Back to Journals » Advances and Applications in Bioinformatics and Chemistry » Volume 15
The Potential of 24-Propylcholestrol as Antibacterial Oral Bacteria of Enterococcus faecalis ATCC 29212 and Inhibitor Biofilms Formation: in vitro and in silico Study
Authors Windaryanti D, Gabriel CS, Hidayat IW, Zainuddin A, Dharsono HDA , Satari MH , Kurnia D
Received 22 June 2022
Accepted for publication 22 November 2022
Published 22 December 2022 Volume 2022:15 Pages 99—111
DOI https://doi.org/10.2147/AABC.S372337
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr James Briggs
Devi Windaryanti,1 Christine Sondang Gabriel,1 Ika Wiani Hidayat,1 Achmad Zainuddin,1 Hendra Dian Adhita Dharsono,2 Mieke Hemiawati Satari,3 Dikdik Kurnia1
1Department of Chemistry, Faculty of Mathematics and Natural Science, Universitas Padjadjaran, Sumedang, Indonesia; 2Department of Conservative Dentistry, Faculty of Dentistry, Universitas Padjadjaran, Bandung, Jawa Barat, Indonesia; 3Department of Oral Biology, Faculty of Dentistry, Universitas Padjadjaran, Bandung, Jawa Barat, Indonesia
Correspondence: Dikdik Kurnia, Jl. Raya Bandung Sumedang km.21, Hegarmanah, Jatinangor, Kabupaten Sumedang, Sumedang, Jawa Barat, 45363, Indonesia, Tel/Fax +022 7794391, Email [email protected]
Introduction: Uncontrolled biofilm can cause several diseases such as dental caries, gingivitis, and periodontitis. Dental caries is a disease caused by the accumulation of plaque-containing pathogenic bacteria, including Enterococcus faecalis. These bacteria infect the root canals of teeth and colonize to form biofilms. Biofilm inhibition is carried out by interfering with cell wall formation metabolism. MurA enzyme has a role in peptidoglycan biosynthesis of cell walls. Enterococcal surface protein (Esp) is the main contributor of E. faecalis to form biofilms. In addition, inhibition of biofilms by interfering with the quorum sensing (QS) system, suppressing gelatinase virulence factors by blocking autoinducers gelatinase biosynthesis-activating pheromone (GBAP).
Purpose: Knowing the potential of Piper betel Linn. compounds as antibacterial in vitro and antibiofilm agents against E. faecalis in silico.
Patients and Methods: The compounds were purified by a bioactivity-guided chromatographic method. Antibacterial activity was tested by disc diffusion method, in vitro studies. In silico study, compound P. betel L. was used as the test ligand and compared with positive control fosfomycin, ambuic acid, quercetin, and taxifolin. The proteins used MurA, Esp, GBAP, and gelatinase were docking with the Autodock Vina PyRx 0.8 followed by the PYMOL program and visualized with the Discovery Studio 2020 program.
Results: An antibacterial compound was identified 24-propylcholesterol which can inhibit the activity of E. faecalis ATCC 29212 with MIC value of 78.1 μg/mL and MBC value of 156.3 μg/mL. Molecular docking results showed the binding affinity of 24-propylcholesterol with MurA, ESP, GBAP, and gelatinase enzymes was − 7.6, − 8.7, − 5.3, and − 7.9 kcal/mol.
Conclusion: 24-propylcholesterol has potential as an antibacterial against E. faecalis and as an antibiofilm through in silico inhibition of QS. However, further research is needed in vitro and in vivo to determine the effectiveness of these compounds as antibacterial and antibiofilm.
Keywords: Piper betel Linn, 24-propylcholesterol, biofilm, MurA, QS
Introduction
Structural collection of bacterial cells that attached to the tooth surface, consisted of a matrix of polysaccharides, lipids, and proteins, and formed soft layer known as biofilm.1,2 Biofilms can form irreversible layers on a variety of surfaces, including living tissue. More than 60% of all bacterial infections are caused by biofilm formation.2 Uncontrolled growth of biofilms on teeth can lead to several diseases such as dental caries, gingivitis, and periodontitis.3,4 Currently, biofilms are considered to be the main mediators of dental infection and increase bacterial metabolism and virulence.5 The formation of biofilms can cause poor conditions and increase resistance to antibacterial treatment6 and the host’s immune system elements by preventing their penetration through the bacterial surfaces.7 Nowadays, it is imperative to develop novel antibacterial agents active against both drug-sensitive and drug-resistant bacterial infections with favorable profiles as high efficacy, low toxicity, and short therapy duration.8
WHO reports dental caries as the fourth most expensive chronic disease to treat in most countries, and given the breadth of this problem, oral health is a public health problem that cannot be ignored.9 One of the resistant species usually found in dental root canal infections is Enterococcus faecalis. E. faecalis bacteria were colonized on the surface to form biofilms on dentin walls.10 Treatment of E. faecalis in tooth root infection can be done through the mechanism of inhibiting bacterial cell wall biosynthesis. The main component of the cell wall is the peptidoglycan layer,11 which plays a role in stabilizing the cell wall as a large cellular macromolecule or the main structural component. Peptidoglycan consists of polysaccharides and polypeptides with many cross-links. These polysaccharides contain N-acetylmuramic and N-acetylglucosamine. UDP-N-acetylglucosamine enolpyruvyl transferase commonly known as MurA play an important role in the first step of biosynthesis by catalyzing its reaction by condensing phosphoenolpyruvate to UDP-N-acetylglucosamine. This condensation process does not occur in mammals so the MurA enzyme is often used as an effective target for the development of antibacterial drugs.12 Deletion and inactivation of the MurA enzyme can be harmful to bacteria due to loss of cell integrity and increased susceptibility to osmotic lysis.13,14
