Back to Journals » Cancer Management and Research » Volume 15
The Pro-Tumor Biological Function of IL-36α Plays an Important Role in the Tumor Microenvironment of HCC
Authors Song Y, Chu H , Liu F, Guo W , Gao N , Chen C , Bao S
Received 6 March 2023
Accepted for publication 18 July 2023
Published 29 August 2023 Volume 2023:15 Pages 895—904
DOI https://doi.org/10.2147/CMAR.S407123
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Bilikere Dwarakanath
Yanmei Song,* Huiyuan Chu,* Fang Liu, Wenjie Guo, Na Gao, Che Chen, Shisan Bao
Department of Clinical Laboratory Diagnostics, School of Public Health Gansu University of Chinese Medicine, Lanzhou, 730000, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Che Chen; Shisan Bao, Department of Clinical Laboratory Diagnostics, School of Public Health Gansu University of Chinese Medicine, 35, Dingxi East Road, Chengguan, Lanzhou, Gansu, 730000, People’s Republic of China, Tel +86 187 09312856 ; +81 180 176 84959, Email [email protected]; [email protected]
Purpose: To investigate the role of IL-36 in the tumorigenesis of hepatocellular carcinoma (HCC). IL-36 composed of a natural antagonist (IL-36Ra) and three agonists (IL-36α, -β, -γ) that stimulate inflammation by binding to a common receptor consisting of IL-36R and IL-1RAcP. HCC is a common malignancy associated with high morbidity and mortality, often diagnosed at later stages. Although the exact role of IL-36α in HCC remains controversial, it is hypothesized that it may play a significant role in the development and progression of this cancer.
Materials and Methods: In the current study, we measured both circulating and intrahepatic levels of IL-36α from HCC patients and healthy controls, using ELISA. The association between IL-36 and the differentiation of HCC was determined. Furthermore, the role IL-36 in both HCC and non-HCC cell lines was evaluated in vitro.
Results: Circulating and intra-hepatic IL-36α was inversely correlated with differentiation of HCC, suggesting that IL-36α contribute to protection during the development of HCC. Based on bioinformatics, miR-27b-3p is closely related to downstream IL-36α. Thus, we determined miR-27b-3p expression in HCC tissues, showing upregulated miR-27b-3p was inversely correlated with IL-36α in HCC, perhaps via CXCL1 in HCC cells. It was confirmed that IL-36α inhibited HCC proliferation, viability and migration in vitro, consistent with reduced the expression of cytokines IL-1β, IL-18, implying that IL-36α inhibited the possible involvement of pyroptosis.
Conclusion: Our data suggests that IL-36α may be a potential therapeutic target and a prediction biomarker for the management of HCC.
Keywords: hepatocellular carcinoma, interleukin-36α, prognosis, biomarker
Introduction
Hepatocellular cancer (HCC) is the fourth most common malignancy with unacceptably high mortality and morbidity,1 due to lack of a sensitive and a reliable biomarker for early diagnosis. Consequently, a large number of HCC patients are diagnosed at advanced stages with distant metastases, and receive only palliative care.1 The 5-year survival rate has been reported to be as low as 9%,1,2 primarily due to high postoperative recurrence rates and resistance to chemotherapy.3 Alpha-fetoprotein (AFP), a routinely used biomarker for HCC, is not sensitive enough for early diagnosis.4 Thus, it is critically important to explore novel, reliable biomarkers for early detection and for predicting prognosis.
IL-36 is a member of the IL-1 family and is produced by a wide range of cells, including leukocytes, epithelial cells, and fibroblasts.5 The crystal structure of IL-36α, IL-36β, IL-36γ, and IL-36Ra has confirmed that, like IL-1β, these molecules share an evolutionarily conserved 12-stranded β-sheet structure.6 Successful activation of IL-36α is initiated by binding to the IL-36R receptor and receptor accessory protein IL-1RAcP from the heterodimeric receptor complex.7 This generates an intracellular signaling cascade, leading to the activation of NF-κB and mitogen-activated protein kinases that induce inflammatory responses.8 To explain the mechanism of IL-36 better, we have drawn a figure for clarification (Figure 1). The activity of IL-36α was determined from its induction of CXCL1 mRNA transcripts by neutrophil chemokine, and CXCL1 was positively correlated with IL-36α.9 Although IL-36α has been reported to be involved in inflammatory diseases, the precise role of IL-36α in the pathogenesis of HCC remains to be explored.
