident in our city. We characterized the drug resistance and virulence factors of S. Enteritidis at both the gene and protein levels, and further revealed the genetic characteristics and evolutionary relationship of the strains through comparative genomics analysis. This study helps to elucidate the pathogenic and drug resistance mechanisms of S. Enteritidis, providing important information for source tracing and the prevention and treatment of related diseases. At the same time, it provides a scientific basis for food safety assurance and public health monitoring.
Materials and Methods
Isolation and Identification of Strain
In May 2021, we investigated thirteen patients with infectious diarrhea admitted in the same batch to a rank A tertiary hospital in Shanxi, China, and collected clinical data using the electronic medical record system. We collected the patient’s feces, inoculated them into SBG enrichment solution, and incubated them in 35°C incubator for 24 hours. Then inoculate the enrichment solution into SS culture medium, and incubate them in 35°C incubator for 24 hours. The suspicious colonies on SS culture medium were selected for identification. The isolates were identified by matrix-assisted laser desorption ionization-time of flight (MALDI-TOF/TOF) mass spectrometry. All the isolates were identified as Salmonella, and strain 27A belonged to S. Enteritidis.
Antimicrobial Susceptibility Testing
The tests were interpreted using Clinical and Laboratory Standards Institute (CLSI) guidelines.16 The minimum inhibitory concentration (MIC) of bacterial strains against 14 antibiotics was detected by VITEK2-compact automatic drug sensitivity analyzer. The antibiotics include ampicillin, piperacillin, ampicillin/sulbactam, piperacillin/sulbactam, cefatriaxone, ceftazidime, cefepime, aztreonam, imipenem, meropenem, compound sulfamethoxazole, furadantin, levofloxacin, and ciprofloxacin. Cefoperazone was tested by K-B method. ATCC25922 and ATCC25923 were used for quality control.
Whole-Genome Sequencing and Comparative Genomics Analysis
The 27A genome was sequenced at the Beijing Genomics Institute (BGI, Shenzhen, China) using a PacBio Sequel II and DNBSEQ platform. The PacBio platform utilized four SMRT cells Zero-Mode Waveguide arrays for sequencing, and the resulting subreads set was generated. Subreads were repaired and the Canu program was employed for self-correction. The assembled contigs were used for predicting genome component, including tRNA,17 rRNA,18 sRNA,19 tandem repeats, minisatellite DNA, microsatellite DNA and CRISPR identification. And for function annotation, these genes were annotated some databases, including VFDB,20 CARD,21 COG,22 GO,23 and KEGG.24 Otherwise, based on the Achtman scheme, WGS data were used to predict in multi-locus sequence type (MLST).
Then analyze the Core/Pan genes of 27A strain and 18 reference strains, as well as the functions of these genes. Cluster these 19 strains based on Core/Pan genes, and construct the phylogenetic tree using the TreeBeST with the NJ method. The iTOL was used to visualize the results.25
Quantitative Proteomic Analysis
Sample Preparation
S. Enteritidis 27A was used as a test strain, S. Typhimurium ATCC14028 was used as a control strain. The strains were inoculated into 10mL LB culture at a rate of 2% and incubated at 37°C at 220 rpm for 12h. The 2mL bacterial solution was transferred to 250mL triangular flask containing 100mL LB culture medium, and continued to be cultured at 37°C, 220 rpm for 12h. After centrifugation at 6500r/min for 5 min, the bacteria were collected and cleaned once with RNAase free water. Add five times the volume of methanol and stand at 4°C for 1h to inactivate the bacteria; the bacteria were collected by centrifugation, frozen them in liquid nitrogen for 30 min and stored at −80°C. Each sample was repeated three times.
Proteomics Bioinformation Analysis
The sequencing of proteomes was carried out by the BGI mass spectrometry platform, including protein extraction, quality control of the samples, protein enzymatic hydrolysis and high pH RP (reversed-phase) separation, and DDA and DIA analysis by nano-LC-MS/MS.
The DDA data was analyzed using the Andromeda search engine in MaxQuant, and the identification results were utilized for constructing a spectral library. Deconvolution of DIA data and the DDA spectrogram library provided qualitative and quantitative information on peptides and proteins. MSstats26 was used to statistically assess the difference significance of all data and to analyze the biological function of differential proteins. Functional annotation was performed by using GO,23 KOG and KEGG24 databases. The STRING software was utilized to predict the potential protein–protein interactions27 and the Cytoscape software was used to map network interactions. The visualization results of heatmaps, cosine plots, etc., were obtained using R language.
Results
Clinical Symptoms and Treatment of Patients
The patient developed diarrhea, watery stools (4 times per day), and fever with the highest body temperature of 39.0°C around 3am on May 29, 2021. The patient took orally phenanthramine, montmorillonite powder, and Huoxiang Zhengqi water, but the patient’s symptoms did not improve. The patient visited the hospital in the afternoon of the same day. On May 30, the patient underwent additional tests, which showed elevated levels of C-reactive protein and procalcitonin, leading to a diagnosis of infectious diarrhea. Take the patient’s feces and isolate the pathogen, which was named as strain 27A. MALDI-TOF-MS identification of 27A strain suggested that it was S. Enteritidis (Figure S1, Table S1). Tracking the patient’s infection history, it was found that the patient consumed unclean sandwiches the day before the onset of disease, and the patient had infectious diarrhea caused by food poisoning. During hospitalization, the patient was treated with levofloxacin and ceftazidime in combination and received symptomatic and supportive treatment through liquid rehydration. After two days of hospitalization, the patient was discharged after clinical recovery with fine mental state, appetite, and sleep (Figure 1).