Inhibition of biofilm formation can also be done by interfering with the QS system. QS is a cell-to-cell communication in which specific signals are activated to coordinate pathogenic behavior and help bacteria adapt to environmental stressors.2 Bacteria communicate through molecules signal called autoinducers. With the appearance of proteomic studies in bacteria, it was revealed that the QS system regulates their specific regulation and controls the expression of many other proteins ranging from surface proteins, transcription factors, virulence, and biofilm formation to metabolic pathways.15 One of the virulence factors identified in E. faecalis strains, Esp induces biofilm formation. Esp is a protein cell wall that plays a role in biofilm production by contributing to the bacteria’s surface colonization.16 In addition, an inhibitor of GBAP synthesis is also a direct strategy to reduce virulence factors and prevent pathological damage from E. faecalis bacteria. GBAP is an autoinducer that interacts with receptors during the QS process.15,17 QS inhibitory agents can cut off QS cell communication consequently inhibiting biofilm formation.18,19
Currently, the development of drugs such as antibiotics continues to be carried out, effectiveness and safety are important points in determining that antibacterial compounds do not have side effects. Indonesia’s plant biodiversity is an asset that showed great potential as an antibacterial active compound.20 Piper betel Linn. is known to have antibacterial activity21 with ethnobotany and ethnopharmacological content that has been identified to developed new antibacterial agents.22 In addition, pharmacological activities have been reported as antiplatelet, anti-inflammatory, antimalarial, antibacterial, antidiabetic, and antioxidant.23 Phytochemical analysis data showed secondary metabolite content of alkaloids, tannins, phenols, flavonoids, tannins, saponins, glycosides, terpenoids, and steroids.24,25 Natural steroids which are characterized by a ring structure, have shown various biological activities and have been shown to benefit as antitumor, antiviral, antibacterial, and antioxidant agents.26
In silico methods can speed up finding the potential compound, reduce initial identification costs and increase the chances of finding compounds as drug.27 Molecular docking is a structure-based in silico method that helps predict interactions that occur between molecules and biological targets.28 This method is used to see the interaction between ligand and protein. This interaction can be seen from the binding site of macromolecule targets.29 This research focuses on determining the active compound isolated from P. betel L. as an antibacterial that can inhibit the E. faecalis biofilm formation through the inhibition pathway of the enzymes MurA, Esp, GBAP, and gelatinase as receptors. The results of this study are expected to provide molecular modeling and informatics-based approaches in explaining the bioactive compound from P. betel L. which is a potential candidate for natural biofilm prevention.
Materials and Methods
Materials
Leaves of P. betel L. were harvested from local farmers in Jatinangor, Sumedang, West Java – Indonesia. Determined (No. 40/HB/02/2019) at the Taxonomy Laboratory, Department of Biology, Faculty of Mathematics and Natural Sciences, Padjadjaran University, Indonesia. Chemicals for extraction and purification using. Organic solvents methanol, n-hexane, ethyl acetate, acetone, ethanol (Merck), and aqudest (Ikapharmindo Putramas). The purification process of column chromatography was performed by Silica G 60 (0.063–0.200 mm and 0.200–0.500 mm, Merck) and ODS RP-18 (0.040–0.063 mm, Merck), while for thin layer chromatography on Silica G 60 F254 and ODS RP-18 F254S (Merck). Compounds spot on TLC plates were visualized under UV light at 254 nm and 365 nm and were sprayed with 10% H2SO4 in ethanol (v/v). In the antibacterial test, E. faecalis ATCC 29212 bacteria, Mueller–Hinton broth medium (Oxoid, CM1135), MuellerHinton agar medium (Oxoid, CM0337), and chlorhexidine (Sigma-Aldrich) as positive control were used for in vitro studies.
Material for in silico assay, 3D structure of proteins were obtained from the Research Collaboratory for Structural Bioinformatics (RCSB) (https://www.rcsb.org/), MurA enzyme (protein data bank ID: 1UAE),3 Esp (protein data bank ID: 6ORI),16 gelatinase (protein data bank ID: 5A3Y), and 3D structure of GBAP G8ADP0 was retrieved from the UniProt Knowledgebase (http://www.uniprot.org/).30 P. betel L. compound as the test ligand, namely 24-propylcholesterol (CID: 91195166). The ligands of positive controls were ambuic acid (CID 11152290), fosfomycin (CID 446987), taxifolin (CID 439533), and quercetin (CID 5280343). Ligand structures were retrieved from the PubChem database (https://www.ncbi.nlm.nih.gov/pccompound).31
Instruments
Determination of active compound structure was done by spectroscopic methods including ultraviolet (UV) by 8452A diode array, infrared (IR) with FTIR Shimadzu 8400, NMR (1H-NMR, 13C-NMR, DEPT 135°, HMQC, 1H-1H COZY, HMBC) with type JEOL ECA 500 MHz, and mass spectrometry (MS) with water acquity UPLC type triquadrupole. The TLC plates were visualized with a UV detector lamp with wavelengths of 254 nm and 365 nm. Ninety-six-well microplates (Iwaki, 3820 024), micropipette (Eppendorf), microtube (Biologix, code 80–0500), filter tips (Biologix, code 22–0019, 22–0200, and 22–1000), parafilm (Sigma-Aldrich P7688-1EA), incubator (Memmert), paper disk 6 mm (Sigma-Aldrich, Z741310), and microplate readers (Biochrom EZ Read 400) were used for antibacterial assays.