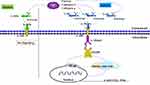 |
Figure 1 The activating process of IL-36α. |
IL-36α expression may contribute to the activation of adaptive T cell-mediated immunity.10 However, the underlying mechanism of IL-36α in HCC remains to be elucidated. It has been suggested that miR-27b-3p, consisting of 19 to 25 base pairs, is involved in the oncogenesis of multiple cancers11 and inhibits IL-36α via CXCL1 in HCC based on bioinformatics database search. The upregulation of miR-27b-3p has been observed in HCC and cell lines.12 The role of IL-36α and the pyroptosis pathway during the development of HCC is unclear. Pyroptosis is an inflammatory cell death accompanied by the activation of inflammasomes and maturation of pro-inflammatory cytokines IL-1β and IL-18.13
The objective of this study was to determine the correlation between IL-36α and HCC differentiation and survival period, and its potential use as a diagnostic biomarker for HCC. The correlation between IL-36α and AFP production was also examined. In addition, the study evaluated the inhibitory effects of IL-36α on HCC proliferation, viability, and migration in vitro, as well as its potential role in inducing pyroptosis through the expression of cytokines IL-1β and IL-18.14 These findings could potentially lead to the development of novel diagnostic and therapeutic targets for the management of HCC.
Material and Methods
Patients and Tissue
The study collected HCC liver tissues and serum samples from Gansu Provincial People’s Hospital. Serum samples were obtained from HCC (n=15), HBV (n=15), cirrhosis (n=15), and healthy control (routine healthy checkup) (n=15) patients. Blood (3 mL) was collected from HCC patients and healthy controls in the early morning prior to breakfast, and serum was collected by centrifugation (1000 g/15 min) and kept at −80°C until used. None of the HCC patients received pre-operative chemotherapy prior to surgery. Hepatic tissues were collected from HCC (n=9) and non-tumor tissue (n=3). Surgical specimens of cancerous tissues and paired adjacent non-tumor tissue were obtained from three HCC cases. The study has been approved by the Ethics Committee of Gansu Provincial People’s Hospital. The patients consent has been obtained. The study complied with the Declaration of Helsinki. Demographic information including patients’ sex, age, liver cirrhosis, hematoxylin-eosin staining of pathological sections, AFP, and CEA level of serum were collected. The sample included 29 men and 31 women, with a mean age of 52 years (range 30–78 years, median 59 years for women, median 49.5 years for men). All tissue samples were stored at −80°C until tested.
Cell Lines
HepG2 (a low-differentiated human HCC cell line) and Lo2 (a normal human liver cell line) were obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). Huh7 (a human high-differentiated HCC cell line) was obtained from The Second People’s Hospital of Lanzhou University (Gansu, China) (ref14). Lo2 and Huh7 cells were cultured in RPMI 1640 medium supplemented with 10% heat-inactivated fetal bovine serum and 1% penicillin-streptomycin, whereas HepG2 cells were cultured in DMEM supplemented with 1% penicillin-streptomycin. The cells were cultured in a humidified atmosphere of 5% CO2 at 37°C incubator.
Bioinformatic Analysis
TARGETSCAN is a widely used database for predicting miRNA targets in vertebrates. It has undergone updates and improvements over time, including the incorporation of new algorithms to increase the accuracy of match predictions. In a search using gene symbols, the results can be displayed for different transcripts of a gene. When searching for the mRNA CXCL1, the results will show the predicted miRNA target(s) for the specific transcript of interest. The TARGETSCAN database can be accessed at https://www.targetscan.org/vert_80/.
