Back to Journals » Research and Reports in Urology » Volume 16
The Prevalence and Association of Different Uropathogens Detected by M-PCR with Infection-Associated Urine Biomarkers in Urinary Tract Infections
Authors Haley E , Luke N , Mathur M, Festa RA, Wang J, Jiang Y, Anderson LA, Baunoch D
Received 6 October 2023
Accepted for publication 16 December 2023
Published 9 January 2024 Volume 2024:16 Pages 19—29
DOI https://doi.org/10.2147/RRU.S443361
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Panagiotis J Vlachostergios
Emery Haley,1 Natalie Luke,1 Mohit Mathur,2 Richard A Festa,3 Jimin Wang,4 Yan Jiang,4 Lori A Anderson,5 David Baunoch3
1Department of Clinical Research, Pathnostics, Irvine, CA, USA; 2Department of Medical Affairs, Pathnostics, Irvine, CA, USA; 3Department of Research and Development, Pathnostics, Irvine, CA, USA; 4Department of Statistical Analysis, Stat4Ward, Pittsburgh, PA, USA; 5L.Anderson Diagnostic Market Access Consulting, San Diego, CA, USA
Correspondence: David Baunoch, Pathnostics, 15545 Sand Canyon Suite 100, Irvine, CA, 92618, USA, Tel +1-714-966-1221, Fax +1-714-966-1231, Email [email protected]
Background: Many emerging uropathogens are currently identified by multiplex polymerase chain reaction (M-PCR) in suspected UTI cases. Standard urine culture (SUC) has significantly lower detection rates, raising questions about whether these organisms are associated with UTIs and truly cause inflammation.
Objective: To determine if microbes detected by M-PCR were likely causative of UTI by measuring inflammatory biomarkers in the urine of symptomatic patients.
Design, Setting, and Participants: Midstream voided urine was collected from subjects ≥ 60 years presenting to urology clinics with symptoms of UTI (n = 1132) between 01/2023 and 05/2023. Microbe detection was by M-PCR and inflammation-associated biomarker (neutrophil gelatinase-associated lipocalin, interleukin 8, and interleukin 1β) was by enzyme-linked immunosorbent assay. Biomarker positivity was measured against individual and groups of organisms, E. coli and non-E. coli cases, emerging uropathogens, monomicrobial and polymicrobial cases.
Outcome Measurements and Statistical Analysis: Distributions were compared using 2-sample Wilcoxon Rank Sum test with 2-tailed p-values < 0.05 considered statistically significant.
Results and Limitations: M-PCR was positive in 823 (72.7%) specimens with 28 of 30 (93%) microorganisms/groups detected. Twenty-six of twenty-eight detected microorganisms/groups (93%) had ≥ 2 biomarkers positive in > 66% of cases. Both non-E. coli cases and E. coli cases had significant biomarker positivity (p < 0.05). Limitations were that a few organisms had low prevalence making inferences about their individual significance difficult.
Conclusion: The majority of microorganisms identified by M-PCR were associated with active inflammation measured by biomarker positivity, indicating they are likely causative of UTIs in symptomatic patients. This includes emerging uropathogens frequently not detected by standard urine culture.
Plain Language Summary: The M-PCR assay is a novel diagnostic assay for UTI.
This study found that most organisms included in the M-PCR assay were:detected in the urine of patients at least 60 years of age with a presumptive UTI diagnosisassociated with biomarkers of infection and inflammation
Thus, the M-PCR assay:is clinically relevanthas a low likelihood of false-positivity for UTI
Keywords: diagnostic testing, IL-8, IL-1β, M-PCR, NGAL, UTI
Introduction
Urinary tract infections (UTIs) are infections of any part of the urinary tract, generally grouped into lower UTI, called cystitis, in which the infection is confined to the bladder, and upper UTI, called pyelonephritis, in which the infection has spread to the kidneys.1 UTIs constitute a significant healthcare burden worldwide.2 A diagnosis of UTI is a leading cause of prescribed antibiotic usage in outpatients,3 with most infections being treated empirically. Most UTIs occur in otherwise healthy, sexually active, young adult females, in whom anatomic and lifestyle factors result in a predisposition to cystitis.4 However, while simple UTIs, particularly cystitis, are typically managed successfully with empirically prescribed antibiotics in an outpatient setting, patients with additional risk factors often require guided treatment. Newborns, children, elderly adults, and persons with diabetes or other comorbidities are at increased risk for recurrent and/or complicated UTIs (r/cUTIs).4,5 These groups, particularly elderly adults, have higher treatment failure rates and poorer outcomes, such as UTI recurrence,6 urosepsis,7 and even death).8 In 2018 r/cUTIs accounted for approximately >600,000 hospitalizations at an estimated mean cost of $70,063 per hospitalization (non-CAUTI related) in the US.9 As the threat of microbial antibiotic resistance continues to increase, providing the correct antibiotic treatment quickly enough to avoid prolonged empiric therapy is a growing concern among healthcare stakeholders.10–13