Extraction and Purification of P. betel L. Leaves
Leaves of P. betel L. (580 g) were macerated 3×24 h with methanol and then concentrated with a rotary evaporator at 40°C. The methanol extract (10.0411 g) then was partitioned with organic solvents, namely n-hexane and ethyl acetate. The fraction of n-hexane, ethyl acetate, and methanol was concentrated using a rotary evaporator at 40°C to obtain n-hexane fraction (0.0156 g), ethyl acetate fraction (2.2684 g), and methanol fraction (7.6722 g). Ethyl acetate extract (2.2684 g) was an active extract against E. faecalis and continued to purify by chromatography process on Silica G 60 (20–230 mesh) eluted with several organic solvents, n-hexane, ethyl acetate, and methanol with a gradient of 10% (v/v) resulted in 11 fractions. The fractions were guided by thin-layer chromatography to determine the target compound. Fractions 9 (0.0212 g) and 10 (0.0352 g) had simple and almost single stain patterns. Fractions 9 and 10 were then purified by decantation using methanol followed by 2D thin layer chromatography with Silica plate G 60 F254 and eluted by n-hexane-ethyl acetate (9:1, v/v) and n-hexane-acetone (9:1, v/v) continued by spraying the plate with 10% H2SO4 in ethanol, resulted purified compound (0.0121 g).
Antibacterial Activity of the Extracts and Active Compounds of P. betel L.
The antibacterial activity of P. betel L. extracts against E. faecalis was examined by the inhibitory zone of the Kirby–Bauer method.32 E. faecalis were grown in Mueller–Hinton broth medium. Bacteria were incubated for 24 h at 37°C. After that, the optical density was determined using a microplate reader with the wavelength of 620 nm. The solution was diluted with Mueller–Hinton broth to form a bacterial solution of 0.5 McFarland. P. betle L. extract was made with variations in concentrations of 2, 10, 20, and 40%. Two negative controls were used in this assay, methanol and water, while positive control in this assay was chlorhexidine 2%. Mueller–Hinton agar was smeared with bacterial suspension (100 µL). Samples, negative control and positive control in the form of a solution were dripped on a paper disk (20 μL). The paper disk containing the test solution was put into solid media which already contained bacteria and then incubated for 24 h at 37°C. The inhibitions zone value (in mm) was measured after the incubation.
MIC (minimum inhibitory concentration) and MBC (minimum bactericidal concentration) assays of the compound against E. faecalis were tested using the 96-well microplate dilution method.30 The measurement of MIC and MBC, the concentration of the compound used was 5000 µg/mL. Those solutions (0.1 mL) were added into the well until 12 concentrations of a compound were obtained by serial twofold dilution. Then, cultured bacteria 0.5 McFarland were loaded into the well. After incubation for 24 h at 37°C, the absorbance of the sample was measured using a microplate reader at 620 nm. MIC is the lowest concentration at which bacterial cells are not visually observed but is determined based on the value of optical density. Then, each solution in the well was smeared on the Mueller–Hinton agar medium surface and incubated for 24 h at 37°C. The minimum concentration of the sample without bacterial growth was determined as the MBC value. As for MIC and MBC assays, fosfomycin was also used as a positive control.
Molecular Docking Material for in silico Study
Chemical structure of compound 24-propylcholesterol was confirmed and retrieved using an online program of PubChem (https://pubchem.ncbi.nlm.nih.gov/compound/91195166). Three-dimensional structure of the compound was obtained by downloading the compound canonical SMILES format (CCCC(CCC(C)C1CCC2C1(CCC3C2CC=C4C3(CCC(C4)O)C) C)C(C)C) and converting canonical format into 3D structure with the OPEN BABEL 2.4.2 program. The 3D structure was saved in PDB format. The 3D structure of MurA and Esp was retrieved from RSCB protein database (https://www.rcsb.org/structure) with PDB format. MEANWHILE, GBAP and gelatinase 3D structural models were built using the SWISS-MODEL server (https://swissmodel.expasy.org/) and the result was saved in PDB format.30 Autodock Vina in the open-source PyRx 0.8 software was used for virtual screening and ligand-protein docking. The compound used as a ligand and four proteins as receptors (MurA, Esp, GBAP, and gelatinase).3,14,28,29 The chosen conformation is the conformation with a bond energy value of less than 1.0Å in the mean square root deviation (RMSD) and showed the lowest bond energy compared by other conformations. The best conformation and the receptor were combined and visualized using PYMOL and then analyzed and visualized by the Discovery Studio 2020 Client program. Ligand-residue interactions and docking positions in 3D molecular form was showed by PYMOL program. The docking positions of each protein-ligand complex were compared with the 3D structure of the ligand-binding protein at the active site, the similarity of the ligase position of the compound and its positive control was determined in this step. All software is used under default conditions.