The Encyclopedia of RNA Interactomes (ENCORI) is an open-source platform that facilitates the study of miRNA-mRNA, RNA-RNA, RBP-ncRNA, and RBP-mRNA interactions using degradome-seq and RNA-RNA interactome data. In our current study, we utilized ENCORI to predict the miRNA (miR-27b-3p) that regulates CXCL1 and to verify its correlation with CXCL1 expression. ENCORI is available at http://starbase.sysu.edu.cn/index.php.
Gene Expression Profiling Interactive Analysis (GEPIA) is a web-based tool used for interactive online gene expression analysis. It is based on data from 9736 tumors and 8587 normal samples obtained from the TCGA and the GTEx databases.15 In our current study, we used GEPIA to explore the correlation between the receptor gene IL-36R and overall survival in HCC. Dot plots and body maps were also generated using this tool. GEPIA is available at http://gepia.cancer-pku.cn/.
Reagents
Recombinant human IL-36α (LC12JU2903) was purchased from Sino Biological, Beijing, China. The CCK-8 kit, including human IL-36α/IL1F6 (IL36A) Kit, human IL-18, and IL-1β, was purchased from Solarbio, Beijing, China. RPMI 1640 and DMEM were obtained from HyClone Laboratories, Logan, UT, USA. Heat-inactivated fetal bovine serum was purchased from Gibco, Grand Island, NY, USA, and penicillin-streptomycin was obtained from Solarbio, Beijing, China.
ELISA
IL-36α levels in HCC tissues, serum, and cell supernatant (without intervention) were measured using the human IL-36α/IL1F6 (IL36A) Kit. The levels of IL-18 and IL-1β in the supernatants from IL-36α-treated HCC cells in vitro were determined using the human IL-18 Kit. All experiments were performed in triplicate.
Cell Growth (CCK-8)
HepG2, Lo2, and Huh7 cells were seeded in 96-well plates at a density of 1.5 × 10^3 cells per well in 200 μL of the respective culture media mentioned previously. These cells were cultured with or without IL-36α (500 ng/mL) for 24 hours in triplicate. Cell proliferation was determined, using the CCK-8 kit according to the manufacturer’s instructions (Bio-Rad Laboratories, Hercules, CA, USA).16
Based on preliminary experiments, HepG2 cells were selected for IL-36α intervention, as their survival rate was the lowest among Lo2, Huh7, and HepG2 cells. HepG2 cells were treated with varying concentrations of IL-36α (62.5, 125, 250, 500, 750 ng/mL) for 0, 24, 48, or 72 hours. The optimal time and concentration of exogenous IL-36α for HepG2 cells were determined.
Wound Scratch
HepG2 and Lo2 cells were treated with or without IL-36α (500 ng/mL) for 12 h until they reached 90% confluency. The monolayer cells were then scraped on the plate using a sterile pipette to create a gap, ensuring that no cells were present in the gap and that it was straight, as previously described.17 The scratch area was imaged under an inverted microscope at 0, 24, and 48 h after IL-36α treatment. The distance of cell migration (wound healing), was measured using ImagePro Plus 9 software, as previously described.18
Statistics
The statistical analysis was performed, using SPSS software (version 25.0). The results are presented as means ± standard deviation of at least three independent experiments. The statistical significance of differences between groups was analyzed by one-way analysis of variance (ANOVA) followed by Student’s t-test. The statistical significance was considered at P < 0.05. The significance levels are represented by asterisks as follows: *P < 0.05, **P < 0.01, and ***P < 0.001.
Results
Circulating Expression of IL-36α, AFP, CEA from HCC Patients and HC
The levels of circulating IL-36α were significantly higher in liver disease patients, in the order of CHB, cirrhosis, and HCC, compared to healthy controls (HC) (Figure 2A). In contrast, the levels of AFP gradually increased from CHB, cirrhosis to HCC patients, compared to HC (Figure 2B). A similar pattern was observed for CEA levels in the serum (Figure 2C).