As a diagnostic test for UTI, standard urine culture (SUC) has been in use for over 60 years with little advancement to accommodate for the identification of more recently discovered emerging uropathogens.14 The standard urine culture method is optimized for the growth of gram-negative bacteria, primarily Escherichia coli (E. coli), the most commonly identified organism in acute UTIs.15,16 Furthermore, the turn-around time for SUC, which includes antimicrobial susceptiblity testing, can be 3–5 days, potentially delaying results-guided antimicrobial treatment even in cases where the causative organism is detected.15 Recent studies have shown that when more sensitive culture techniques such as enhanced-quantitative urine culture (EQUC) are used, many additional clinically relevant microbial species including several gram-positive organisms, fastidious microbes, and fungi have been isolated from symptomatic subjects.17
Similarly, previous studies have demonstrated that multiplex-PCR (M-PCR) is superior for detecting non-E. coli and polymicrobial infections in urine specimens compared to SUC.12,13,18,19 Polymicrobial infections, which have been reported in up to 39% of suspected UTI cases in older adult populations,17,20,21 have specifically been associated with poorer outcomes.22 Additionally, M-PCR has the benefit of faster turnaround times to reported results, allowing for a more rapid transition to directed antimicrobial therapy or avoiding empiric therapy altogether.12,13,18,19
Despite these advantages, the clinical validity of identifying additional organisms by M-PCR in the urine of symptomatic subjects with UTIs has been questioned in terms of relevance to causing UTIs. Recently, the urine biomarkers neutrophil gelatinase-associated lipocalin (NGAL), interleukin 8 (IL-8), and interleukin 1 beta (IL-1β) have demonstrated a positive correlation and high specificity for active UTI infections.23–27 These biomarkers become elevated in urine as resident and recruited immune cells rapidly mount a pro-inflammatory response to pathogens detected within the urinary tract.28,29
The purpose of this study was to validate the relevance of individual microbial species or groups using three infection-associated biomarkers, NGAL, IL-1β, and IL-8, as an indicator of the state of the immune system in conjunction with a unique M-PCR assay for detection and quantification of microorganisms in patients with lower urinary tract symptoms and diagnosed presumptively with UTIs in a specialty setting.
Materials and Methods
Study Design
This study utilized banked urine specimens from a randomly collected cross-section of 1132 subjects, at least 60 years old, presenting at urology clinics in 22 US states between 01/17/2023 and 05/16/2023 with clinical presentations consistent with UTI, and for which there was enough specimen to effectively conduct M-PCR and biomarker studies. The samples included in the biobank and used for this analysis are intended to be representative of the samples that would routinely be sent for urine microorganism identification and quantification testing as part of the diagnosis and management of cases seen in outpatient urologic specialty settings. Since this study utilized urine samples from a biobank in which the samples were de-identified and associated only with the assigned ICD-10-CM code(s) and the subject’s age and sex, the study was exempted from review from the Western Institutional Review Board- Copernicus Group (WCG), an external independent agency that reviews and approves industry-sponsored clinical trials.
All urine samples utilized in this study were collected via the midstream voided “clean catch” method which is standard practice for busy clinical offices. Samples were transferred to gray-top boric acid (for M-PCR) and yellow-top (for P-AST and biomarker analysis) Vacutainer Tubes (Becton Dickinson, Franklin Lakes, NJ) and shipped overnight at ambient temperature for evaluation at a central testing laboratory (Pathnostics, Irvine CA). Urine samples were processed for M-PCR/P-AST and for urinary biomarkers (NGAL, IL-1β, and IL-8) by enzyme-linked immunosorbent assay (ELISA). Only samples where microbes were detected above a positivity threshold ≥10,000 cells/mL for bacteria/bacterial groups and >0 cells/mL for yeasts by M-PCR were included in the biomarker analysis.
Specimen Testing
Biomarker Quantitation by ELISA – ELISAs for NGAL, (human Lipocalin-2/NGAL Quantikine ELISA Kit (Catalog number SLCN20), human IL-1β/IL-1F2 Quantikine ELISA kit (Catalog number SLB50), and human IL-8/CXCL8 Quantikine ELISA Kit (Catalog number S8000C), purchased from R&D Systems/Bio-Techne (Minneapolis, MN) were performed using the manufacturer’s instructions-for-use with a TECAN microplate reader (Infinite M Nano+) taking OD measurement readings at 450nm and 540nm. Biomarker positivity was defined by using threshold values previously published (Table 1).30,31 This study defined biomarker consensus as any combination of at least two of the three biomarkers positive at or above the cutoff levels.