Results
Determination of Compound Chemical Structure
The compound isolated from P. betle L was in white solid form with the molecular formula C30H52O commonly known as 24-propylcholesterol, with Rf value of 1.46 on Silica G 60 F254 thin layer chromatograph eluted by n-hexane-ethyl acetate 9:1, v/ v; and n-hexane-acetone 9:1, v/v. Spectrum data of the compound are 1H-NMR (CD3OD): δH 0.66 (3H, s), 0.79 (3H, d, 6.9 Hz), 0.81 (3H, d, 6 Hz), 0, 83 (3H, t, 7.5 Hz), 0.90 (3H, d, 6.3 Hz), 0.98 (3H, s), 3.50 (1H, m), 5.33 (1H, d, 5.1 Hz). The 1H-NMR spectrum showed six methyl signals at H 0.6–1.0 ppm, one multiple signal at H 3.50 ppm, and one doublet signal at H 5.33 ppm. These signals are typical characteristics of sterol group steroid compounds.33 13C-NMR (CD3OD): δC 37.3 (C-1), 31.7 (C-2), 71.9 (C-3), 42.2 (C-4), 104.8 (C- 5), 121.8 (C-6), 32.0 (C-7), 32.0 (C-8), 50.3 (C-9), 36.6 (C-10), 21, 2 (C-11), 39.8 (C-12), 42.2 (C-13), 56.8 (C-14), 24.4 (C-15), 28.3 (C-16), 56.1 (C-17), 11.5 (C-18), 19.5 (C-19), 36.2 (C-20), 18.9 (C-21), 34.0 (C-22), 26.1 (C-23), 45.9 (C-24), 29.1 (C-25), 19.9 (C-26), 19.1 (C-27), 23.1 (C-28), 29.8 (C-29), 12.1 (C-30). 13C-NMR showed 30 carbons. A total of six carbons are primary carbons, 12 carbons are secondary carbons, nine carbons are tertiary carbons and three carbons are quaternary carbons. 13C-NMR signals at δC 71.9 (C-3) and 121.8 (C-6) are –OH groups and double bonds. This compound molecular weight is 428.37292 g/mol. IR spectrum showed absorption band of 3417.60 cm−1 (-OH), 2931.59 cm−1 (CH sp2), 1674.08 cm−1 (C=C), 2862.66 cm−1 (CH sp3) and absorption at 1375.43 and 1366.31 cm−1 are characteristic of dimethyl gems (CH gem-dimethyl). According to the data above, the following structure is shown in Figure 1.
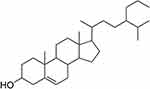 |
Figure 1 Structure of 24-propylcholesterol. |
Antibacterial Activity of P. betel L Extracts
Based on the data of extracts of antibacterial activity in Table 1, it shows that ethyl acetate extract was active against E. faecalis at all variation concentrations of 2, 10, 20 and 40% with the highest inhibition zone at 40% with inhibition zone of 15.7 mm. Similar to chlorhexidine as positive control by having inhibition zone value of 18.8 mm. Based on the reference value of the inhibition zone, the ethyl acetate extract was considered as an antibacterial agent with strong activity with inhibition zone range of 11.85–25.5 mm. According to the reference, the strength of sample antibacterial activity was divided by three based on inhibition zone values antibacterial agent (ie, 6 mm: strong, 3–6 mm: moderate, 3 mm: weak).34 These results indicate that the ethyl acetate extract contained more active components than n-hexane and methanol extracts. Thus, ethyl acetate extract of P. betel L. was selected for further purification.
 |
Table 1 The Results of the Inhibitory Zone of the Kirby–Bauer Method from P. Betel L. Extracts Against E. faecalis |
Antibacterial activity of the the compound was further tested by MIC assay. Based on Table 2, the MIC value of the compound is 78.1 µg/mL and MBC is 156.3 µg/mL. Fosfomycin and chlorhexidine as positive controls had MIC values of 62.5 µg/mL and 3.12 µg/mL and MBC value for chlorhexidine is 6.25 µg/mL. Based on the reference, MIC value of the compound showed strong activity against bacteria. MIC value of 100 µg/mL: strong, 100–625 µg/mL: moderate, and 625 µg/mL: weak.35
 |
Table 2 MIC and MBC Values of 24-Propylcholesterol Against E. faecalis |
Antibacterial Activity Prediction of P. betel L Through Molecular Docking
Based on in vitro analysis data of 24-propylcholesterol compounds against E. faecalis, an in silico inhibition mechanism was predicted. In this study, molecular docking used proteins which involved in inhibition of cell wall biosynthesis and QS, namely MurA, Esp, GBAP, and gelatinase enzymes. The docking analysis results are displayed in Tables 3 and 4.
 |
Table 3 The Binding Affinity of 24-propylcholesterol and Positive Controls |
 |
Table 4 Hydrogen Bond in 24-Profoilkolesterol and Positive Controls |
The docking results showed interaction of 24-propylcholesterol and the target protein was evaluated through value of binding affinity and intermolecular interactions, such as hydrogen bonding. Table 3 showed binding affinity of 24-propylcholesterol with the enzymes MurA, ESP, GBAP, and gelatinase are −7.6, −8.7, −5.3, and −7.9 kcal/mol. Quercetin for the enzyme MurA and gelatinase showed the strongest affinity, −8.3 and −8.1 kcal/mol. In GBAP, 24-propylcholesterol showed the best affinity of −5.3 kcal/mol. However, in Esp, taxifolin is the strongest ligand with an affinity value of −9.9 kcal/mol.