IL-36α Expression of HCC and Non-Tumor Tissues
The histopathological differentiation of HCC and non-cancer tissues is presented in Figure 3A, showing non-cancer, high, moderate, and poorly differentiated HCC. Constitutive expression of IL-36α was observed in non-cancer tissues, while IL-36α expression was significantly elevated in HCC tissues (Figure 3B). Interestingly, IL-36α expression was highest in well-differentiated HCC tissues and gradually and significantly reduced in moderately and poorly differentiated HCC tissues (P<0.05) (Figure 3B).
Bioinformatic Tools for microRNA-27b-3p Target Prediction
In the search for CXCL1 target miRNAs on TARGETSCAN, 766 potential matches were found according to the transcript. Among them, miR-27b-3p showed a perfect match to the seed region, ie nucleotides 2 to 7, which have perfect Watson-Crick pairing with the 3’ UTR of CXCL1 (Figure 4A). To investigate whether miR-27b-3p can regulate CXCL1, we used the ENCORI database (Figure 4B) and found an inverse correlation between miR-27b-3p and CXCL1 expression, as calculated from 370 HCC samples in the database (Figure 4C).
Linkage Between IL-36α and Prognosis
The Expression of the Only Receptor IL-36R via GEPIA
To investigate the prognostic value of IL-36α in HCC, we used the GEPIA database. Since IL-36α was not available in the database, we used IL-36R as a substitute to explore any potential relationship between IL-36α and cancer in general (Figure 5A). We further focused on HCC and found that IL-36R was highly expressed in normal (peri-tumor) tissue but lowly expressed in HCC based on TPM maps (Figure 5B). We generated a survival plot using the Cox proportional hazard ratio and the 95% confidence interval information. The overall survival of HCC based on IL-36R expression is shown in Figure 5C, indicating an inverse correlation between IL-36R expression and HCC survival.
The Expression of the mRNA CXCL1 by Using ENCORI Database
Using ENCORI database, we compared the transcriptional levels CXCL1 in HCC (n=374) with normal liver samples (n=50) (Figure 6A), showing a correlation between CXCL1 and IL-36R at the transcriptional levels. The data generated from ENCORI, showing that constitutive expression of CXCL1 in peri-tumor, and such expression was significantly suppressed in HCC. Then we further determined the correlation between overall survival and CXCL1 expression, showing CXCL1 expression was associated with prognosis of overall survival (P< 0.0012) (Figure 6B).
IL-36α Inhibited Viability and Migration in Cells Lines
Constitutive levels of IL-36α were detected in non-HCC cells in vitro. Huh7 or HepG2 HCC cells showed 1.4 or 1.7-fold higher expression of IL-36α (*P<0.5) compared to non-HCC cells, indicating upregulation of IL-36α in HCC cells that depended on differentiation (Figure 7A). To investigate the effect of exogenous IL-36α on HCC cells, HepG2 cells were treated with 750, 500, 250, 125, and 62.5 ng/mL IL-36α for 24 h. Results revealed a dose-dependent inhibition of cell viability by IL-36α (Figure 7B), with the highest inhibition observed at 500 ng/mL. Wound scratch assay was conducted to evaluate the effect of IL-36α (500 ng/mL) on the migration ability of HepG2 cells at 24 or 48 h post-treatment (Figure 7C), while DMEM only was used as control. Results showed that IL-36α suppressed wound healing by 10% or 20% at 24 or 48 h post-treatment, respectively.
IL-36α Suppressed IL-1β and IL-18 in HepG2 Cells in vitro
IL-18 and IL-1β have been reported to be produced by gastrointestinal cells during pyroptosis. Therefore, we measured the levels of IL-18 and IL-1β in HepG2 cells in response to IL-36α stimulation with 500 ng/mL for 24 h, using ELISA. We found that the expression of IL-18 and IL-1β was suppressed by approximately 40% and 10%, respectively, in HepG2 cells in the presence of IL-36α (500 ng/mL) for 24 h, compared to mock-treated cells (Figure 8).