 |
Table 1 Biomarker Positivity Cutoffs |
Multiplex- Polymerase Chain Reaction (M-PCR) and Pooled Antibiotic Susceptibility Testing (P-AST) – The M-PCR/P-AST assay (Guidance® UTI, Pathnostics, Irvine, CA) analyzes 27 individual uropathogens, three bacterial groups, 32 antibiotic-resistance genes, phenotypic Extended-Spectrum Beta-Lactamase (ESBL), and pooled phenotypic susceptibility testing against 19 antibiotics. It is intended for use as a diagnostic test in symptomatic patients suspected of having active complicated, persistent, recurrent, and elevated-risk urinary tract infections. Testing was performed as previously described; however, results of antibiotic resistance gene detection, ESBL phenotype, and P-AST were not considered in this study.12,13,32
Statistical Analysis
Participant demographics and ICD-10-CM code breakdown were described by summary statistics (eg, mean and standard deviation (SD) for continuous variables such as age, number, and percentage for categorical variables such as sex and ICD-10-CM). Summary statistics (n, median, mean) of all three biomarker levels were provided. Among all M-PCR positive cases, the number and percentage of cases positive for biomarkers and consensus biomarkers were listed for each of the organisms and for combinations of the organisms. Statistical comparisons of biomarkers were compared using subgroup median values via the Wilcoxon test. All hypothesis tests were 2-sided, and a p-value < 0.05 was considered statistically significant. All data analyses were performed using R 4.2.2 (https://www.r-project.org/).
Results
Subject Demographics
The study included 1132 subjects presenting to urology clinics with symptoms of r/cUTI. The median subject age was 76.3 (range 60.0–103 years), and the mean was 76.6 (standard deviation = 8.72). Female patients comprised the majority of the cohort, 66.4% (n = 752), and males accounted for 33.6% (n = 380) (Table 2). Many specimens were associated with 2 or more ICD-10-CM (https://www.icd10data.com) codes. The most prevalent of these ICD-10-CM codes was N39.0 “Urinary tract infection, site not specified” [76.0% (n = 977)]; followed by R30.0 “Dysuria” [8.1% (n = 104)]; R31.0 “Gross hematuria” [3.3% (n = 42)]; Z87.440 for “Personal history of urinary (tract) infections” [1.8% (n = 23)]; and R31.9 for “Hematuria, unspecified” [1.2% (n = 16)]. All other r/cUTI-related ICD-10-CM codes, each with a prevalence of <1% of subjects, were grouped under “Other” (Supplemental Table S1).
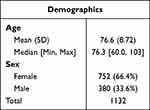 |
Table 2 Demographics of the Study Cohort |
Microbial Prevalence with Detection and Identification by M-PCR/P-AST Assay
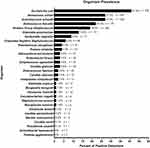
All 1132 specimens were tested for the presence of microbes by M-PCR; of those, 823 (72.7%) were positive. Within these positive specimens, M-PCR identified 1589 microorganisms, with a significant fraction of the total cases being polymicrobial infections (n = 522, 46.1%). Of the 27 species and three groups of microorganisms included in the M-PCR assay, only two (A. baumannii and P. agglomerans) were not detected in any specimen (Figure 1). Two-thirds of the microorganisms (20 of 30) accounted for approximately 99% of all positive results at the case level (Supplemental Table S4).
We analyzed the levels of biomarkers based on the classification groups of the detected microorganisms. The list of classifications and references is provided in Supplemental Table S2. Among the top five most prevalent organisms, we observed a diverse representation: one belonged to the classical gram-negative category (E. coli), one to the classical gram-positive type (E. faecalis), and three belonged to the emerging and/or fastidious uropathogen group (A. urinae, A. schaalii, and Viridans Group Streptococcus [VGS]). Gram-negative bacteria were detected in 581 (51.3%) specimens with over half of those (57.8%, n = 336) identified as E. coli. Gram-positive bacteria were detected in 438 (38.7%) specimens, of which 40.4% (n = 177) were identified as E. faecalis. Fastidious organisms were detected in 570 (50.4%) of total cases. A. urinae was the predominant species identified in 224 (39.3%) cases with fastidious organisms detected. Yeasts were detected in 40 cases (3.5%), and C. glabrata accounted for over half of the detected yeasts (n = 22, 55%). Additionally, we found that two organisms traditionally considered contaminants from the skin, VGS [(n = 160), 14.1% and Coagulase Negative Staphylococcus (CoNS) [(n = 49), 4.3%], were among the top 10 most prevalent organisms detected in the study specimens.