Table 4 shows the amino acid residues bound to the target protein. All ligands in MurA and gelatinase enzymes are bound to different residues, yet still in the same pocket (Figures 2 and 3). In the MurA enzyme, 24-propylcholesterol and ambuic acid bound to the same residue, Asn23. While fosfomycin, quercetin, and taxifolin are not bound to it. In gelatinase, 24-propylcholesterol and quercetin bound to the same residue of Asn35.
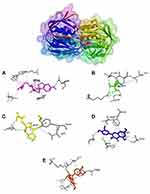 |
Figure 2 Active site of MurA for 24-propylcholestrol (A), fosfomycin (B), ambuat acid (C), quersetin (D) and taxifolin (E). |
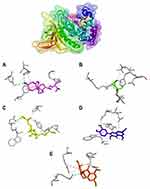 |
Figure 3 Active site of Gelatinase for 24-propylcholesterol (A), fosfomycin (B), ambuic acid (C), quersetin (D) and taxifolin (E). |
Different from MurA and gelatinase ligation positions, Esp (Figure 4) has two ligation positions with fosfomycin, quercetin, and taxifolin bound to Asn64 and Asn149. While 24-propylcholesterol and ambuic acid do not bind them. Meanwhile, GBAP showed three ligation positions (Figure 5). First ligation position belongs to fosfomycin, the second is 24-propylcholesterol, ambuic acid, taxifolin, and the last is quercetin.
 |
Figure 4 Active site of Esp for 24-propylcholesterol (A), fosfomycin (B), ambuic acid (C), quersetin (D) and taxifolin (E). |
 |
Figure 5 Active site of GBAP for 24-propylcholesterol (A), fosfomycin (B), ambuic acid (C), quersetin (D) and taxifolin (E). |
Discussion
The process of finding potential compounds as oral pathogenic antibacterial agents from natural sources of the herbal plant P. betel L. is guided by bioactivity. The results of the isolation and structural elucidation showed the compound is a secondary metabolite compound of the steroid group, which is characterized by a four-fusion ring structure, shown various biological activities, and shown to have benefits as an antitumor, antiviral, antibacterial, and antioxidant agents.26 Steroids with a propyl group attached to C-24, the compound has the name 24-propylcholesterol. The results of the MIC test on E. faecalis showed that at a concentration of 78.1 µg/mL and MBC 156.3 µg/mL, 24-propylcholesterol was able to inhibit bacterial growth. MIC is the lowest concentration of antibacterial agents that can inhibit the growth of microorganisms36 while MBC is the lowest concentration of antibacterial agents that can kill bacteria after incubation.37 Thus, 24-propylcholesterol is a compound with strong activity against E. faecalis bacteria.
The molecular docking approach can be used to see the mechanism of interaction between ligands and proteins.38 In addition, the binding affinity can help predict the potential of a compound as a drug with a higher affinity value, the more potential it would have.39 Hydrogen bonds formed between amino acids and ligands indicate specific molecular interactions.40 It should be noted that the distance between the ligand and the interacting amino acid residues affects the bond, the closer the distance, the stronger and more stable the interaction. The distance between the donor and acceptor bonds also determines the strength of the hydrogen bonds. The closer the hydrogen bond, the stronger the bond energy formed.41 Stability of the complex protein-ligand was contributed by hydrogen bond between ligand and protein. Hydrogen bonds represented in green bonds showed in Figures 1–4, while purple bonds represented hydrophobic bonds.
The docking results in Table 3 showed that 24-propylcholesterol-MurA, 24-propylcholesterol-Esp, and 24-propylcholesterol-gelatinase have good binding affinities, but the lowest binding affinity was shown by 24-propylcholesterol-GBAP of −5.3 kcal/mol. Fosfomycin and ambuic acid as positive controls had a lower binding affinity than 24-propylcholesterol in MurA and Esp. Therefore, 24-propylcholesterol was considered a potential inhibitor of MurA and Esp with binding affinities of −7.6 kcal and −8.7 kcal/mol which showed better results than the positive controls. 24-propylcholesterol in GBAP had the highest binding affinity compared to four positive controls including quercetin, taxifolin, fosfomycin, and ambuic acid of −5.3 kcal/mol. Furthermore, 24-propylcholesterol with gelatinase had a higher binding affinity value than the three positive controls, namely ambuic acid, taxifolin, and fosfomycin of −7.9 kcal/mol. Consequently, 24-propylcholesterol showed potential to inhibit GBAP and gelatinase.
