Back to Journals » OncoTargets and Therapy » Volume 11
The miR-503 cluster is coordinately under-expressed in endometrial endometrioid adenocarcinoma and targets many oncogenes, cell cycle genes, DNA repair genes and chemotherapy response genes
Authors Devor EJ, Cha E , Warrier A, Miller MD , Gonzalez-Bosquet J , Leslie KK
Received 20 July 2018
Accepted for publication 30 August 2018
Published 18 October 2018 Volume 2018:11 Pages 7205—7211
DOI https://doi.org/10.2147/OTT.S180921
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Federico Perche
Eric J Devor,1,2 Elizabeth Cha,1 Akshaya Warrier,1 Marina D Miller,1 Jesus Gonzalez-Bosquet,1,2 Kimberly K Leslie1,2
1Department of Obstetrics and Gynecology, University of Iowa Carver College of Medicine, Iowa City, IA 52246, USA; 2Holden Comprehensive Cancer Center, University of Iowa Hospitals and Clinics, Iowa City, IA 52246, USA
Background: The miR-503 miRNA cluster, located at Xq23.1, is composed of six miRNAs; miR-424, miR-503, miR-542, miR-450a-1, miR-450a-2 and miR-450b. Numerous studies have focused on the relationship of one or two members of the cluster and various human cancers. Here, we suggest that the entire cluster as a single coordinately expressed polycistron transcribed from a single promoter in endometrial endometrioid adenocarcinoma (EEA).
Subjects and methods: A tissue panel composed of twenty histologically confirmed endometrial endometrioid adenocarcinomas (EEA) and four benign endometrium was assembled under informed consent. Expression of each member of the miR-503 cluster was determined by quantitative PCR and differences in expression between EEA and benign tissues were assessed via the standard ΔΔCt method. In addition, the role of promoter methylation status in miRNA expression was examined in Ishikawa H cells following exposure to the cytidine analog Decitabine.
Results: Expression of each member of the miR-503 cluster is significantly down-regulated in EEA in our tumor sample. Both in our tumor sample and in The Cancer Genome Atlas (TCGA) there is evidence of highly correlated expression further supporting the idea that the miR-503 cluster is a polycistron. Looking at each member of the miR-503 cluster we were able to identify 55 unique experimentally validated target genes which include a substantial number of genes involved in carcinogenesis, DNA damage response, cell cycle regulation and chemotherapeutic response. We also found preliminary evidence that regulation of the miR-503 cluster is governed by methylation of the promoter in EEA.
Conclusion: The totality of the data presented here strongly suggest that the miR-503 cluster as a whole merits further investigation as an important potential therapeutic target in EEA.
Keywords: microRNA-503 cluster, polycistron, endometrial cancer, methylation, decitabine, TCGA, correlated expression
Introduction
Endometrial cancer is the most common gynecologic cancer and the fourth most common cancer among women worldwide.1 In 2017, there were >60,000 new cases in the USA and nearly 11,000 deaths.2 It is estimated that, at any one time, in excess of 700,000 women are living with endometrial cancer in the USA alone.2 Although the overall prognosis for endometrial cancer is relatively favorable, particularly in the developed world, there are disturbing indications that endometrial cancer incidence is on the rise as is the recurrence risk.3 Indeed, one estimate projects a staggering 55% increase in endometrial cancer by the year 2030.3 In addition, data collected since the turn of the twenty-first century indicate that there is a growing disparity in outcomes among racial/ethnic groups and that African-American women suffer nearly twice the mortality rate of other women.4 Thus, it is important to expand our understanding of the underlying mechanisms of endometrial carcinogenesis and recurrence in order to develop more effective prevention and treatment.
We have reported that expression of PLAC1, located on chromosome Xq23.1, is highly expressed in gynecologic cancers and is an indicator of both advanced disease and poor outcomes.5–8 Recently, a cluster of miRNAs collectively referred to as the miR-503 cluster was shown not only to be located very close to the PLAC1 gene locus but also to have expression patterns very similar to PLAC1.9 The miR-503 cluster is home to long noncoding RNAs, linc00629 and miR503HG, and six miRNAs; miR-424, miR-503, miR-542, miR-450a-1, miR-450a-2, and miR-450b. In the human genome, miR-450a-1 and miR-450a-2 are identical and will be referred to here collectively as miR-450a. Given the proximity of the miR-503 cluster to PLAC1 and the expression pattern of its members, we have assessed expression levels of the cluster member miRNAs in endometrial endometrioid adenocarcinoma (EEA), the most common type of endometrial cancer. We observed that all five unique miRNAs in the cluster are significantly underexpressed in EEA compared with benign endometrium. In addition, expression of cluster members is highly and significantly correlated across samples suggesting that the miR-503 cluster may be a polycistron transcribed from a single source in a manner similar to the classic miR-17 polycistron.10 A compilation of experimentally validated miR-503 cluster target genes includes a number of well-known oncogenes as well as several loci implicated in DNA repair, DNA damage response, cell cycle, and chemotherapy response. Thus, down-regulation of the miR-503 cluster frees expression of a variety of genes involved in the initiation and maintenance of endometrial carcinogenesis. Finally, consistent with evidence from other cancers, we suggest that the mechanism of miR-503 cluster suppression in endometrial cancer is hypermethylation. Taken together, these observations nominate the miR-503 cluster as a locus of interest in endometrial carcinogenesis and an inviting therapeutic target.