Infection-Associated Biomarkers in M-PCR-Positive Urine Samples
In order to comprehensively assess the presence of infection-associated biomarkers (NGAL, IL-8, and IL-1β), we analyzed the same urine specimens in which microorganisms were detected by M-PCR. By comparing biomarker positivity based on the thresholds outlined in Table 1, we examined the rate of biomarker positivity among different groups of organisms. In Table 3, the 30 detectable microorganisms are presented in groups of 5, by descending order of prevalence, starting with the five most frequently detected organisms, followed by the next most prevalent 5, and ending with the five organisms detected with the least frequency.
 |
Table 3 Biomarker Positivity in Groups of Five Organisms by Prevalence |
Urine samples with detected organisms exhibited high percentages of biomarker positivity. Specifically, NGAL showed a positivity rate of 81%, IL-8 showed a positivity rate of 86%, and IL-1β exhibited a positivity rate of 64%. Furthermore, the simultaneous positivity rate of two or more biomarkers was observed in 80% of cases. To provide a more granular analysis, we delved into the biomarker positivity rates for each individual organism (refer to Supplemental Table S3) and sub-grouped the organisms starting with the top five most detected organisms, gradually expanding in groups of five (refer to Supplemental Table S4).
Considering the remarkable sensitivity of M-PCR in detecting a diverse array of organisms extending beyond E. coli and classical uropathogens,12,13,18,19 we examined the biomarker positivity in all (both positive and negative) M-PCR specimens (Table 4, Supplemental Table S5A) and stratified cases into different groups (Table 5, Supplemental Table S5B). These groups comprised cases with and without E. coli detection, cases with solely classical uropathogens detected, and cases exhibiting exclusively emerging uropathogens (Table 5, Supplemental Table S5B). Furthermore, given the substantial capability of M-PCR to identify a significantly higher number of polymicrobial infections we also compared biomarker levels between specimens with monomicrobial and polymicrobial samples (Table 5, Supplemental Table S5B).12,13,18,19 Across all groups of M-PCR positive specimens, all three biomarkers (NGAL, IL-8, and IL-1β) had a significantly higher percent positivity (p < 0.0001) than in M-PCR negative cases.
 |
Table 4 Biomarker Positivity in M-PCR Positive versus Negative Cases |
 |
Table 5 Biomarker Positivity in M-PCR Positive Cases Grouped by Organism and Infection Characteristics |
We examined the biomarker consensus percent positivity for each subgroup (Table 5). All subgroups of M-PCR-positive specimens had a biomarker consensus positivity >70%, ranging from 74% for cases with only emerging uropathogens to 86% for cases with E. coli detected. The biomarker consensus positivity for each subgroup was also significantly higher than that of the M-PCR-negative cases (p < 0.0001).
We also examined the biomarker consensus positivity for individual organisms to confirm their status as uropathogenic organisms (Figure 2). Biomarker consensus positivity rates were independent of microorganism prevalence in the study cohort. For example, Gardnerella vaginalis, the seventh most prevalent microorganism detected (n = 71) had a 66% biomarker consensus percent positivity, while Mycoplasma hominis, detected in only 5 specimens, had 100% biomarker consensus percent positivity (Figure 2 and Supplemental Table 3). Of the detected organisms, all but 2 [Providencia stuartii (n = 1) and Serratia marcescens (n = 2)] had >66% consensus biomarker positivity. Overall, 80% (n = 661) of the 823 M-PCR-positive specimens were positive for biomarker consensus.
Discussion
To guide antimicrobial selection for UTI patients, clinicians currently rely on microbial identification and quantitation by SUC and associated antibiotic susceptibility testing. Failure to detect and correctly identify those organisms missed by SUC can result in many UTIs remaining undiagnosed and untreated or being sub-optimally treated with empiric broad-spectrum antibiotics which potentially prolongs symptoms or results in serious complications.33 Though it is evident that M-PCR has greater sensitivity than SUC there are some questions about the value of detecting these organisms, and whether they are associated with UTIs or are incidental findings.
We found that of the 30 organisms/organism groups included in the assay for this study just two (A. baumannii and P. agglomerans) were not detected in these symptomatic presumed UTI cases, though those 2 organisms have previously been shown to be uropathogenic.34–41 Additionally, we also found that two organisms traditionally considered contaminants from skin, VGS and CoNS, were among the top 10 most prevalent organisms detected in the study specimens.42 Other studies identified VGS and CoNS in both midstream voided and catheter-collected specimens at similar prevalence and densities, further indicating the organisms’ likely pathogenic nature.43 Therefore, the approach of detecting only classical uropathogens may result in missed cases, as emerging or less common pathogens can cause infections and pose significant health threats.
Having shown that these organisms were found within a significant number of presumed UTI cases, we then examined their association with urinary biomarkers associated with UTIs. Biomarker percent positivity was significantly higher for all 3 biomarkers in M-PCR positive specimens, compared to M-PCR negatives overall (p < 0.001). The small number of M-PCR-negative specimens with elevated biomarkers may represent UTIs caused by viruses, yeast, or bacteria not targeted by the M-PCR test, or by non-infectious bladder inflammation, such as interstitial cystitis.