 |
Figure 1 Clinical Diagnosis, Treatment and Epidemiological Investigation of the Patient. |
Drug Resistance Characteristics
The results of the drug sensitivity test showed that 27A strain was a multi-drug resistant strain, resistance to ampicillin, piperacillin, ampicillin/sulbactam, levofloxacin and ciprofloxacin, sensitive to piperacillin/tazobactam, cefoperazone/sulbactam, ceftriaxone, ceftazidime, cefepime, aztreonam, imipenem, meropenem and compound sulfamethoxazole, and moderately sensitive to furantoin (Table 1). The 27A strain had high resistance to β-lactam and quinolone antibiotics, but was sensitive to carbapenems and sulfonamides.
 |
Table 1 Drug Resistance of 27A to Various Antimicrobial Agents |
Whole-Genome Sequencing and Analysis
Basic Information of the Genome
A total of 8,764,210 reads were obtained from whole-genome sequencing of 27A, and after quality control 8,647,646 valid reads were retained, with an effective rate of 98.67%. The size of 27A genome was 4748869bp (Table 2), including a circular chromosome with a size of 4,679,690 bp and a GC content of 52.17%, as well as two circular plasmids. Plasmid 1 had a size of 64327bp and a GC content of 51.76%, while plasmid 2 had a size of 4852bp and a GC content of 59.70%. Chromosome and plasmid information were drawn separately as a genome circle map (Figure 2). The 27A genome predicted 4660 coding genes and 174 non-coding genes. The non-coding genes only existed in chromosomes, including 22 rRNAs, 68 sRNAs, and 84 tRNAs. There were 71 tandem repeat sequences distributed in chromosomes, and 2 tandem repeat sequences distributed in plasmid 1, all of which were small satellite DNA. Plasmid 2 did not contain tandem repeat sequences. A total of two CRISPR structures were identified in 27A, repeated 5 and 8 times respectively, with sequences as follows: GTGTTCCCCGCGCCAGCGGGGATAAACCG.
 |
Table 2 Basic Information of 27A |
Functional Annotation
To further analyze the genes functions of strain 27A, this study annotated its genome with GO, COG, KEGG, and CAZY databases, with a total of 3893 genes annotated (Figure 3A). The COG database predicted 3682 genes and annotated 4287 pieces of information, of which 44.67% were related to metabolism, including 414 genes related to carbohydrate transport and metabolism, 381 genes related to amino acid transport and metabolism, 312 genes related to energy metabolism, and 228 genes related to inorganic ion transport and metabolism (Figure 3B). According to GO analysis, 3088 genes were annotated into 35 GO subclasses, accounting for 66.27% of all coding genes. The genes of 27A were relatively active in biological processes, accounting for 52.09%. Among the major categories of biological processes, the cellular process subclass had the highest number of genes, with 1939, followed by metabolic processes with 1714 (Figure 3C). In the KEGG database, a total of 3260 genes in 27A were annotated on 40 entries of 6 functional categories, accounting for 69.95% of the coding genes. The metabolic process was annotated with a total of 12 entries in KEGG, which was the highest number of annotated genes among the 6 functional categories (2226 genes), accounting for 68.28% of the total annotated genes. Among them, the top three were global and overview maps (852 genes), carbohydrate metabolism (355 genes), and amino acid metabolism (219 genes) (Figure 3D). The strain 27A had 132 genes encoding carbohydrate active enzymes (CAZY). Among them, CBM50 had the most genes, also known as LysM domains, which attached to various enzymes from families GH18, GH19, GH23, GH24, GH25 and GH73 (Figure 3E).
Analysis of Drug Resistance Genes and Virulence Genes
The 27A isolate was annotated with resistance and virulence genes using CARD, ARDB, VFDB, and T3SS databases. Table 3 summarizes the annotated 33 resistance and 386 virulence genes. Drug resistance genes were mainly divided into seven categories: resistance-nodulation-cell division transporter (RND), major facilitator superfamily transporter (MFS), potassium reverse transport system, β-lactamase, small multidrug resistance transporter (SMR), VanA promoter and other types. Three drug resistance determinants related to antibiotics included blaTEM-194 (β-lactamase genes), aac(6)-Ib, aac(6)-If, aph(3”)-Ib, aph(6)-Id (aminoglycoside resistance genes), tetA, tetR, tet34 (tetracycline resistance genes). Three efflux pump systems included RND (acrA, acrB, tolC, oprM, mexE, mexF, macB), MFS (mdtG, mdtH, mdtL, mdtM, mdtK, rosA, tetA, tetR, emrA, emrR), SMR (ykkc). Virulence genes were mainly divided into eight categories, including Salmonella pathogenicity island, plasmid-related virulence factors, adhesins, flagella, phage encoded virulence factors, fimbriae, lipopolysaccharide and capsule. Among them, Salmonella pathogenic islands were divided into SPI-1 to SPI-5. SPI-1 and SPI-2 played major roles in Salmonella invasion and infection, and most of virulence factors existed as secretion systems (Figure 4B). 27A strain contained two plasmids, and plasmid 1 contained one resistance gene blaTEM-194 related to β-lactam antibiotics, 9 virulence genes and 18 secretion system-related genes. Apart from them, it contained toxin gene ccdB and binding transfer fimbriae assembly protein genes traL, traE, traK, traB, traV. There were only two plasmid replicons in strain 27A, IncFIB(S) and IncFII(S), both of which were on plasmid 1 and contained T3SS related genes. Plasmid 2 contained two resistance genes (aph(6)-Id, aph(3”)-Ib) associated with aminoglycosides, two virulence genes (tetA, tetR) related to efflux pumps, and one gene (trfA) associated with plasmid replication initiation protein (Figure 4A).