Subjects and methods
Tissue procurement
A screening panel composed of 20 histology-confirmed EEAs and four benign endometrial tissues was assembled for this study. Tissues were obtained under written informed consent from patients undergoing surgery at the University of Iowa Hospitals and Clinics and who were enrolled in the Gynecologic Tissue Bank (Institutional Review Board [IRB]#200209010) that is part of the Women’s Health Tissue Repository (WHTR; IRB#200910784) maintained in the Department of Obstetrics and Gynecology of the University of Iowa Carver College of Medicine.11 In addition to the archived tissue samples, the WHTR provides complete clinical information and outcomes for each patient (Table 1).
Nucleic acid purification
Total cellular RNA was purified from 20 tumor and four control tissue samples using the mirVana miRNA isolation kit also according to manufacturer’s (Thermo Fisher, Waltham, MA, USA) instructions. RNA yield and quality were assessed using an Agilent Model 2100 bioanalyzer and a Trinean DropSense 16 spectrophotometer. Only RNAs with an RNA integrity number (RIN)12 >7.0 were used in these studies. The average RIN was 8.2 with a range from 7.0 to 9.5.
Cell culture
For in vitro experiments, we selected the well-known and validated endometrial adenocarcinoma model cell line Ishikawa H.13 Cells were authenticated with CODIS VNTR markers by Bio-Synthesis (Lewisville, TX, USA) and comparison with published data.14 CODIS typing confirmed that our cells are from the 3-H-4 subline established in 1993 and distributed between 1993 and 1996.15,16 In addition, these cells are ER+, PR+, contain a TP53 mutant of unknown significance (M247V, rs483352695), a terminal PTEN mutant (E91fs ter), a nonactivating POLE mutant (P102S), and a PIK3R1 mutant (L570P) also of uncertain significance. This combination of molecular features as well as the endometrioid histology of mouse explants makes these cells a prime model of a Type I endometrial cancer.13 However, their placement within the new four class system17,18 is less certain as they display features of both Class 2 and 3 tumors. Cells were grown in DMEM media supplemented with 10% FBS and 1% antibiotic (Sigma, St Louis, MO, USA; Pen-Strep). All cell culture experiments were carried out in triplicate. Whole-cell RNA purifications and quality control (QC) were performed as previously stated.
miRNA expression
miRNA-specific expression assays were carried out on fixed mass RNA inputs of 250 ng. Total cellular RNAs were reverse transcribed using miRNA-specific RT primers (Thermo Fisher) in the presence of MultiScribe Reverse Transcriptase (Thermo Fisher). Resulting cDNAs were then amplified in miRNA-specific TaqMan fluorescence assays (Thermo Fisher). For miRNA assays, the standard, well-validated endogenous RNA control RNU48 (Thermo Fisher) was used for normalizing. miRNA expression (Ct) was normalized (ΔCt) and tumor miRNA expression compared with benign tissue expression via the standard ΔΔCt method19,20 where ΔΔCt=ΔCttumor–ΔCtcontrol and fold change is 2−ΔΔCt. Statistical significance was assessed by a two-tailed t-test with unequal variance.21 A P-value<0.05 was taken as statistically significant.
miRNA expression correlations were carried out pairwise across all five unique members of the miR-503 cluster on all 24 tissues in the sample using Pearson product–moment correlation. Normalized expression values (ΔCt) were the input data. Statistical significance was assessed using a standard look-up table with n–2 degrees of freedom. A P-value<0.05 was taken as statistically significant.
Target validation
Experimentally validated targets were culled from the extant literature as well as from the miRTarBase database, Version 7.0 (http://miRTarBase.mbc.nctu.edu.tw/).22 Only those targets identified in miRTarBase by reporter assay, Western blot, quantitative PCR (qPCR), or any combination of these methods were accepted by us as experimentally validated.
Promoter de-methylation
In order to assess the methylation status of the miR-503 cluster promoter, Ishikawa H cells were treated with the cytidine analog, 5-aza-2′ deoxycytidine (Sigma, Decitabine). Cells were grown in optimum media for 24 hours and then treated for 72 hours with 1 μM Decitabine in dimethyl sulfoxide (DMSO). Decitabine-containing media was replaced every 24 hours and cells were harvested on day 5 for RNA purification. Control cells were treated in parallel with vehicle (DMSO) only. The entire process was carried out in triplicate.
Total cellular RNA was purified and QC validated as previously. MiR-503 cluster member expression levels between treated and un-treated cells were assessed via miR-specific TaqMan assays as previously. Again, differences in expression between Decitabine-treated and un-treated cells were evaluated via the ΔΔCt method described previously with a P-value<0.05 taken as statistically significant.