Using biomarker consensus percent positivity, we then examined whether the detection of a spectrum of organisms present in this assay was associated with positive consensus biomarkers. Overall, 80% of M-PCR-positive specimens were positive for biomarker consensus. Further, of the 28 detected organisms, all but two [P. stuartii (n = 1) and S. marcescens (n = 2)] had >66% of cases with consensus biomarker positivity. When the organisms were categorized into groups of 5 by decreasing prevalence, all groups showed an association with positive consensus and individual markers. Polymicrobial cases, monomicrobial cases, cases with only classical uropathogens, cases with only emerging uropathogens, cases with E. coli, and those without E. coli all had elevated biomarkers. These results strongly indicate that the microbes detected by this assay, many of which are fastidious and emerging pathogens that will likely be missed by SUC, are causative of the UTIs in these cases and not incidental findings. These results make it important to question whether SUC is underdiagnosing due to low sensitivity when the clinical diagnosis from a urology specialty setting, and the urine inflammatory biomarkers agree a UTI is present.
A subset of specimens exhibited outlier data points with low inflammatory biomarker levels despite high microbial densities detected by M-PCR. These outliers with low inflammatory biomarker levels may reflect scenarios where immune responses are compromised due to medications or underlying health conditions, or instances of a resolving UTI.44–48 They may also be the result of natural variation in the ability of different individual microorganisms and collections of microorganisms to elicit an immune response. Future work could explore such microorganism-specific biomarker thresholds.
Additionally, in a related recent study,49 both standard urine culture and culture-free M-PCR methods were used to characterize microbial densities of urine specimens from symptomatic presumptive UTI patients for correlation with detected levels of immune response biomarkers NGAL, IL-8, and IL-1β. A significantly higher percentage of SUC-negative specimens were biomarker-positive compared to M-PCR-negative specimens.49 This suggests that M-PCR has higher sensitivity and specificity for detecting microbes that are causing a UTI, challenging the sensitivity of the current “gold standard” test, SUC, for the identification of uropathogens, and raising questions about false negatives in culture-based testing.
Building upon prior studies,49,50 this paper shows that symptomatic cases involving non-E. coli and emerging uropathogens, along with VGS and CoNS were associated with substantially higher levels of all three biomarkers than in cases where no organisms were detected. These findings suggest that the assay size should not be limited to E. coli and a small set of highly prevalent and longstanding uropathogens, which would lead to missed UTI diagnosis and potentially poorer treatment outcomes. Almost all of the organisms present here would be important to include in a UTI assay in order to be confident that most UTI cases could have the causative pathogen identified.
The biggest strength of this study was the comparison of the immune response according to biomarkers NGAL, IL-1β, and IL-8 in a large number of samples obtained from UTI symptomatic patients from a urology specialty setting, against the identification of microorganisms detected by M-PCR from the same urine sample. This approach allowed us to directly associate the presence of microorganisms identified by the M-PCR assay to infection-associated immune responses in the urinary tract of each subject. However, a few organisms in the assay had low detection percentages or were not detected at all, making inferences about their individual clinical relevance uncertain. Future studies with a larger cohort would be helpful to interpreting the individual clinical relevance for these low-prevalence organisms. Additionally, future studies will examine the relationship between these infection-associated urine biomarkers and clinical outcomes data, such as healthcare utilization and the resolution of symptoms upon treatment with antibiotics appropriate for the organism(s) detected by M-PCR.
Conclusions
Measuring microbial detection by M-PCR against positivity rates of three UTI-associated biomarkers (NGAL, IL-8, and IL-1β), we sought to determine whether these 30 microorganisms were clinically relevant for UTI diagnostics. In the study cohort of 1132 individuals >60 years of age who were symptomatic for r/cUTI, 28 of 30 microorganisms included in the M-PCR assay were detected. Additionally, 80% of M-PCR positive specimens were positive for biomarker consensus, which in symptomatic patients diagnosed in a specialty setting are a measure of inflammation with high sensitivity and specificity for UTI. Together, these findings demonstrate that the majority of organisms detected by the M-PCR assay are likely clinically relevant with high specificity and that their detection by M-PCR has a low likelihood of false-positivity for UTI.