Molecular docking can be used to predict the mechanism of 24-propylcholesterol and MurA interaction. To get stronger and more stable bond, binding affinity with lower value is necessary.42 Fosfomycin as native ligand of the MurA enzyme was used in virtual screening as a positive control since its ability to inhibit biosynthesis of bacteria cell wall.43 The active site of the MurA located at amino acid residues of Asp305, Arg93, Val327, and Cys115.30,44 In Figure 2, 24-propylcholesterol test ligand has the same hydrogen bonds as positive control of ambuic acid, while fosfomycin has hydrogen bonds with Arg232, Asp305, Glu188, Thr304. In MurA enzyme, Asp305 bound to fosfomycin which acted as MurA inhibitor, but has the lowest binding affinity compared to other ligands. All ligands occupy the same site but have different amino acid residues. Therefore, inhibition of the ligand in active site would affect the normal function of the enzyme.
Gelatinase-24-propylcholesterol complex (Figure 3) has hydrogen-bonded residues on Asn351 and His332. 24-propylcholesterol has the same residue as quercetin in Asn351. Ambuic acid and fosfomycin have the same residue on Glu329. Fosfomycin and taxifolin have the same residue on Arg384. Positive control of ambuic acid, fosfomycin, and taxifolin showed lower affinity than quercetin and 24-propylcholeterol because both quercetin and 24-propylcholeterol were not bound to Asn351 or His322 which plays an important role in protein gelatinase. These results indicate that there is a competitive reaction mechanism between the 24-propylcholeterol and quercetin in the enzyme caused they were bound in the same pocket of the enzyme.41
24-propylcholesterol-Esp complex (Figure 4) bound to amino acid residue of Thr44. In Esp, it has two different ligation positions, the first is fosfomycin, quercetin, and taxifolin ligation, while the second is 24-propylcholesterol and ambuic acid ligation. The first ligase position bound to the same amino acid residues of Asn64 and Asn149. While 24-propylcholesterol did not bind to the same amino acid residues as the four positive controls. However, the binding affinity value of 24-propylcholeterol and quercetin were same of −8.7 kcal/mol. Thus, this test ligand has potential to inhibit Esp but noncompetitive to other positive controls.
24-propylcholesterol -GBAP complex (Figure 5) has the highest affinity value compared to the four positive controls of −5.3 kcal/mol, and the hydrogen bond was bound to the same residue as ambuic acid and taxifolin of Arg15. Based on the data obtained, 24-propylcholesterol has potential as an anti-QS agent since it showed as a competitive ligand to positive controls on GBAP and gelatinase. Gelatinase inhibition by 24-propylcholesterol will disrupt the fsr (two-component signal transduction system) regulation system and reduce the virulence factor of gelatinase. Thus, signaling molecules (GBAP) will inhibit QS system and GBAP formation was indirectly disrupted.30
Based on all the research data above, 24-propylcholesterol has the potential to act as a noncompetitive inhibitor of MurA, Esp, GBAP, and competitive to gelatinase. The mechanism of action of this compound is through the inhibition of the enzymes MurA and Esp in cell wall biosynthesis and the QS pathway of E. faecalis. The combination of 24-propylcholesterol and the positive controls are mutually supportive to each other. On one side, these compounds can damage the outermost layer of bacteria (cell wall) and weaken the bacteria. Meanwhile on the other side, these compounds can reduce virulence factors by destroying the quorum-sensing system in E. faecalis bacteria. Furthermore, inhibition of bacteria will occur more easily.
Conclusion
Based on the research results, 24-propylcholesterol compound from P. betel L. has good activity to inhibit E. faecalis and has potential as an antibacterial agent, in vitro. Based on an in silico study the binding affinity of 24-propylcholesterol was stronger than that of positive controls (fosfomycin for MurA, and ambuic acid for GBAP). This study can be used as basic data to support further analysis and development of new antibacterial agents through further studies that are specific and need to be tested in vitro, in vivo, and clinically to determine their effectiveness.
Acknowledgments
The authors are grateful to Academic Leadership Grant (ALG) Prof. Dikdik Kurnia, MSc, PhD, Indonesia (2203/UN6.3.1/PT.00/2022; 20 May 2022) and to Universitas Padjadjaran for all research facilities.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Perry AD. Plaque control for the periodontal patient. Carranza’s Clin Periodontol. 2012:452–460. doi:10.1016/B978-1-4377-0416-7.00044-5
2. Zhou L, Zhang Y, Ge Y, Zhu X, Pan J. Regulatory mechanisms and promising applications of quorum sensing-inhibiting agents in control of bacterial biofilm formation. Front Microbiol. 2020;11:1–11. doi:10.3389/fmicb.2020.589640
3. Kurnia D, Hutabarat GS, Windaryanti D, Herlina T, Herdiyati Y, Satari MH. Potential allylpyrocatechol derivatives as antibacterial agent against oral pathogen of S. sanguinis ATCC 10556 and as inhibitor of MurA enzymes: in vitro and in silico study. Drug Des Devel Ther. 2020;14:2977–2985. doi:10.2147/dddt.s255269
4. Najafi S, Ghasempour M, Davoodabadi A, Kazemi S. Effect of arginine, protamine, and aqueous extracts of Green Tea and Aloe Vera against Enterococcus faecalis. J Islam Dent Assoc IRAN. 2019;31(1):8–13. doi:10.30699/jidai.31.1.2
5. Noviyandri PR, Andayani R, Rizky E. Potensi ekstrak alga Merah Gracilaria verrucosa sebagai penghambat perkembangan pembentukan biofilm Enterococcus faecalis pada infeksi saluran akar gigi. J Syiah Kuala Dent Soc. 2018;1(3):6–15.