Discussion
In the current study, it was demonstrated that circulating IL-36α levels were significantly upregulated in patients with CHB, cirrhosis, and HCC, compared to healthy controls. The hepato-protective role of IL-36 in CHB has been well documented,19 as IL-36 can be produced by both hepatocytes20 and peripheral leukocytes.21 However, the protective role of IL-36 may be compromised in susceptible individuals, resulting in persistent liver chronic inflammation.22 Additionally, cirrhosis often causes persistent liver damage without effective intervention,23 and the host immunity in cirrhotic patients is often compromised, contributing to incomplete protection from IL-36. The lower IL-36 levels in cirrhotic patients than in CHB patients may be related to the severity of liver damage, with more severe hepatic damage in cirrhosis than in CHB. Similarly, the lowest levels of IL-36 in HCC patients, where the liver is most severely damaged among these three liver diseases, could be explained by this phenomenon. These findings suggest that IL-36α could be a novel biomarker for the detection of HCC.
In our study, we observed that IL-36α expression was mainly detected in well-differentiated HCC in vitro, while it was weakly expressed in poorly differentiated HCC. We also found a correlation between the expression of mir-27b-3p and differentiation status, where higher levels of mir-27b-3p were associated with poorer differentiation. Therefore, the reduced expression of IL-36α in HCC could be attributed to the upregulation of mir-27b-3p. Moreover, given the consistent upregulation of IL-36α and CXCL1 in HCC cells, suggesting that mir-27b-3p may negatively regulate IL-36α via CXCL1 in HCC cells.
On the other hand, IL-36R (IL-1R6), is initiated by binding to IL-36 ligands.24 We performed bioinformatics analysis to investigate the overall survival of HCC based on IL-36R expression and found an inverse correlation between IL-36R expression and HCC survival. Given the secretory nature of the IL-36 ligands and the distribution patterns of the IL-36R, we assigned an important role for IL-36α cytokines in binding with IL-36R. Using bioinformatics, we also found that down-regulated expression of CXCL1 was associated with a better prognosis of overall survival, suggesting that CXCL1 contributes to the activity of IL-36α, which is consistent with other studies showing that IL-36α is regulated by CXCL1 in neutrophil chemokine.25 These findings suggest that IL-36α may play a protective role during the development of HCC, and that up-regulated IL-36α may serve as an early predictor of prognosis.
In our study, we observed that IL-36α inhibited HCC proliferation, viability, and migration in vitro, which correlated with the reduced expression of IL-1β and IL-18. Pyroptosis is an inflammatory cell death process that is accompanied by the activation of inflammasomes and the maturation of pro-inflammatory cytokines, including IL-1β and IL-18.13 Although we do not have firm evidence, our data suggest that IL-36α may inhibit IL-1β and IL-18-mediated pyroptosis during the development of HCC, which will require further investigation.
The incidence of HCC is highest in China,26 and the currently routinely used biomarker for its diagnosis is AFP, which is both economic and versatile.27 However, the sensitivity and specificity of AFP in diagnosing HCC need improvement.28 Our current data suggest that IL-36α is rather sensitive and specific in HCC and may be used as a biomarker for early diagnosis and prediction of prognosis. Our speculation is supported by others, indicating that IL-36 plays a critical role in tumor treatment and prognosis.29 Furthermore, our data suggest that IL-36α may be a potential therapeutic target in the management of HCC, which is in line with the findings in colorectal cancer during the early stages of the disease.30 Therefore, we believe that IL-36α is a novel prognostic marker for HCC.
Finally, our data is relatively in small sample size, and future study will extend to large sample size from multi-centers, particularly in exploring pyroptosis during the development of HCC at molecular and cellular level. In the current study, we focused on investigating the role of a single gene, which may not fully represent the complexity of the potential pathways involved in the development of HCC studies. We plan to explore the involvement of additional genes using gene sets to better understand the underlying pathogenesis of HCC. Framework method for the analysis of qualitative data has been used by many researchers, which could boost the credit of the study. We will perform our study to clarify our hypothesis in future.