Disclosure
Drs Natalie Luke and Emery Haley report they are employees of Pathnostics, outside the submitted work; In addition, Dr Natalie Luke has a patent US 10,160,991 issued to Pathnostics, a patent US 11,053,532 issued to Pathnostics, a patent US 17/178,091 pending to Pathnostics, a patent US 17/335,767 pending to Pathnostics, a patent US 17/830,227 pending to Pathnostics, a patent US 18/351,385 pending to Pathnostics, a patent US 18/351,286 pending to Pathnostics, a patent US 63/493,416 pending to Pathnostics, a patent US 63/493,416 pending to Pathnostics, a patent US 63/514,785 issued to Pathnostics, a patent AU 2018254514 B2 issued to Pathnostics, a patent BR112019021943-9 B1 issued to Pathnostics, a patent NZ 759292 issued to Pathnostics; Dr Mohit Mathur reports is an employee of Pathnostics, outside the submitted work; Dr Richard Festa reports is an employee of Pathnostics, during the conduct of the study. Dr David Baunoch reports is an employee and hold stocks from Pathnostics, outside the submitted work; In addition, Dr David Baunoch has a patent US10,160,991 issued to PATHNOSTICS, a patent US11,053,532 issued to PATHNOSTICS, a patent US17/178,091 pending to PATHNOSTICS, a patent US17/335/767 issued to PATHNOSTICS, a patent US17/830/227 pending to PATHNOSTICS, a patent US18/351,286 pending to PATHNOSTICS, a patent PCT/US22/16816 pending to PATHNOSTICS, a patent PCT/US22/77477 pending to PATHNOSTICS, a patent AU2018254514 B2 issued to PATHNOSTICS, a patent BR112019021943-9 B1 issued to PATHONSTICS, a patent NZ759292 pending to PATHNOSTICS, a patent EP3612638 pending to PATHNOSTICS, a patent JP2022-042545 pending to PATHNOSTICS, a patent CA 3,175,879 issued to PATHNOSTICS, a patent CA 3,176,586 issued to PATHNOSTICS, a patent CA 3,061,015 issued to PATHNOSTICS, a patent HK 62020014337.3 issued to PATHNOSTICS, a patent CN 201880039956.9 issued to PATHNOSTICS, a patent IL 294577 issued to PATHNOSTICS. The authors report no other conflicts of interest in this work.
References
1. Kolman KB. Cystitis and pyelonephritis: diagnosis, treatment, and prevention. Primary Care. 2019;46(2):191–202. doi:10.1016/j.pop.2019.01.001
2. Tandogdu Z, Wagenlehner FM. Global epidemiology of urinary tract infections. Curr Opin Infect Dis. 2016;29(1):73–79. doi:10.1097/QCO.0000000000000228
3. Grigoryan L, Nash S, Zoorob R, et al. Qualitative Analysis of Primary Care Provider Prescribing Decisions for Urinary Tract Infections. Antibiotics. 2019;8(2):84. doi:10.3390/antibiotics8020084
4. Storme O, Tirán Saucedo J, Garcia-Mora A, Dehesa-Dávila M, Naber KG. Risk factors and predisposing conditions for urinary tract infection. Ther Adv Urol. 2019;11:1756287218814382. doi:10.1177/1756287218814382
5. Harb A, Yassine V, Ghssein G, Salami A, Fakih H. Prevalence and Clinical Significance of Urinary Tract Infection among Neonates Presenting with Unexplained Hyperbilirubinemia in Lebanon: a Retrospective Study. Infect Chemother. 2023;55(2):194–203. doi:10.3947/ic.2022.0117
6. Siff LN. Recurrent Urinary Tract Infections. Deckermed Obstetrics Gynecol. 2021. doi:10.2310/obg.19160
7. Wagenlehner FM, Lichtenstern C, Rolfes C, et al. Diagnosis and management for urosepsis. Int J Urol. 2013;20(10):963–970. doi:10.1111/iju.12200
8. Complicated Urinary Tract Infections - StatPearls - NCBI Bookshelf. Available from: https://www.ncbi.nlm.nih.gov/books/NBK436013/.
9. Zilberberg MD, Nathanson BH, Sulham K, Shorr AF. Descriptive Epidemiology and Outcomes of Hospitalizations With Complicated Urinary Tract Infections in the United States, 2018. Open Forum Infect Dis. 2022;9(1):ofab591. doi:10.1093/ofid/ofab591
10. Sabih A, Leslie SW. Complicated urinary tract infections. StatPearls. 2022. https://www.ncbi.nlm.nih.gov/books/NBK436013/.
11. Balasubramanian S, Wang X, Sahil S, Cheng A, Sutkin G, Shepherd JP. Risk factors for the development of acute pyelonephritis in women with a positive urine culture. Neurourol Urodynam. 2022;41(7):1582–1589. doi:10.1002/nau.25005
12. Wojno KJ, Baunoch D, Luke N, et al. Multiplex PCR Based Urinary Tract Infection (UTI) Analysis Compared to Traditional Urine Culture in Identifying Significant Pathogens in Symptomatic Patients. Urology. 2019;136:119–126. doi:10.1016/j.urology.2019.10.018
13. Vollstedt A, Baunoch D, Wojno K, et al. Multisite prospective comparison of multiplex polymerase chain reaction testing with urine culture for diagnosis of urinary tract infections in symptomatic patients. J Sur Urol. 2020;JSU–102.