 |
Table 3 Resistance Genes and Virulence Genes of 27A |
 |
Figure 4 Resistance Genes and Virulence Genes Distribution Diagram of 27A. (A) Distribution of drug resistance and virulence genes on 27A plasmid. (B) Salmonella pathogenic island SPI-1 and SPI-2. |
Comparative Genomics Analysis
MLST typing was performed on strain 27A, and the results showed that its sequence type was ST11 (Table S2). We selected 17 ST11 and 1 ST3632 S. Enteritidis isolates from the NCBI website (Table S3), and conducted core and pan genomic analysis on 27A and these reference strains. The core genome of these 19 isolates consisted of 3445 genes, with 0–324 non-essential genes distributing among different strains (Figure 5A). 193 non-essential genes and 37 core genes were selected for cluster analysis on 19 isolates, which were divided into two branches. The strain 27A and ASM130523v1 were located in the same branch, while the other 17 isolates were located in another branch. 27A had the closest genetic relationship with ASM130523v1 (Figure 5B). According to COG database analysis, 3445 core genes and 509 non-essential genes were enriched into 24 and 22 subcategories, respectively, with 45.1% of core genes and 44.2% of non-essential genes involved in metabolism. For core genes, 10.0% were involved in carbohydrate metabolism, 9.0% in amino acid metabolism, and 6.9% in energy metabolism; For non-essential genes, 13.2% were involved in Mobile: phases, transitions, 8.1% in carbohydrate metabolism, and 7.3% in amino acid metabolism (Figure 5C). Also, COG functional annotation was performed on the specific genes of 19 strains, and 115 specific genes were annotated in 8 strains. The specific genes in 27A were mainly enriched in energy generation and conversion, intracellular transport, secretion, and vesicular transport, as well as mobile: phases, transports. The ASM130523v1 isolate only had 2 specific genes, one participating in the resistance mechanism, and one with unknown function. The ASM276113v1 isolate had the most number of specific genes, with 41, of which 11 were involved in Mobile: phases, transitions (Figure 5D).
Based on the CorePan results of 19 isolates, a phylogenetic tree was constructed using TreeBeST (Figure 6). These 19 isolates formed a branch, with 27A strains clustered near the isolates ASM2413794v1 and ASM130523v1. 27A, ASM2413794v1, and ASM130523v1 are all ST11 S. Enteritidis strains from Asia, originating from clinical, animal, and food, respectively. This suggested a consistent relationship between Salmonella causing human diseases and Salmonella isolated from farms or food. These strains from different sources contained common core genes, with small genomic differences, and could be transmitted between different hosts. For resistance genes, except for strain 27A, all contained aminoglycoside resistance gene aac(6’)-Iaa. And other aminoglycoside resistance genes (aph(3”)-Ib, aph(6)-Id), β-lactam resistance gene (blaTEM) were the most abundant resistance genes in all strains. For the plasmid replicon, 27A contained the plasmid replicon IncFIB(S) and IncFII(S), which appeared in pairs in the other 12 isolates. ASM130523v1 and ASM276095v1 contained only one of these two. ASM130523v1 contained IncFIB(S), while ASM276095v1 contained IncFII(S). Besides, the plasmid replicon IncX4 only existed in ASM331256v1.
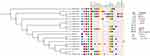 |
Figure 6 Phylogenetic Tree Based on CorePan Results and the Distribution of Resistance Genes, Plasmid Replicons. |
Quantitative Proteomics Analysis
Identification and Functional Annotation of Proteins
This study identified a total of 37,221 peptides and 3604 proteins, with an average of 3214 proteins identified by the reference strain and 3603 proteins identified by the 27A strain. Among the identified proteins, the number of unique peptides was mostly 1 (16.5%) or more than 11 (18.4%) (Figure S2A). 98.9% of the protein coverage was concentrated within 70%, among which the protein coverage of less than 10% was the most distributed, containing 1198 proteins, followed by the protein coverage of 10–20%, containing 676 proteins (Figure S2B). The molecular weight of the identified protein basically conformed to the normal distribution in the range of 0–100kDa, and the proteins in the range of 20–40kDa was the most, accounting for 43.2% (Figure S2C). Principal component analysis suggested that 27A strain and the reference strain had significant differences in the expression levels (Figure S2D).
All identified proteins were annotated with GO, KOG, and KEGG database, resulting in a total of 3267 proteins being annotated. Based on GO functional annotation, intracellular processes (54.8%) and metabolic processes (52.5%) was mainly annotated in biological process, catalysis (63.5%) and binding (50.2%) in molecular function. Cell composition showed that they were mainly localized within the cell (95.6%) and on the cell membrane (44.6%) (Figure 7A). The KOG functional annotation indicated that proteins were involved in transport metabolism, translation, ribosome structure and biogenesis, energy production and transformation, post-translation modification, protein turnover, signal transduction, RNA processing and modification. 6.6% proteins were involved in amino acid transport and metabolism, 4.8% in translation, ribosome structure and biogenesis, and 4.7% in energy production and transformation (Figure 7B). The KEGG pathway annotation showed that proteins were involved in six major types of pathways. Among them, metabolism accounted for 64.9%. The main metabolic pathways were carbohydrate metabolism, amino acid metabolism, cofactor and vitamin metabolism, and energy metabolism (Figure 7C).