Conclusion
We conclude that IL-36α is constitutively expressed and upregulated in HCC, and its expression is inversely correlated with mir-27b-3p, which is associated with HCC progression and prognosis. IL-36α exerts a protective role by inhibiting HCC proliferation, viability, and migration. In addition, we found that reduced IL-36R and CXCL1 expression is associated with overall survival in HCC patients. Our data suggest that IL-36α could be a novel prognostic marker and indicator in HCC.
Acknowledgments
This work was supported by The Natural Science Foundation of Gansu Province (No. 20JR10RA311), The Science and Technology Department Project of Lanzhou District of Gansu Province (No. 2020JSCX0084), and The Postgraduate Innovation Fund, Gansu University of Chinese Medicine (No. 2021CX66). We also acknowledge the staffs from The Second People’s Hospital of Lanzhou University and Gansu Provincial People’s Hospital for their assistance.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Wang H, Liu Y, Shen K, et al. A comparison between radiofrequency ablation combined with transarterial chemoembolization and surgical resection in hepatic carcinoma: a meta-analysis. J Cancer Res Ther. 2019;15(7):1617–1623. doi:10.4103/jcrt.JCRT503_19
2. Clark T, Maximin S, Meier J, et al. Hepatocellular carcinoma: review of epidemiology, screening, imaging diagnosis, response assessment, and treatment. Curr Probl Diagn Radiol. 2015;44(6):479–486. doi:10.1067/j.cpradiol.2015.04.004
3. Singal AG, Tiro JA, Marrero JA, et al. Mailed outreach program increases ultrasound screening of patients with cirrhosis for hepatocellular carcinoma. Gastroenterology. 2017;152(3):608–615. doi:10.1053/j.gastro.2016.10.042
4. Cao C, Sun J, Zhang D, et al. The long intergenic noncoding RNA UFC1, a target of MicroRNA 34a, interacts with the mRNA stabilizing protein HuR to increase levels of β-catenin in HCC cells. Gastroenterology. 2015;148(2):415–426. doi:10.1053/j.gastro.2014.10.012
5. Garlanda C, Dinarello CA, Mantovani A. The interleukin-1 family: back to the future. Immunity. 2013;39(6):1003–1018. doi:10.1016/j.immuni.2013.11.010
6. Dinarello CA. IL-1: discoveries, controversies and future directions. Eur J Immunol. 2010;40(3):599–606. doi:10.1002/eji.201040319
7. Smith DE, Renshaw BR, Ketchem RR, et al. Four new members expand the interleukin-1 superfamily. J Biol Chem. 2000;275(2):1169–1175. doi:10.1074/jbc.275.2.1169
8. Towne JE, Garka KE, Renshaw BR, et al. Interleukin (IL)-1F6, IL-1F8, and IL-1F9 signal through IL-1Rrp2 and IL-1RAcP to activate the pathway leading to NF-kappaB and MAPKs. J Biol Chem. 2004;279(14):13677–13688. doi:10.1074/jbc.M400117200
9. Johnston A, Xing X, Wolterink L, et al. IL-1 and IL-36 are dominant cytokines in generalized pustular psoriasis. J Allergy Clin Immunol. 2017;140(1):109–120. doi:10.1016/j.jaci.2016.08.056
10. Pan QZ, Pan K, Zhao JJ, et al. Decreased expression of interleukin-36α correlates with poor prognosis in hepatocellular carcinoma. Cancer Immunol Immun. 2013;62(11):1675–1685. doi:10.1007/s00262-013-1471-1
11. Zhuo L, Liu J, Wang B, et al. Differential miRNA expression profiles in hepatocellular carcinoma cells and drug-resistant sublines. Oncol Rep. 2013;29(2):555–562. doi:10.3892/or.2012.2155
12. Zhu HT, Liu RB, Liang YY, et al. Serum microRNA profiles as diagnostic biomarkers for HBV-positive hepatocellular carcinoma. Liver Int. 2017;37(6):888–896. doi:10.1111/liv.13356
13. Wei Q, Zhu R, Zhu J, et al. E2-induced activation of the NLRP3 inflammasome triggers pyroptosis and inhibits autophagy in HCC cells. Oncol Res. 2019;27(7):827–834. doi:10.3727/096504018X15462920753012
14. Li L, Ye T, Zhang Q, et al. The expression and clinical significance of TPM4 in hepatocellular carcinoma. Int J Med Sci. 2021;18(1):169–175. doi:10.7150/ijms.49906
15. Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–W102. doi:10.1093/nar/gkx247
16. Wu B, Hu K, Li S, et al. Dihydroartiminisin inhibits the growth and metastasis of epithelial ovarian cancer. Oncol Rep. 2012;27(1):101–108. doi:10.3892/or.2011.1505