6. Cuenca M, Sánchez MC, Diz P, et al. In vitro anti-biofilm and antibacterial properties of Streptococcus downii sp. Nov. Microorganisms. 2021;9(2):1–16. doi:10.3390/microorganisms9020450
7. Okba MM, El-Shiekh RA, Abu-Elghait M, Sobeh M, Ashour RMS. HPLC-PDA-ESIMS/MS Profiling and anti-biofilm potential of Eucalyptus sideroxylon flowers. Antibiotics. 2021;10:761. doi:10.3390/antibiotics10070761
8. Nassar O, Desouky SE, El-Sherbiny GM, Abu-Elghait M. Correlation between phenotypic virulence traits and antibiotic resistance in Pseudomonas aeruginosa clinical isolates. Microb Pathog. 2022;162. doi:10.1016/j.micpath.2021.105339
9. Zhu B, Macleod LC, Kitten T, Xu P. Streptococcus sanguinis biofilm formation & interaction with oral pathogens. Future Microbiol. 2018;13(8):915–932. doi:10.2217/fmb-2018-0043
10. Pasril Y, Yuliasanti A, Umy GF. Daya antibakteri ekstrak daun sirih Merah (Piper Crocatum) terhadap bakteri Enterococcus Faecalis sebagai bahan medikamen saluran akar dengan metode dilusi anti-bacterial power of Red Batel leaves (Piper Crocatum) to Enterococcus Faecalis Bacteria as. Idj. 2014;3(1):88–95.
11. Walsh CT, Wencewicz TA. Prospects for new antibiotics: a molecule-centered perspective. J Antibiot (Tokyo). 2014;67(1):7–22. doi:10.1038/ja.2013.49
12. Musmade DS. MUR-A: a critical target behind new antibacterial drug discovery. Indo Am J Pharmaceutical Res. 2014;4:1.
13. Chang CM, Chern J, Chen MY, et al. Avenaciolides: potential MurA-targeted inhibitors against peptidoglycan biosynthesis in methicillin-resistant Staphylococcus aureus (MRSA). J Am Chem Soc. 2015;137(1):267–275. doi:10.1021/ja510375f
14. Hrast M, Rožman K, Jukič M, Patin D, Gobec S, Sova M. Synthesis and structure–activity relationship study of novel quinazolinone-based inhibitors of MurA. Bioorganic Med Chem Lett. 2017;27(15):3529–3533. doi:10.1016/j.bmcl.2017.05.064
15. Ali L, Goraya MU, Arafat Y, Ajmal M, Chen JL, Yu D. Molecular mechanism of quorum-sensing in Enterococcus faecalis: its role in virulence and therapeutic approaches. Int J Mol Sci. 2017;18(5):960. doi:10.3390/ijms18050960
16. Ghameshlouei S, Zarrabi Ahrabi N, SarveAhrabi Y. In vitro and in silico evaluation of biological activity of a new series of oxadiazole compounds against Esp gene expression in Enterococcus faecalis biofilm. Gene, Cell Tissue. 2021;8(2):6. doi:10.5812/gct.112403
17. Jiang Q, Chen J, Yang C, Yin Y, Yao K, Song D. Quorum sensing: a prospective therapeutic target for bacterial diseases. Biomed Res Int. 2019;2019:1–15. doi:10.1155/2019/2015978
18. Augustine N, Kumar P, Thomas S. Inhibition of Vibrio cholerae biofilm by AiiA enzyme produced from Bacillus spp. Arch Microbiol. 2010;192(12):1019–1022. doi:10.1007/s00203-010-0633-1
19. Chatterjee M, D’Morris S, Paul V, et al. Mechanistic understanding of Phenyllactic acid mediated inhibition of quorum sensing and biofilm development in Pseudomonas aeruginosa. Appl Microbiol Biotechnol. 2017;101(22):8223–8236. doi:10.1007/s00253-017-8546-4
20. Thomford NE, Senthebane DA, Rowe A, et al. Natural products for drug discovery in the 21st century: innovations for novel drug discovery. Int J Mol Sci. 2018;19(6):6. doi:10.3390/ijms19061578
21. Kursia S, Lebang JS, Taebe B, Burhan A, Rahim WO. Uji aktivitas antibakteri ekstrak etilasetat daun sirih Hijau (Piper betle L.) terhadap bakteri Staphylococcus epidermidis. Indones J Pharm Sci Technol. 2016;3(2):72–77.
22. Bustanussala DA, Suhardi DJ. Efektivitas antibakteri ekstrak daun Sirih (Piper betle Linn) terhadap Staphylococcus aureus ATCC 25923. Fitofarmaka. 2015;5(2):58–64.