14. Anger JT, Bixler BR, Holmes RS, Lee UJ. Updates to Recurrent Uncomplicated Urinary Tract Infections in Women: AUA/CUA/SUFU Guideline. J Urol. 2022, Aug 9;:. doi:10.1097/JU.0000000000002860
15. Price TK, Hilt EE, Dune TJ, Mueller ER, Wolfe AJ, Brubaker L. Urine trouble: should we think differently about UTI? Int Urogynecol J. 2018;29(2):205–210. doi:10.1007/s00192-017-3528-8
16. Sokhn ES, Salami A, El Roz A, Salloum L, Bahmad HF, Ghssein G. Antimicrobial Susceptibilities and Laboratory Profiles of Escherichia coli, Klebsiella pneumoniae, and Proteus mirabilis Isolates as Agents of Urinary Tract Infection in Lebanon: paving the Way for Better Diagnostics. Med Sci. 2020;8(3):32. doi:10.3390/medsci8030032
17. Price TK, Dune T, Hilt EE, et al. The Clinical Urine Culture: enhanced Techniques Improve Detection of Clinically Relevant Microorganisms. J Clin Microbiol. 2016;54(5):1216–1222. doi:10.1128/jcm.00044-16
18. Vollstedt A, Baunoch D, Wolfe A, et al. Bacterial Interactions as Detected by Pooled Antibiotic Susceptibility Testing (P-AST) in Polymicrobial Urine Specimens. J Surg Urol. 2020;1:67.
19. Daly A, Baunoch D, Rehling K, et al. Utilization of M-PCR and P-AST for diagnosis and management of urinary tract infections in home-based primary care. JOJ Uro Nephron. 2020;7(2):555707.
20. Hilt EE, McKinley K, Pearce MM, et al. Urine Is Not Sterile: use of Enhanced Urine Culture Techniques To Detect Resident Bacterial Flora in the Adult Female Bladder. J Clin Microbiol. 2014;52(3):871–876. doi:10.1128/jcm.02876-13
21. Older persons | UNHCR. Available from: https://emergency.unhcr.org/protection/persons-risk/older-persons.
22. McCann E, Sung AH, Ye G, Vankeepuram L, Tabak YP. Contributing Factors to the Clinical and Economic Burden of Patients with Laboratory-Confirmed Carbapenem-Nonsusceptible Gram-Negative Urinary Tract Infections. Clinicoecon Outcomes Res. 2020;12:191–200. doi:10.2147/ceor.s234840
23. Price JR, Guran L, Lim JY, et al. Neutrophil Gelatinase-Associated Lipocalin Biomarker and Urinary Tract Infections: a Diagnostic Case-Control Study (NUTI Study). Female Pelvic Medicine Reconstr Surg. 2017;23(2):101–107. doi:10.1097/spv.0000000000000366
24. Gadalla AAH, Friberg IM, Kift-Morgan A, et al. Identification of clinical and urine biomarkers for uncomplicated urinary tract infection using machine learning algorithms. Sci Rep. 2019;9(1):19694. doi:10.1038/s41598-019-55523-x
25. Martino FK, Novara G. Asymptomatic Bacteriuria or Urinary Tract Infection? New and Old Biomarkers. Int J Transl Med. 2022;2(1):52–65. doi:10.3390/ijtm2010006
26. Rodhe N, Löfgren S. primary … SJ of. Cytokines in urine in elderly subjects with acute cystitis and asymptomatic bacteriuria. Scandinavian Journal of Primary Health Care. 2009;27(2):74–79. doi:10.1080/02813430902757634
27. Horváth J, Wullt B, Naber KG, Köves B. Biomarkers in urinary tract infections – which ones are suitable for diagnostics and follow-up? GMS Infect Dis. 2020;8(Doc24). doi:10.3205/id000068
28. Abraham SN, Miao Y. The nature of immune responses to urinary tract infections. Nat Rev Immunol. 2015;15(10):655–663. doi:10.1038/nri3887
29. Li L, Li Y, Yang J, Xie X, Chen H. The immune responses to different Uropathogens call individual interventions for bladder infection. Front Immunol. 2022;13:953354. doi:10.3389/fimmu.2022.953354
30. Valdimarsson S, Jodal U, Barregård L, Hansson S. Urine neutrophil gelatinase-associated lipocalin and other biomarkers in infants with urinary tract infection and in febrile controls. Pediatr Nephrol. 2017;32(11):2079–2087. doi:10.1007/s00467-017-3709-1
31. Tyagi P, Tyagi V, Qu X, Chuang YC, Kuo HC, Chancellor M. Elevated CXC chemokines in urine noninvasively discriminate OAB from UTI. Am J Physiol-Renal. 2016;311(3):F548–F554. doi:10.1152/ajprenal.00213.2016
32. Baunoch D, Luke N, Wang D, et al. Concordance Between Antibiotic Resistance Genes and Susceptibility in Symptomatic Urinary Tract Infections. Infect Drug Resist. 2021;14:3275–3286. doi:10.2147/idr.s323095