 |
Figure 7 Functional Annotation of the Proteins Based on Database Searches. (A) GO functional annotation. (B) KOG functional annotation. (C) KEGG pathway annotation. |
Identification and Functional Annotation of Differential Proteins
S. Typhimurium ATCC14028 was used as a control. Two filtration criteria (Fold change > 2 and P value < 0.05) were used to get significant differential proteins. A total of 279 differentially expressed proteins were detected, with 119 proteins showing significant up-regulation and 160 proteins exhibiting significant down-regulation (Figure 8A). Of these differential proteins, 33.3% were related to drug resistance and virulence. A total of 5 drug-resistant proteins, 43 T3SS-related proteins and 45 virulence factor were screened, with 3, 19 and 14 proteins significantly up-regulated and 2, 24 and 21 proteins significantly down-regulated respectively (Figure 8B). These differential proteins were annotated in KOG, GO, and KEGG databases. Based on the KOG functional annotation, differential proteins mainly participated in metabolism and intracellular signal transduction, with only one protein, DusA, involved in information storage and processing. Proteins Yici and HutH were simultaneously involved in metabolism and intracellular signal transduction (Figure 8C). After functional annotation in the GO database, these differential proteins were enriched into three major categories and 18 subcategories. During molecular function, metal ion binding (GO: 0046872), heme binding (GO: 0020037), and oxidoreductase activity (GO: 0016491) were annotated into 4, 4 and 5 proteins, respectively, and these proteins were significantly downregulated. And, 4, 5 and 6 proteins were annotated with the phosphoenolpyruvate-dependent sugar phosphotransferase system (GO: 0009401) in biological processes, extracellular membrane (GO: 0009279) in cell components, and transferase activity (GO: 0016740) in molecular functions, respectively, all of which were significantly upregulated (Figure 8D). KEGG enrichment showed that differential proteins were mainly involved in human diseases, cellular processes, organismal systems, metabolic processes, signal transduction, and membrane transport. There were 13 proteins involved in signal transduction, of which 61.5% were significantly downregulated; 12 proteins were involved in carbohydrate metabolism, of which 63.6% were significantly upregulated. Among these proteins, 67.7% of them only participated in one biological process, while two proteins were functionally rich and could participate in four different biological processes. Protein FliC and G0L88_ 22505 were significantly downregulated and could participate in human diseases, cellular processes, organismal systems, and signal transduction (Figure 8E).
Time Series Analysis of Differential Proteins
According to the protein expression, 3604 identified proteins can be grouped into time-associated protein clusters, and proteins with the same expression pattern will be clustered into the same cluster. These proteins were grouped into nine clusters, and the protein expression level changed significantly with the change of time (Figure 9). In cluster 1, 5 and 9, protein expression descended gradually as time went; in clusters 2 and 8, protein expression showed an upward trend.
 |
Figure 9 Time Series Analysis of Differential Proteins. CG: The reference strain, EG: 27A. TheX-axis represents each time point, theY-axis represents the expression level after normalization. |
Interactions Among Differential Proteins
A protein–protein interaction (PPI) analysis was conducted on 93 screened resistant and virulent proteins, and 45 proteins formed an association network (Figure 10). The PPI network had 45 nodes and 135 edges, including 25 virulent proteins and 20 T3SS proteins. The outer membrane protein PagN was the most widely used protein and was related to T3SS proteins PipB2, SipB, SptP, SseL, YshA, and virulence proteins PagC, SpaO, SsaJ, SsaQ, SsrB, SteC, and YncJ; Next was the virulence membrane protein PagC, which interacted with T3SS proteins PipB2, SipB, SseL, and virulence proteins PagN, SpaO, SsaJ, SsaQ, SsrB, SteC, and YncJ. Combined with the GO functional annotation in Figure 8, it was shown that these proteins were mainly enriched in biological processes, including pathogenesis, T3SS protein secretion, etc.; in cellular components, including cell membrane, intracellular, cytoplasmic, and extracellular regions; also in molecular functions, including kinase activity, hydrolase activity, transferase activity, etc.
 |
Figure 10 Interaction Network of Differentially Expressed Proteins. |
Discussion
In order to investigate the molecular characteristics of S. Enteritidis in a food poisoning incident, we collected a strain of S. Enteritidis 27A from a tertiary A hospital in Shanxi, China. Combining the patient’s clinical information, we studied its drug resistance, genome and proteome information.
S. Enteritidis was the main serotype causing human infectious diarrhea in the United States and European countries, while in China, the main serotype in the southern and southwestern regions is S. Typhi and in the eastern, northern, and northwestern regions, it is S. Enteritidis.28 Additionally, the period from May to October is the peak season for outbreaks of S. Enteritidis.29 The S. Enteritidis isolated in this study originated from a food poisoning incident in northern China in May. According to the reports, patients infected with Salmonella had a history of consuming contaminated food (90.5%), abdominal pain (58.05%), diarrhea (≥5 times) (50.44%), moderate fever (24.96%), and increased fecal leukocytes (41.42%). Most patients showed clinical symptoms within 1 to 72 hours after consuming contaminated food.29 This was consistent with the clinical manifestations of the patients in this study, who developed significant clinical symptoms, such as diarrhea and fever within 24 hours of eating dirty sandwiches.