23. Ghosh R, Darin K, Nath PDP. An overview of various Piper species for their biological activities. Int J Pharma Res Rev. 2014;3(1):67–75.
24. Patel N, Mohan J. Isolation and characterization of potential bioactive compounds from Piper betle varieties Banarasi and Bengali leaf extract. Int J Herb Med. 2017;5(5):182–191.
25. Mihalovits LM, Ferenczy GG, Keserü GM. Catalytic Mechanism and Covalent inhibition of UDP-N-Acetylglucosamine Enolpyruvyl Transferase (MurA): implications to the design of novel antibacterials. J Chem Inf Model. 2019;59(12):5161–5173. doi:10.1021/acs.jcim.9b00691
26. Vollaro A, Esposito A, Antonaki E, et al. Steroid derivatives as potential antimicrobial agents against staphylococcus aureus planktonic cells. Microorganisms. 2020;8(4):1–14. doi:10.3390/microorganisms8040468
27. Pinzi L, Rastelli G. Molecular docking: shifting paradigms in drug discovery. Int J Mol Sci. 2019;20:18. doi:10.3390/ijms20184331
28. Kitchen DB, Decornez H, Furr JR, Bajorath J. Docking and scoring in virtual screening for drug discovery: methods and applications. Nat Rev Drug Discov. 2004;3(11):935–949. doi:10.1038/nrd1549
29. Zhang L, Lin D, Sun X, et al. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved a-ketoamide inhibitors. Science. 2020;368(6489):409–412. doi:10.1126/science.abb3405
30. Kurnia D, Ramadhanty ZF, Ardani AM, Zainuddin A, Dharsono HDA, Satari MH. Bio-mechanism of catechin as pheromone signal inhibitor: prediction of antibacterial agent action mode by in vitro and in silico study. Molecules. 2021;26(21):6381. doi:10.3390/molecules26216381
31. Apriyanti E, Satari MH, Kurnia D. Potential of MurA enzyme and GBAP in Fsr quorum sensing system as antibacterial drugs target: in vitro and in silico study of antibacterial compounds from Myrmecodia pendans. Comb Chem High Throughput Screen. 2020;24(1):109–118. doi:10.2174/1386207323666200628111348
32. Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Disk Susceptibility Tests: Approved Standard - Eleventh Edition. Vol. 32; 2012.
33. Goad LJ, Akihisa T. Nomenclature and biosynthesis of sterols and related compounds. Anal Sterols. 1997;1–42. doi:10.1007/978-94-009-1447-6_1
34. Ravipati AS, Reddy N, Koyyalamudi SR. Biologically Active Compounds from the Genus Uncaria (Rubiaceae). Vol. 43.
35. Kuete V. Potential of Cameroonian plants and derived products against microbial infections: a review. Planta Med. 2010;76(14):1479–1491. doi:10.1055/s-0030-1250027
36. Balouiri M, Sadiki M, Ibnsouda SK. Methods for in vitro evaluating antimicrobial activity: a review. J Pharm Anal. 2016;6(2):71–79. doi:10.1016/j.jpha.2015.11.005
37. Mogana R, Adhikari A, Tzar MN, Ramliza R, Wiart C. Antibacterial activities of the extracts, fractions and isolated compounds from canarium patentinervium miq. Against bacterial clinical isolates. BMC Complement Med Ther. 2020;20(1):1–11. doi:10.1186/s12906-020-2837-5
38. Meng X-Y, Zhang H-X, Mezei M, Cui M. Molecular docking: a powerful approach for structure-based drug discovery. Curr Comput Aided-Drug Des. 2012;7(2):146–157. doi:10.2174/157340911795677602
39. Kumar A, Saranathan R, Prashanth K, Tiwary BK, Krishna R. Inhibition of the MurA enzyme in Fusobacterium nucleatum by potential inhibitors identified through computational and in vitro approaches. Mol Biosyst. 2017;13(5):939–954. doi:10.1039/c7mb00074j
40. Hubbard RE, Kamran Haider M. Hydrogen bonds in proteins: role and strength. eLS. 2010. doi:10.1002/9780470015902.a0003011.pub2
41. Kapitan OB, Ambarsari L, Falah S. Inhibition docking simulation of zerumbone, gingerglycolipid b, and curzerenone compound of Zingiber zerumbet from Timor Island Against MurA Enzyme. J Appl Chem Sci. 2016;279–288. doi:10.22341/jacs.on.00301p279
42. Gupta A, Chaudhary N, Kakularam KR, Pallu R, Polamarasetty A. The augmenting effects of desolvation and conformational energy terms on the predictions of docking programs against mPGES-1. PLoS One. 2015;10(8):1–16. doi:10.1371/journal.pone.0134472
43. Popovic M, Steinort D, Pillai S, Joukhadar C. Fosfomycin: an old, new friend? Eur J Clin Microbiol Infect Dis. 2010;29(2):127–142. doi:10.1007/s10096-009-0833-2
44. Zhu JY, Yang Y, Han H, et al. Functional consequence of covalent reaction of phosphoenolpyruvate with UDP-N-acetylglucosamine 1-carboxyvinyltransferase (MurA). J Biol Chem. 2012;287(16):12657–12667. doi:10.1074/jbc.M112.342725
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
