Back to Journals » International Journal of General Medicine » Volume 16
The Prognostic Significance of the TEAD4 in Hepatocellular Carcinoma
Authors Lei L, Yang J, Peng H, Huang R, Liang L, Liu R, Li J
Received 19 September 2023
Accepted for publication 4 December 2023
Published 20 December 2023 Volume 2023:16 Pages 6005—6013
DOI https://doi.org/10.2147/IJGM.S440973
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Scott Fraser
Liping Lei,1,2,* Jingjing Yang,1,* Hao Peng,1 Ruiyan Huang,1 Lichun Liang,1 Ruifang Liu,1 Jiangfa Li1
1Department of Hepatobiliary and Pancreatic Surgery, The Affiliated Hospital of Guilin Medical University, Guilin, 541001, People’s Republic of China; 2Department of Geriatric Medicine, The Affiliated Hospital of Guilin Medical University, Guilin, 541001, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Jiangfa Li, Department of Hepatobiliary and Pancreatic Surgery, The Affiliated Hospital of Guilin Medical University, Guilin, 541001, People’s Republic of China, Tel +86-7732824373, Email [email protected]
Background: Abnormal expression of genes causes tumorigenesis, tumor progression, and poor prognosis in hepatocellular carcinoma (HCC). Therefore, the aims of this study were to explore the transcription enhancer domain factor 4 (TEAD4) in patients with liver cancer and its relationship with prognosis.
Methods: HTSeq-FPKM data and corresponding clinical data of HCC patients were obtained from The Cancer Genome Atlas (TCGA). Difference in TEAD4 expression between normal and tumor and the correlation with clinical characteristics were analyzed by the chi-squared test based on UALCAN. HepG2 cell lines were used to study the effect of TEAD4 on HCC cell lines. The expression and clinical significance of TEAD4 in HCC were detected in clinical cases.
Results: The transcription and post-transcription levels of TEAD4 were higher in HCC tumors than normal illustrated different expressed transcription of TEAD4 in gender, nodal metastasis status, tumor grades, and individual cancer stages. The high TEAD4 expression was significantly associated with tumor grades. The high expression of TEAD4 was significantly correlated to shorter 2– 5 years overall survival. Inhibition of TEAD4 expression in HepG2 cells resulted in significantly decreased cell proliferation and invasion.
Conclusion: TEAD4 was identified as an independent prognostic factor, and inhibition of TEAD4 expression in HepG2 cells resulted in significantly decreased cell proliferation and invasion.
Keywords: TCGA, TEAD4, hepatocellular carcinoma
Introduction
Liver cancer is the sixth most frequently diagnosed and fourth resulting tumor-related death in the world, and it has been reported in approximately 841,000 new cases and 782,000 deaths every year.1 Hepatocellular carcinoma (HCC) is the most common subtype of liver cancer worldwide, and more than 75% of liver cancer patients were diagnosed with HCC. Hepatitis B virus (HBV) and hepatitis C virus (HCV) infection, aflatoxin contamination, excessive drinking, obesity, smoking, and type 2 diabetes.2,3 Although diagnosis and treatment of HCC have improved, the prognosis of HCC patients remains poor.4,5 Abnormal expression genes are significantly associated with tumorigenesis, development, and prognosis.6–9 Therefore, the biomarkers and underlying mechanism for tumorigenesis, development and prognosis should be investigated.
The transcription enhancer domain (TEAD) is a downstream factor of the Hippo pathway and a key factor in this pathway.10,11 TEAD family of transcription factors that regulate YAP/TAZ includes four members of the mammalian family (TEAD1-TEAD4).12 TEAD4 is an important gene associated with blastocyst development and trophoblast differentiation.13 It participates in Hippo signaling pathway for modulating organ size and tissue homeostasis and dysregulation of Hippo signaling pathway associated with tumorigenesis.14 TEAD expression was higher in cancer than in normal tissues, and higher TEAD4 levels tended to have worse overall survival (OS).15 High level of TEAD4 may also lead to excessive transcription and phosphorylation of ERK proteins as well as Yes-associated proteins; therefore, these can accelerate the process of tumor development.16,17 Increasing evidences have demonstrated that TEAD4 plays a critical role in HCC by promoting tumorigenesis by mediating Yes-associated protein oncogenic activity,18 or modulating chromosomal instability in liver cancer patients.19 Overexpression of TEAD4 activates YAP and is associated with shorter survival in HCC patients. However, the role and molecular mechanism of TEAD4 in HCC remain unclear. In addition, TEAD4 and YAP interact with IRF3 to regulate inflammatory in gastric cancer.20 However, whether TEAD4 modulates its target genes to affect HCC tumorigenesis and development is unknown.