The strain 27A exhibited multidrug resistance, being resistant to β-lactam drugs (ampicillin, piperacillin, ampicillin/sulbactam) and quinolones (levofloxacin, ciprofloxacin), but sensitive to cephalosporins (ceftriaxone, ceftazidime, cefepime), which was consistent with domestic reports on multidrug resistant Salmonella.30 β-lactam and quinolone antibiotics were commonly used drugs for clinical treatment of Salmonella infections. Previous studies have shown that the resistance rates of Salmonella isolated from clinical diarrhea patients to ampicillin could reach 63.93%, the resistance rate of ampicillin/sulbactam was 55.74%, and the resistance rates of ciprofloxacin and levofloxacin were low at 4.92% and 1.64%, respectively.31 But in some developing countries, the resistance rate to ciprofloxacin could reach 90.9%.32 As a zoonotic pathogen, Salmonella can spread among humans, animals and the environment. There were similarities in antibiotic resistance phenotypes of Salmonella from different sources. Analysis of Salmonella from pig farms showed that S. Enteritidis was the most common serotype, with high resistance rates to nalidixic acid (100.0%), streptomycin (100.0%), ampicillin (98.4%) and erythromycin (93.7%).33 A study on the resistance of S. Enteritidis in ducks, chickens, pig farms and retail markets in eastern China showed that 75.26% isolates were multidrug-resistant, and the majority were resistant to tetracycline (76.6%) and ampicillin (67.2%).34 Jeamsripong et al analyzed aquatic animals and estuarine environments, and found that Salmonella had the highest resistance to sulfamethoxazole at 95.2%, followed by trimethoprim (37.3%) and ampicillin (36.5%).35
The 90% resistance of Salmonella to ampicillin was caused by the blaTEM gene, which encoded β-lactamase, an enzyme that breaks down β-lactam molecules and mediates resistance to β-lactam antibiotics. This study annotated the blaTEM-194 gene from the 27A genome, which is located on plasmid and was derived from mutations of classic blaTEM (blaTEM-1 and blaTEM-2) genes. The blaTEM-194 was initially discovered in Acinetobacter baumannii.36 A total of 18 efflux pump genes belonging to three major categories were detected in strain 27A, including RND, MFS and SMR. Among them, RND was the most important efflux pump in pathogens, which was a secondary transporter that could efflux various structurally different antibiotics.37 Moreover, the ermA and ermR genes promoted increased resistance to quinolones, such as ciprofloxacin, while the mdtK gene conferred resistance to tetracycline, chloramphenicol, norfloxacin and doxorubicin in Salmonella.38 These resistance genes were mostly involved in multiple resistance pathways and could serve as potential drug targets for therapeutic strategies.
The virulence factors of Salmonella are mainly encoded on SPIs, which can help Salmonella escape from the attack of host immune system, thereby making Salmonella infect, reproduce and spread in complex host environments.31 SPI-1 contributed to Salmonella invade into host cells and regulate host immune responses. The invA gene was an important structural component of SPI-1 and was associated with invasion of human and animal intestinal epithelial tissues. It was highly conserved in Salmonella with high positive rate and could be used as a specific biomarker for identification of Salmonella. The absence of the invA gene in Salmonella isolates may indicate non-invasiveness or the presence of alternative invasion mechanisms.35 SPI-2 was associated with systemic infection and intracellular accumulation of Salmonella. It had 7 core effectors SseF, SseG, PipB, SteA, SifA, SteD and PipB2, which existed in all serotypes and could exert virulence effects on all hosts. For S. Enteritidis, it had a specific set of effectors SseL, SifB, SopD2, SseJ, SteB, SteC, SlrP and SseK2, which only exerted virulence on hosts within the intestine and had no effect on hosts outside the intestine.39 According to the research, the presence of both SPI-1 and SPI-2 in Salmonella was positively related to its pathogenicity,31 which indicated that strain 27A had strong virulence. Unlike the T3SS invasion mechanism mediated by SPI-1 and SPI-2, there was also an invasion mechanism mediated by the outer membrane protein PagN in strain 27A. PagN induced bacterial invasion through the zipper mechanism, that was, PagN interacted with heparan sulfate proteoglycans (HSPG) to activate phosphatidylinositol 3-kinase and phosphorylate tyrosine protein, leading to actin polymerization and membrane rearrangement, thereby leading to bacterial internalization.40 In this study, PagN was significantly upregulated and was found to interact with SPI-1 T3SS protein SpaO, SPI-2 T3SS proteins PipB2, SsaQ and SseL to jointly mediate the invasion of S. Enteritidis. Furthermore, the strain 27A also predicted the fimbriae virulence gene sef14, which only existed in a small number of S. Enteritidis and its related serotypes, and could affect virulence traits related to serotypes. SEF14 fimbriae were encoded by a sef operon, which consisted of sefABCD genes that encoded different subunits, and their transcription was activated by an AraC-like regulatory protein encoded by sefR. SEF14 fimbriae did not participate in the primary attachment of bacteria to host intestinal epithelial cells. They mainly survived within macrophages by binding to surface receptors, thereby enhancing the adsorption of S. Enteritidis.41