17. Martinotti S, Ranzato E. Scratch wound healing assay. Methods Mol Biol. 2020;2109:225–229.
18. Wang M, Liang Y, Zhao C, et al. AZT sensitizes hepatocellular carcinoma cells to As2O3 by up-regulating the arsenic transporter aquaglyceroporin 9. Transl Cancer Res. 2018;7(6):1439–1448. doi:10.21037/tcr.2018.11.08
19. Gong Y, Tingxi Z, Qing L, et al. Elevated production of IL-36α in chronic hepatitis B virus-infected patients correlates with viral load. Microb Pathog. 2017;113:412–415. doi:10.1016/j.micpath.2017.11.023
20. Wang X, Liang Y, Wang H, et al. The protective role of IL-36/IL-36R signal in Con A-induced acute hepatitis. J Immunol. 2022;208(4):861–869.
21. Hiz P, Kanbur E, Demir N, et al. Roles of novel IL-1 family (IL-36, IL-37, and IL-38) members in chronic brucellosis. Cytokine. 2020;135:155211. doi:10.1016/j.cyto.2020.155211
22. Neurath MF. IL-36 in chronic inflammation and cancer. Cytokine Growth Factor Rev. 2020;55:70–79. doi:10.1016/j.cytogfr.2020.06.006
23. Moon AM, Singal AG, Tapper EB. Contemporary epidemiology of chronic liver disease and cirrhosis. Clin Gastroenterol Hepatol. 2020;18(12):2650–2666. doi:10.1016/j.cgh.2019.07.060
24. Scheibe K, Backert I, Wirtz S, et al. IL-36R signalling activates intestinal epithelial cells and fibroblasts and promotes mucosal healing in vivo. Gut. 2017;66(5):823–838. doi:10.1136/gutjnl-2015-310374
25. Girbl T, Lenn T, Perez L, et al. Distinct compartmentalization of the chemokines CXCL1 and CXCL2 and the atypical receptor ACKR1 determine discrete stages of neutrophil diapedesis. Immunity. 2018;49(6):1062–1076. doi:10.1016/j.immuni.2018.09.018
26. Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi:10.3322/caac.21262
27. Luo P, Wu S, Yu Y, et al. Current status and perspective biomarkers in AFP negative HCC: towards screening for and diagnosing hepatocellular carcinoma at an earlier stage. Pathol Oncol Res. 2020;26(2):599–603. doi:10.1007/s12253-019-00585-5
28. Liu K, Ding Y, Wang Y, et al. Combination of IL-34 and AFP improves the diagnostic value during the development of HBV related hepatocellular carcinoma. Clin Exp Med. 2022;23(2):397–409. doi:10.1007/s10238-022-00810-7
29. Hu M, Tong Y, Fang H, et al. IL36 indicating good prognosis in human Hepatocellular Carcinoma. J Cancer. 2020;11(21):6248–6255. doi:10.7150/jca.47106
30. Chen F, Qu M, Zhang F, et al. IL-36 s in the colorectal cancer: is interleukin 36 good or bad for the development of colorectal cancer? BMC Cancer. 2020;20(1):92. doi:10.1186/s12885-020-6587-z
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.