Therefore, in this study, we investigated the differential expression of TEAD4 in HCC and explored the correlation with clinical characteristics. HCC patients were distributed into high-expression and low-expression TEAD4 groups. The differentially expressed genes of TEAD4 associated with immune cells were identified, and the biological function of these genes was investigated. Our results identified independent prognostic factors associated with immune cells in HCC.
Methods
Data Acquiring, Processing, and TEAD4 Expression Analysis
HTSeq-FPKM data and corresponding clinical data of liver hepatocellular carcinoma (LIHC), including 50 normal and 374 tumors were obtained from The Cancer Genome Atlas (TCGA). The different expression of TEAD4 between normal and tumors, gender, nodal metastasis status, cancer stages, tumor grades were detected by Student’s t-test or one-way ANOVA test. The correlation between TEAD4 expression and the clinicopathological parameters was analyzed using chi-squared test. Moreover, TEAD4 protein level between normal and tumors was calculated by immunohistochemistry in Human Protein Atlas (HPA).
Analysis of the Correlation Between TEAD4 and Overall Survival
According to the median expression levels of TEAD4, the HCC patients were distributed into high and low TEAD4 expression groups. The Kaplan–Meier plotter was used to analyze the effect of TEAD4 on survival in HCC patients. The log-rank P value and hazard ratio (HR) with 95% confidence intervals were evaluated.
Univariate and Multivariate Cox Analysis
The expression data of TEAD4-target genes and corresponding clinical data were combined, and the prognostic variates were identified by a univariate proportional hazards model using survival R package. Next, the prognostic variates with P value < 0.05 were enrolled in the multivariate proportional hazards model to identify the independent variates for the prognosis of HCC patients. Furthermore, combination-independent variates and survival times were used to analyze the overall survival rate of HCC patients.
Clinical Case Analysis
Thirty patients with hepatocellular carcinoma treated by hepatobiliary surgery at Affiliated Hospital of Guilin Medical University were enrolled, and five patients who underwent partial hepatectomy due to hemangioma were selected. Clinical data were collected, and TEAD4 expression was detected in postoperative pathological tissues. The Ethics Committee of the Affiliated Hospital of Guilin Medical University approved the study. Written informed consent was obtained from each participant. Paraffin sections of liver tissue and liver tumor tissue were taken and dewaxed for TEAD4 staining. Tissues were removed and put into 4% paraformaldehyde fixing solution, allowed to set for 1 day, and then immersed in 70% ethanol for approximately 10 hr. The tissue was embedded and cut into 5-mm slices using a rotary microtome (Leica, Mannheim, Germany). The slide to be dyed was placed into a dyeing rack and then placed it in an oven at 60°C for 1 to 2 hr. Turpentine I dewaxing for 15 min was followed by turpentine II dewaxing for 15 min, anhydrous ethanol I for 10 min, anhydrous ethanol II for 10 min, 95% ethanol I for 10 min, 95% ethanol II for 10 min, 80% ethanol for 10 min, and washing in tap water for 1 min. The dehydrated paraffin sections were then tested for TEAD4 according to the instructions.
Cell Experiment
Cell Culture
HepG2 cell lines were used to study the effect of TEAD4 on HCC cell lines. HepG2 was purchased from Cellcook (Jinglai Biological Co. LTD, China). HepG2 cells were cultured in a DMEM medium containing 10% fetal bovine serum, 100U/mL penicillin, 100U/mL streptomycin, under the condition of 37°C, 5%CO2, and saturated humidity in a constant temperature incubator. The liquid was changed every 2 to 3d, and when the cell was 90% full, it was then purchased for digestion. SiRNA plasmids targeting TEAD4 were constructed and transfected into HepG2 cells using lentivirus. SiRNA3 was the most effective knockout, which was used in cell proliferation and invasion experiments. Cell proliferation was detected by MTT assay, and cell invasion assays were performed with Transwell chambers.