Two plasmid replicons, IncFIB(S) and IncFII(S), were detected in strain 27A. They were the main plasmid replicons in S. Enteritidis, belonging to the IncF family, and widely distributed in Enterobacteriaceae, especially S. Enteritidis and E. coli. These IncF plasmid replicons promoted bacterial infection and drug resistance by carrying virulence and resistance determinants (including drug resistance genes, bacteriocins, iron carriers, cytotoxics and adhesion factors).42 Plasmids could encode virulence and drug resistance genes, and promoted bacterial diversity and adaptation through horizontal gene transfer. Served as epidemiological markers of bacterial strains, they contributed to monitor and investigate outbreaks of bacterial infections. Among them, the presence of virulence genes on plasmids enhances bacterial adhesion and colonization in host cells, potentially contributing to the increased pathogenicity of Salmonella.43 In this study, spv genes (spvA, spvB, spvC, spvD and spvR) related to plasmid transmission were predicted. spv was a highly conserved sequence located on the plasmid. The spvABCD genes were neatly arranged in the operon and were positively regulated by upstream spvR gene. The spv gene inhibited the type I interferon response and neutrophil chemotaxis by inhibiting autophagy, and it could also disrupt the integrity of intestinal epithelial cells to increase intestinal permeability, thereby achieving Salmonella translocation.44 SpvC deactivated mitogen activated protein kinase (MAPK) to inhibit intestinal inflammation through β elimination, and also inhibited host cell pyrosis to promote bacterial transmission in the body.45
Multi-drug resistant Salmonella can spread among humans, animals, and the environment, and its resistance can be transmitted to humans through poultry production chains or other pathways. Whole-genome sequencing and multilocus sequence typing (MLST) can be used to characterize these pathogens and determine their cloning and distribution in various environments and hosts.46 ST11 was the main epidemic type of S. Enteritidis in China and was distributed in all populations.47,48 The 27A strain in this study also belonged to ST11 S. Enteritidis. The phylogenetic tree showed that human-derived 27A strain was closely related to animal-derived ASM2413794v1 and food-derived ASM130523v1, which showed similar resistance genes and plasmid replicons, indicating that S. Enteritidis from different sources had similar genetic relationships. Studies have shown that ST11 S. Enteritidis has been detected in various hosts, including humans, poultry and food, with a wide geographical distribution spanning Asia, Africa, the Americas and Europe.49 The phylogenetic analysis in this study also showed that these ST11 S. Enteritidis strains isolated from different geographical distributions and hosts have similar genetic relationships. These strains clustered together and shared similar resistance genes, which strongly supported the potential transmission of Salmonella among humans, animals, food and the environment. Animals infected with Salmonella can transmit the infection to humans through the food chains, production chains and environment to cause human Salmonella infection.
Conclusion
This study screened and isolated S. Enteritidis from the feces of food poisoning patients and conducted resistance testing, whole-genome and proteomic analysis on strain 27A, revealing its resistance, virulence and molecular evolution characteristics. The strain 27A was a multidrug-resistant ST11 S. Enteritidis, carrying multiple virulence and resistance genes, which highlighted the pathogenic potential of 27A. These virulence proteins changed significantly, and PPI analysis found that PagN and T3SS jointly mediated the invasion of 27A to human body. This information will be useful for studying the drug resistance mechanisms and pathogenic mechanisms of S. Enteritidis. In addition, we explored its genetic evolution characteristics through phylogenetic trees. It was found that there was potential transmission among S. Enteritidis strains isolated from humans, food and animals. The epidemiology of Salmonella is characterized by both diversity and close relationships with different hosts, which can enhance our understanding of the phylogeny of Salmonella and help develop new strategies for the prevention and treatment of Salmonella infections. Combining the patient’s epidemiological investigation, the patient consumed contaminated sandwiches the day before the onset of the disease. To determine the source of this food poisoning incident, further sampling and analysis of the sandwiches are needed to explore the epidemiological situation and track and prevent Salmonella diseases.
Nucleotide Sequence Accession Number
The nucleotide sequence of the chromosome and plasmids of S. Enteritidis isolate 27A have been deposited in GenBank under accession number CP122301-CP122303.
Data Sharing Statement
All raw data and supporting materials related to this paper can be obtained from the corresponding author.
Ethical Approval
The study was approved by the Ethics Committee at Fenyang College of Shanxi Medical University. Written informed consent has been provided by the patient to have the case details published. All methods were performed in accordance with relevant guidelines and regulations. All investigators adhered to the principles expressed in the Declaration of Helsinki.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This study was supported by Fundamental Research Program of Shanxi Province (Grant no. 20210302123397; 202203021212351), Key R&D Projects of Introducing High-Level Scientific and Technological Talents in Lvliang City (Grant no. 2021RC-1-4), the Project of Lvliang City Science and Technology Program (Grant no. 2020SHFZ29), Science and Technology Innovation Project of Colleges and Universities in Shanxi Province (Grant no. 2020L0749), the National College Students’ Innovation and Entrepreneurship Training Program (Grant no. 20221569), Projects of Innovation and Entrepreneurship Training Program for College Students of Fenyang College of Shanxi Medical University (Grant no. FDC202209; FDC202214; FDC202215), and Special Fund for Key Disciplines of Fenyang College of Shanxi Medical University (Grant no. 2022B14).
Disclosure
The authors have no conflict of interest to declare.