33. Haley E, Luke N, Korman H, et al. Improving Patient Outcomes While Reducing Empirical Treatment with Multiplex-Polymerase-Chain-Reaction/Pooled-Antibiotic-Susceptibility-Testing Assay for Complicated and Recurrent Urinary Tract Infections. Diagnostics. 2023;13(19):3060. doi:10.3390/diagnostics13193060
34. Howard A, O’Donoghue M, Feeney A, Sleator RD. Acinetobacter baumannii. Virulence. 2012;3(3):243–250. doi:10.4161/viru.19700
35. Cerqueira GM, Peleg AY. Insights into Acinetobacter baumannii pathogenicity. Iubmb Life. 2011;63(12):1055–1060. doi:10.1002/iub.533
36. Pour NK, Dusane DH, Dhakephalkar PK, Zamin FR, Zinjarde SS, Chopade BA. Biofilm formation by Acinetobacter baumannii strains isolated from urinary tract infection and urinary catheters. Fems Immunol Med Microbiol. 2011;62(3):328–338. doi:10.1111/j.1574-695x.2011.00818.x
37. Bagińska N, Cieślik M, Górski A, Jończyk-Matysiak E. The Role of Antibiotic Resistant A. baumannii in the Pathogenesis of Urinary Tract Infection and the Potential of Its Treatment with the Use of Bacteriophage Therapy. Antibiotics. 2021;10(3):281. doi:10.3390/antibiotics10030281
38. Bian X, Liu X, Zhang X, et al. Epidemiological and genomic characteristics of Acinetobacter baumannii from different infection sites using comparative genomics. BMC Genomics. 2021;22(1):530. doi:10.1186/s12864-021-07842-5
39. Al-Dahmoshi HOM, Al-Khafaji NSK, Al-Alaq FT. Urinary Tract Infections - the Imbalance Between the Pathogen Virulence and the Host Defense [Working Title]. Int J Med. 2020. doi:10.5772/intechopen.94508
40. Cruz AT, Cazacu AC, Allen CH. Pantoea agglomerans, a Plant Pathogen Causing Human Disease. J Clin Microbiol. 2007;45(6):1989–1992. doi:10.1128/jcm.00632-07
41. Wasfi R, Hamed SM, Amer MA, Fahmy LI. Proteus mirabilis Biofilm: development and Therapeutic Strategies. Front Cell Infect Mi. 2020;10:414. doi:10.3389/fcimb.2020.00414
42. Kranz J, Schmidt S, Lebert C, et al. The 2017 Update of the German Clinical Guideline on Epidemiology, Diagnostics, Therapy, Prevention, and Management of Uncomplicated Urinary Tract Infections in Adult Patients. Part II: therapy and Prevention. Urol Int. 2018;100(3):271–278. doi:10.1159/000487645
43. Wang D, Haley E, Luke N et al . Emerging and fastidious uropathogens were detected by M-PCR with similar prevalence and cell density in catheter and Midstream voided urine indicating the importance of these microbes in causing UTIs. Infect Drug Resist. 2023;16:7775–7795. doi: 10.2147/IDR.S429990
44. Hooton TM, Roberts PL, Stapleton AE. Asymptomatic Bacteriuria and Pyuria in Premenopausal Women. Clin Infect Dis. 2020;72(8):1332–1338. doi:10.1093/cid/ciaa274
45. Trautner BW. Urinary tract infection as a continuum—implications for diagnostic and antibiotic stewardship. Clin Infect Dis. 2020;72(8):1339–1341. doi:10.1093/cid/ciaa280
46. Taha AS, Grant V, Kelly RW. Urinalysis for interleukin-8 in the non-invasive diagnosis of acute and chronic inflammatory diseases. Postgrad Med J. 2003;79(929):159. doi:10.1136/pmj.79.929.159
47. Devarajan P. Neutrophil gelatinase‐associated lipocalin (NGAL): a new marker of kidney disease. Scand J Clin Laboratory Investigation. 2008;68(sup241):89–94. doi:10.1080/00365510802150158
48. Sharif-Askari FS, Sharif-Askari NS, Guella A, et al. Blood Neutrophil-to-Lymphocyte Ratio and Urine IL-8 Levels Predict the Type of Bacterial Urinary Tract Infection in Type 2 Diabetes Mellitus Patients. Infect Drug Resist. 2020;13:1961–1970. doi:10.2147/idr.s251966
49. Parnell LKD, Luke N, Mathur M, et al. Elevated UTI Biomarkers in Symptomatic Patients with Urine Microbial Densities of 10,000 CFU/mL Indicate a Lower Threshold for Diagnosing UTIs. MDPI. 2023;13(16):1–15. doi:10.3390/diagnostics13162688
50. Korman HJ, Baunoch D, Luke N, et al. A Diagnostic Test Combining Molecular Testing with Phenotypic Pooled Antibiotic Susceptibility Improved the Clinical Outcomes of Patients with Non-E. coli or Polymicrobial Complicated Urinary Tract Infections. Res Reports Urol. 2023;15:141–147. doi:10.2147/rru.s404260
 © 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.