Transfection
SiRNAs were transfected by Lipofectamine 3000 kit (Invitrogen, Carlsbad, USA), and transfection according to instructions. TEAD4: siRNA-1, sense: 5ʹ- CCGCCAAAUCUAUGACAAATT-3ʹ, anti sense: 5ʹ- UUUGUCAUAGAUUUGGCGGTT-3ʹ. TEAD4 siRNA-2, sense: 5ʹ- CCACGAAGGUCUGCUCUUUTT-3ʹ, anti sense: 5ʹ- AAAGAGCAGACCUUCGUGGTT-3ʹ, TEAD4 siRNA-3sense: 5ʹ- GAGACAGAGUAUGCUCGCUAUTT-3ʹ, anti sense: 5ʹ- AUAGCGAGCAUACUCUGUCUCTT-3ʹ, NC siRNA:sense: 5ʹ- GUGAGCGUCUAUAUACCAUdTdT-3ʹ, anti sense: 5ʹ- AUGGUAUAUAGACGCUCACdTdT-3.
Real-Time Polymerase Chain Reaction (PCR)
RNA expression of TEAD4 was measured using real-time PCR. The total RNA was isolated with TRIzol reagent (R1200, solarbio, CN) according to the manual. Reverse transcription was performed with Universal reverse transcription kit (11141ES60, Yeasen Biotech, CN). Real-time PCR was performed with Realtime PCR fluorescence Quantitative kit (11201ES08, Yeasen Biotech, CN) on an MA-6000 Touch Real-Time system (Molarray) using primers specific for TEAD4 (forward, 5ʹ- AAGCAGGTCTCCAGCCACATCC-3ʹ; reverse, 5ʹ- TTGTCCTTAGCTGCCTGGTCCTT-3ʹ). Melting curve analysis was performed to ensure the specificity of primers. The expression of the target gene was normalized against GAPDH (forward, 5′-GGAGTCCACTGGCGTCTTCAC-3′; reverse, 5′-GGCCATCCACAGTCTTCTGGG-3′) and calculated with the ΔΔCT method.
Results
High Expression of TEAD4 in HCC
The transcription levels of TEAD4 were analyzed using UALCAN. The TEAD4 transcription level significantly increased in tumors compared to normal (Figure 1A). There showed obvious TEAD4 transcription level between male and female (Figure 1B). The TEAD4 transcription level is elevated in N1 than N0 based on nodal metastasis status (Figure 1C). TEAD4 transcription level also gradually increased in progressive tumor grades (Figure 1D). In addition, TEAD4 transcription level is also gradually elevated in gradual individual cancer stages (Figure 1E). We further revealed the TEAD4 mRNA expression level was strongly upregulated in HCC tumors compared to normal (Figure 1F). In addition, the protein level of TEAD4 was increased in HCC tumors compared to normal (Figure 1G). Our findings suggest that the transcription and post-transcription levels of TEAD4 were higher in HCC tumors than normal and here illustrated different expressed transcription of TEAD4 in gender, nodal metastasis status, tumor grades, and individual cancer stages.
Correlation Between TEAD4 and Clinical Characteristics in Poor HCC Patients
HCC patients were divided into high TEAD4 expression group and low TEAD4 expression group to further study the relationship between TEAD4 expression and clinical features. The high TEAD4 expression was significantly associated with tumor grades. However, it was not obviously associated with other clinical parameters. The prognostic value of TEAD4 expression in HCC is analyzed based on Kaplan–Meier curves and log-rank methods. As shown in Figure 2A–E, high expression of TEAD4 was significantly correlated to shorter 2–5 years overall survival. It has also been demonstrated that high expression of TEAD4 significantly associated with shorter overall survival in male, but not in female (Figure 2F and G). In addition, the high expression of TEAD4 strongly associated with shorter overall survival in alcohol-free HCC patients and with or without hepatitis virus infection but not in alcohol HCC patients (Figure 2H–K). These data indicated that TEAD4 acted as a worse prognostic independent factor for HCC patients.
Clinical Case Outcome
The expression of TEAD4 in para-cancerous tissues of liver cancer patients was significantly higher than that in normal liver tissues of liver hemangioma patients, as shown in Figure 3. The expression of TEAD4 in patients with recurrence 2 years after surgery was significantly higher than that in patients without recurrence 2 years after surgery (p<0.05).
Cell Experiment
The results suggested that TEAD4 was inhibited in HepG2 after transfection, as shown in Figure 4. HepG2 cell proliferation, migration, and invasion ability were significantly reduced after TEAD4 inhibition, as shown in Figures 5 and 6.