References
1. Wojcicki M, Chmielarczyk A, Swider O, et al. Bacterial pathogens in the food industry: antibiotic resistance and virulence factors of Salmonella enterica strains isolated from food chain links. Pathogens. 2022;11(11):1323. doi:10.3390/pathogens11111323
2. Ikejiri K, Suzuki K, Ito A, et al. Invasive Salmonella enteritidis infection complicated by bacterial meningitis and vertebral osteomyelitis shortly after influenza A infection in an immunocompetent young adult. J Infect Chemother. 2020;26(2):269–273. doi:10.1016/j.jiac.2019.08.001
3. Lapierre L, Cornejo J, Zavala S, et al. Phenotypic and genotypic characterization of virulence factors and susceptibility to antibiotics in Salmonella infantis strains isolated from chicken meat: first findings in Chile. Animals. 2020;10(6):1049. doi:10.3390/ani10061049
4. Founou LL, Founou RC, Essack SY. Antibiotic resistance in the food chain: a developing country-perspective. Front Microbiol. 2016;7:1881. doi:10.3389/fmicb.2016.01881
5. Dai W, Zhang Y, Zhang J, et al. Analysis of antibiotic-induced drug resistance of Salmonella enteritidis and its biofilm formation mechanism. Bioengineered. 2021;12(2):10254–10263. doi:10.1080/21655979.2021.1988251
6. Dong N, Li Y, Zhao J, et al. The phenotypic and molecular characteristics of antimicrobial resistance of Salmonella enterica subsp. enterica serovar Typhimurium in Henan Province, China. BMC Infect Dis. 2020;20(1):511. doi:10.1186/s12879-020-05203-3
7. Melo RT, Galvao NN, Guidotti-Takeuchi M, et al. Molecular characterization and survive abilities of Salmonella Heidelberg strains of poultry origin in Brazil. Front Microbiol. 2021;12:674147. doi:10.3389/fmicb.2021.674147
8. Lou L, Zhang P, Piao R, Wang Y. Salmonella Pathogenicity Island 1 (SPI-1) and its complex regulatory network. Front Cell Infect Microbiol. 2019;9:270. doi:10.3389/fcimb.2019.00270
9. Lyu N, Feng Y, Pan Y, et al. Genomic characterization of Salmonella enterica isolates from retail meat in Beijing, China. Front Microbiol. 2021;12:636332. doi:10.3389/fmicb.2021.636332
10. Aljahdali NH, Sanad YM, Han J, Foley SL. Current knowledge and perspectives of potential impacts of Salmonella enterica on the profile of the gut microbiota. BMC Microbiol. 2020;20(1):353. doi:10.1186/s12866-020-02008-x
11. Pornsukarom S, van Vliet AHM, Thakur S. Whole genome sequencing analysis of multiple Salmonella serovars provides insights into phylogenetic relatedness, antimicrobial resistance, and virulence markers across humans, food animals and agriculture environmental sources. BMC Genomics. 2018;19(1):801. doi:10.1186/s12864-018-5137-4
12. McDermott PF, Tyson GH, Kabera C, et al. Whole-genome sequencing for detecting antimicrobial resistance in nontyphoidal Salmonella. Antimicrob Agents Chemother. 2016;60(9):5515–5520. doi:10.1128/AAC.01030-16
13. Zhang D, Zhu L, Wang F, Li P, Wang Y, Gao Y. Molecular mechanisms of eukaryotic translation fidelity and their associations with diseases. Int J Biol Macromol. 2023;242(Pt 1):124680. doi:10.1016/j.ijbiomac.2023.124680
14. Li J, Smith LS, Zhu HJ. Data-independent acquisition (DIA): an emerging proteomics technology for analysis of drug-metabolizing enzymes and transporters. Drug Discov Today Technol. 2021;39:49–56. doi:10.1016/j.ddtec.2021.06.006
15. Tarbeeva S, Kozlova A, Sarygina E, Kiseleva O, Ponomarenko E, Ilgisonis E. Food for thought: proteomics for meat safety. Life. 2023;13(2):255. doi:10.3390/life13020255
16. Cockerill FR. Performance Standards For Antimicrobial Susceptibility Testing: Twenty-Third Informational Supplement. Clinical and Laboratory Standards Institute; 2013.
17. Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25(5):955–964. doi:10.1093/nar/25.5.955
18. Lagesen K, Hallin P, Rodland EA, Staerfeldt HH, Rognes T, Ussery DW. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007;35(9):3100–3108. doi:10.1093/nar/gkm160
19. Gardner PP, Daub J, Tate JG, et al. Rfam: updates to the RNA families database. Nucleic Acids Res. 2009;37(Database issue):D136–140. doi:10.1093/nar/gkn766
20. Chen L, Zheng D, Liu B, Yang J, Jin Q. VFDB 2016: hierarchical and refined dataset for big data analysis--10 years on. Nucleic Acids Res. 2016;44(D1):D694–697. doi:10.1093/nar/gkv1239
21. Alcock BP, Raphenya AR, Lau TTY, et al. CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020;48(D1):D517–D525. doi:10.1093/nar/gkz935
22. Galperin MY, Makarova KS, Wolf YI, Koonin EV. Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Res. 2015;43(Database issue):D261–269. doi:10.1093/nar/gku1223
23. Ashburner M, Ball CA, Blake JA, et al. Gene Ontology: tool for the unification of biology. Nat Genet. 2000;25(1):25. doi:10.1038/75556
24. Jones P, Binns D, Chang HY, et al. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014;30(9):1236–1240. doi:10.1093/bioinformatics/btu031
25. Nandi T, Ong C, Singh AP, et al. A genomic survey of positive selection in Burkholderia pseudomallei provides insights into the evolution of accidental virulence. PLoS Pathog. 2010;6(4):e1000845. doi:10.1371/journal.ppat.1000845
26. Choi M, Chang CY, Clough T, et al. MSstats: an R package for statistical analysis of quantitative mass spectrometry-based proteomic experiments. Bioinformatics. 2014;30(17):2524–2526. doi:10.1093/bioinformatics/btu305
27. Franceschini A, Szklarczyk D, Frankild S, et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41(Database issue):D808–815. doi:10.1093/nar/gks1094
28. Wang Y, Liu Y, Lyu N, et al. The temporal dynamics of antimicrobial-resistant Salmonella enterica and predominant serovars in China. Natl Sci Rev. 2023;10(3):nwac269. doi:10.1093/nsr/nwac269
29. Qi X, Li P, Xu X, Yuan Y, Bu S, Lin D. Epidemiological and molecular investigations on Salmonella responsible for gastrointestinal infections in the Southwest of Shanghai From 1998 to 2017. Front Microbiol. 2019;10:2025. doi:10.3389/fmicb.2019.02025
30. Xu D, Ji L, Yan W, Chen L, Shabbir MAB. Characterization of Clinical Salmonella entericas Trains in Huzhou, China. Can J Infect Dis Med Microbiol. 2022;2022:7280376. doi:10.1155/2022/7280376
31. Yue M, Li X, Liu D, Hu X. Serotypes, antibiotic resistance, and virulence genes of Salmonella in children with diarrhea. J Clin Lab Anal. 2020;34(12):e23525. doi:10.1002/jcla.23525
32. Fardsanei F, Soltan Dallal MM, Zahraei Salehi T, Douraghi M, Memariani M, Memariani H. Antimicrobial resistance patterns, virulence gene profiles, and genetic diversity of Salmonella enterica serotype enteritidis isolated from patients with gastroenteritis in various Iranian cities. Iran J Basic Med Sci. 2021;24(7):914–921. doi:10.22038/ijbms.2021.54019.12142
33. Yang J, Gao S, Chang Y, Su M, Xie Y, Sun S. Occurrence and characterization of Salmonella isolated from large-scale breeder farms in Shandong Province, China. Biomed Res Int. 2019;2019:8159567. doi:10.1155/2019/8159567
34. Tang B, Elbediwi M, Nambiar RB, Yang H, Lin J, Yue M. Genomic characterization of antimicrobial-resistant salmonella enterica in duck, chicken, and pig farms and retail markets in Eastern China. Microbiol Spectr. 2022;10(5):e0125722. doi:10.1128/spectrum.01257-22
35. Jeamsripong S, Kuldee M, Thaotumpitak V, Chuanchuen R, Karunasagar I. Antimicrobial resistance, extended-spectrum β-lactamase production and virulence genes in Salmonella enterica and Escherichia coli isolates from estuarine environment. PLoS One. 2023;18(4):e0283359. doi:10.1371/journal.pone.0283359
36. Ur Rahman S, Ali T, Ali I, Khan NA, Han B, Gao J. The growing genetic and functional diversity of extended spectrum beta-lactamases. Biomed Res Int. 2018;2018:9519718. doi:10.1155/2018/9519718
37. Zwama M, Nishino K. Ever-adapting RND efflux pumps in gram-negative multidrug-resistant pathogens: a race against time. Antibiotics. 2021;10(7):774. doi:10.3390/antibiotics10070774
38. Vilela FP, Rodrigues DDP, Allard MW, Falcao JP, Lianou A. Prevalence of efflux pump and heavy metal tolerance encoding genes among Salmonella enterica serovar Infantis strains from diverse sources in Brazil. PLoS One. 2022;17(11):e0277979. doi:10.1371/journal.pone.0277979
39. Jennings E, Thurston TLM, Holden DW. Salmonella SPI-2 type III secretion system effectors: molecular mechanisms and physiological consequences. Cell Host Microbe. 2017;22(2):217–231. doi:10.1016/j.chom.2017.07.009
40. Barilleau E, Vedrine M, Koczerka M, et al. Investigation of the invasion mechanism mediated by the outer membrane protein PagN of Salmonella Typhimurium. BMC Microbiol. 2021;21(1):153. doi:10.1186/s12866-021-02187-1
41. Quan G, Xia P, Zhao J, et al. Fimbriae and related receptors for Salmonella Enteritidis. Microb Pathog. 2019;126:357–362. doi:10.1016/j.micpath.2018.10.025
42. Mansour MN, Yaghi J, El Khoury A, et al. Prediction of Salmonella serovars isolated from clinical and food matrices in Lebanon and genomic-based investigation focusing on enteritidis serovar. Int J Food Microbiol. 2020;333:108831. doi:10.1016/j.ijfoodmicro.2020.108831
43. Wang W, Chen J, Shao X, Huang P, Zha J, Ye Y. Occurrence and antimicrobial resistance of Salmonella isolated from retail meats in Anhui, China. Food Sci Nutr. 2021;9(9):4701–4710. doi:10.1002/fsn3.2266
44. Sun L, Yang S, Deng Q, et al. Salmonella effector SpvB disrupts intestinal epithelial barrier integrity for bacterial translocation. Front Cell Infect Microbiol. 2020;10:606541. doi:10.3389/fcimb.2020.606541
45. Zhou L, Li Y, Gao S, et al. Salmonella spvC gene inhibits autophagy of host cells and suppresses NLRP3 as well as NLRC4. Front Immunol. 2021;12:639019. doi:10.3389/fimmu.2021.639019
46. Zakaria Z, Hassan L, Sharif Z, et al. Analysis of Salmonella enterica serovar enteritidis isolates from chickens and chicken meat products in Malaysia using PFGE, and MLST. BMC Vet Res. 2020;16(1):393. doi:10.1186/s12917-020-02605-y
47. Chen J, Ed-Dra A, Zhou H, Wu B, Zhang Y, Yue M. Antimicrobial resistance and genomic investigation of non-typhoidal Salmonella isolated from outpatients in Shaoxing city, China. Front Public Health. 2022;10:988317. doi:10.3389/fpubh.2022.988317
48. Yan S, Zhang W, Li C, et al. Serotyping, MLST, and core genome MLST analysis of Salmonella enterica from different sources in China during 2004–2019. Front Microbiol. 2021;12:688614. doi:10.3389/fmicb.2021.688614
49. Shen X, Yin L, Zhang A, et al. Prevalence and characterization of salmonella isolated from chickens in Anhui, China. Pathogens. 2023;12(3):465. doi:10.3390/pathogens12030465
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.