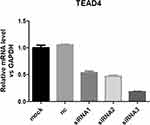 |
Figure 4 The knock down of TEAD4 in HepG2. Notes: The results suggested that TEAD4 was inhibited in HepG2 after transfer and siRNA3 was the most significant. |
Discussion
HCC is a common malignancy, which is prevalent worldwide, and HBV/HCV infection, liver cirrhosis, and aflatoxin contaminated food are the main risk factors that cause HCC.21 Previous study has been demonstrated TEAD4 plays an important role in HCC, it has indicated YAP binds TEAD4 induces HCC cell proliferation and stem cells expansion.22 Inhibition of the YAP-TEAD4 significantly suppressed tumor formation in HCC.23 In this study, we demonstrated TEAD4 mRNA expression and protein levels increased in HCC tumors than normal, and high expression of TEAD4 significantly associated with gender, nodal metastasis status, tumor grades, and cancer stages. Moreover, high expression of TEAD4 correlated with shorter survival time in HCC. Increasing evidences have demonstrated high TEAD4 expression promotes tumor initiation, progression and metastasis in laryngeal cancer,14 our findings confirm the role of TEAD4 in HCC.
In addition, TEAD4 is a key downstream transcription factor of Hippo signaling pathway,24 and Hippo signaling pathway has been demonstrated to be associated with the tumor immune microenvironment and affect tumor secretome and immune infiltrates.25,26 In HCC, activation of the Hippo signaling pathway promotes macrophage recruitment by tumor-initiating cells.27
Conclusion
In summary, we demonstrated that high TEAD4 expression was associated with clinical characteristics and poor prognosis in HCC.
Data Sharing Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethical Approval
This study was approved by the Ethics Committee of the Affiliated Hospital of Guilin Medical University. This study complied with the Declaration of Helsinki. HepG2 was purchased from Cellcook (Jinglai Biological Co. LTD, China).
Consent
Written informed consent was obtained from the patient for the publication of this study.
Funding
This study was supported in part by the Project to Improve the Scientific Research Ability of Middle-aged and Young Teachers (2018glmcy044), and Self-funded Project of Guangxi Zhuang Autonomous Region Health Commission (Z20200013, Z20200261), and Innovation and entrepreneurship project for college students (S202210601063).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Ca a Cancer J Clinicians. 2018;68(6):394–424. doi:10.3322/caac.21492
2. Tang A, Hallouch O, Chernyak V, et al. Epidemiology of hepatocellular carcinoma: target population for surveillance and diagnosis. Abdom Radiol. 2018;43(1):13–25. doi:10.1007/s00261-017-1209-1
3. Ganne-Carrié N, Nahon P. Hepatocellular carcinoma in the setting of alcohol-related liver disease. J Hepatol. 2019;70(2):284–293. doi:10.1016/j.jhep.2018.10.008
4. Xia Y, Zhang J, Ni X. Diagnosis, treatment and prognosis of hepatocellular carcinoma with inferior vena cava/right atrium tumor thrombus. Oncol Lett. 2020;20(4):101. doi:10.3892/ol.2020.11962
5. Li JJ, Liang Q, Sun GC. Traditional Chinese medicine for prevention and treatment of hepatocellular carcinoma: a focus on epithelial-mesenchymal transition. J Integr Med. 2021;19(6):469–477. doi:10.1016/j.joim.2021.08.004
6. Ji Y, Yin Y, Zhang W. Integrated bioinformatic analysis identifies networks and promising biomarkers for hepatitis B virus-related hepatocellular carcinoma. Int J Genomics. 2020;2020:2061024. doi:10.1155/2020/2061024
7. Liu T, Wu H, Qi J, et al. Seven immune-related genes prognostic power and correlation with tumor-infiltrating immune cells in hepatocellular carcinoma. Cancer Med. 2020;9(20):7440–7452.
8. Vogel A, Meyer T, Sapisochin G, et al. Hepatocellular carcinoma. Lancet. 2022;400(10360):1345–1362. doi:10.1016/S0140-6736(22)01200-4
9. Cao X, Shao Y, Meng P, et al. Nascent proteome and glycoproteome reveal the inhibition role of ALG1 in hepatocellular carcinoma cell migration. Phenomics. 2022;2(4):230–241. doi:10.1007/s43657-022-00050-5
10. Pobbati AV, Kumar R, Rubin BP, et al. Therapeutic targeting of TEAD transcription factors in cancer. Trends Biochem Sci. 2023;48(5):450–462. doi:10.1016/j.tibs.2022.12.005
11. Fan M, Lu W, Che J, et al. Covalent disruptor of YAP-TEAD association suppresses defective hippo signaling. eLife. 2022;11:e78810. doi:10.7554/eLife.78810
12. Dey A, Varelas X, Guan KL. Targeting the hippo pathway in cancer, fibrosis, wound healing and regenerative medicine. Nat Rev Drug Discov. 2020;19(7):480–494. doi:10.1038/s41573-020-0070-z
13. Tomikawa J, Takada S, Okamura K, et al. Exploring trophoblast-specific Tead4 enhancers through chromatin conformation capture assays followed by functional screening. Nucleic Acids Res. 2020;48(1):278–289. doi:10.1093/nar/gkz1034
14. Tsinias G, Nikou S, Mastronikolis N, et al. Expression and prognostic significance of YAP, TAZ, TEAD4 and p73 in human laryngeal cancer. Histol Histopathol. 2020;2020:18228.
15. Chi M, Liu J, Mei C, et al. TEAD4 functions as a prognostic biomarker and triggers EMT via PI3K/AKT pathway in bladder cancer. J Exp Clin Cancer Res. 2022;41(1):175. doi:10.1186/s13046-022-02377-3
16. Im JY, Kim DM, Park H, et al. VGLL1 phosphorylation and activation promotes gastric cancer malignancy via TGF-β/ERK/RSK2 signaling. Biochim Biophys Acta Mol Cell Res. 2021;1868(1):118892. doi:10.1016/j.bbamcr.2020.118892
17. Tschaharganeh DF, Chen X, Latzko P, et al. Yes-associated protein up-regulates Jagged-1 and activates the Notch pathway in human hepatocellular carcinoma. Gastroenterology. 2013;144(7):1530–1542 e1512. doi:10.1053/j.gastro.2013.02.009
18. Zhang J, Liu P, Tao J, et al. TEA domain transcription factor 4 is the major mediator of yes-associated protein oncogenic activity in mouse and human hepatoblastoma. Am J Pathol. 2019;189(5):1077–1090. doi:10.1016/j.ajpath.2019.01.016
19. Weiler S. YAP induziert chromosomale Instabilität in Leberkrebspatienten [YAP induces chromosomal instability in liver cancer patients]. Pathologe. 2018;39(Suppl 2):185–188. German. doi:10.1007/s00292-018-0494-y
20. Jiao S, Guan J, Chen M, et al. Targeting IRF3 as a YAP agonist therapy against gastric cancer. J Exp Med. 2018;215(2):699–718. doi:10.1084/jem.20171116
21. Li SK, Tang HC, Man-Hon Leung M, et al. Centrosomal protein TAX1BP2 inhibits centrosome-microtubules aberrations induced by hepatitis B virus X oncoprotein. Cancer Lett. 2020;492:147–161. doi:10.1016/j.canlet.2020.08.005
22. Cai WY, Lin LY, Hao H, et al. Yes-associated protein/TEA domain family member and hepatocyte nuclear factor 4-alpha (HNF4α) repress reciprocally to regulate hepatocarcinogenesis in rats and mice. Hepatology. 2017;65(4):1206–1221. doi:10.1002/hep.28911
23. Feng X, Lu T, Li J, et al. The tumor suppressor interferon regulatory factor 2 binding protein 2 regulates hippo pathway in liver cancer by a feedback loop in mice. Hepatology. 2020;71(6):1988–2004. doi:10.1002/hep.30961
24. Shi Z, He F, Chen M, et al. DNA-binding mechanism of the hippo pathway transcription factor TEAD4. Oncogene. 2017;36(30):4362–4369. doi:10.1038/onc.2017.24
25. White SM, Murakami S, Yi C. The complex entanglement of Hippo-Yap/Taz signaling in tumor immunity. Oncogene. 2019;38(16):2899–2909. doi:10.1038/s41388-018-0649-6
26. Yang W, Yang S, Zhang F, et al. Influence of the hippo-YAP signalling pathway on tumor associated macrophages (TAMs) and its implications on cancer immunosuppressive microenvironment. Ann Transl Med. 2020;8(6):399. doi:10.21037/atm.2020.02.11
27. Guo X, Zhao Y, Yan H, et al. Single tumor-initiating cells evade immune clearance by recruiting type II macrophages. Genes Dev. 2017;31(3):247–259. doi:10.1101/gad.294348.116
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.