Results
Coordinated under-expression of the miR-503 cluster in EEA
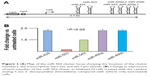
Expression of miR-503 cluster members in endometrioid adenocarcinoma relative to benign endometrium is presented in Figure 1A. As can be seen, each cluster member is significantly under-expressed in the tumors compared with benign endometrium. The magnitude of under-expression ranges from 5-fold in miR-503 to nearly 25-fold in miR-424. Using normalized expression values (ΔCt) for each miRNA in all 24 individuals in the panel, including the benign tissues, pairwise correlation among all five members of the cluster reveals a highly significant pattern of coordinated expression across the entire cluster. As can be seen, correlation coefficients range from 0.76 to 0.94 m and all are statistically significant at P<0.001 (Figure 1B).
Coordinate expression is confirmed in The Cancer Genome Atlas (TCGA) where expression of the five cluster members is also uniformly significant at P<0.001 with correlation coefficients ranging from 0.56 to 0.88. Moreover, comparing the pairwise correlation coefficients between our sample and TCGA is also statistically significant (r=0.62, P<0.05, df=8). Thus, the pattern of expression is the same in the two analyses.
Catalog of experimentally validated miR-503 cluster targets
Using the miRTarBase database (Version 7.0) and the extant literature, a total of 55 unique experimentally validated targets were identified for members of the miR-503 cluster (Table 2). Among these are numerous genes commonly regarded as oncogenes, such as FGFR1, MYB, BCL2, PI3K, and MYCN. In addition, genes involved in DNA repair, such as CHK1 and WEE1; cell cycle, including several cyclins, RUNX2, and CDC25A; anti-apoptosis genes IGF1R and BIRC5 (survivin); and chemotherapy response inhibitors, such as TRAF5 (cisplatin), ZNF217 (paclitaxel), and ERBB3 (gefitinib) are also present. Overall, the targets so far validated for miR-503 cluster members represent a rich array of genes whose dysregulation in cancer would be fortuitous for carcinogenesis, recurrence, and metastasis.
  | Table 2 Experimentally validated targets of members of the miR-503 cluster |
Promoter de-methylation
Changes in expression of miR-503 cluster members as a result of treating Ishikawa cells with Decitabine are shown in Figure 2. Though the fold changes are modest, three of the five cluster members did reach statistical significance. Moreover, the pattern of alteration of expression due to de-methylation in vitro is exactly the inverse of the expression pattern of the cluster members seen in Figure 1 from the primary tumor tissues.
Discussion
The miR-503 miRNA cluster, located at Xq23.1 between hypoxanthine-guanine phosphoribosyltransferase and PLAC1, consists of six miRNAs (miR-424, miR-503, miR-542, miR-450a-1, miR-450a-2, and miR-450b). We have provided evidence that this cluster is transcribed as a polycistron and that the entire cluster is significantly down-regulated in endometrial adenocarcinoma. Considering the cluster as a whole and assembling experimentally validated targets reveals a wide range of loci involved in carcinogenesis and the consequences of carcinogenesis, which includes DNA repair, DNA damage response, cell cycle maintenance, and chemotherapy response. Several studies of individual members of the cluster reported elsewhere have linked down-regulation to carcinogenesis and poor prognosis. Suppression of miR-503 expression in cervical cancer has been linked to significantly reduced progression-free and overall survival in a case–control study.23 An examination of endometrial cancer progression from normal tissue to hyperplasia and finally, to endometrioid carcinomas showed consistently decreasing levels of miR-503 expression.24 Moreover, in that same study, overall survival among the cancer patients was positively correlated with miR-503 expression levels.24 Similarly, miR-424 expression has been shown to be suppressed in endometrial cancers and that de-repression of miR-424 expression inhibits the growth of endometrial cancer cells in vitro.25 An important observation involving miR-542 is that, not only is it significantly down-regulated in endometrial cancer26 but also that down-regulation enhances morphological transformation of endometrial stromal cells,27 which might contribute to the rarer endometrial sarcomas. Finally, while there are fewer studies involving the miR-450 family, their down-regulation in carcinogenesis has also been confirmed albeit not in endometrial cancers until now.28,29
We have provided evidence that down-regulation of the cluster in endometrial adenocarcinomas is accomplished via methylation. We acknowledge that the in vitro reactivation of the miR-503 cluster by the cytidine analog Decitabine in Ishikawa cells is, at best, modest but it is consistent with similar suggestions offered in studies in other cancers.30 Even provisional acceptance of this mechanism opens the possibility of restoring the tumor suppression function of the miR-503 cluster through employing agents, such as cytidine analogs as adjuvants to chemotherapy.
The subtle in vitro data are limitation of this study as is the relatively small patient sample size that was determined by our self-imposed RNA quality threshold. In spite of this, however, further investigation of the miR-503 cluster is warranted by the data presented here. We sought additional support in TCGA and found that expression of four of the five miR-503 cluster members, specifically miR-424, miR-503, miR-450a, and miR-450b, are significantly inversely associated with endometrial cancer grade (OR=0.61, 0.81, 0.75, and 0.81, respectively) and that miR-450b is negatively associated with survival (OR=0.57). The significantly lower expression of miR-503 cluster members seen in our patients compared with controls is consistent with a prediction supported by TCGA data in which both survival and recurrence, implied from tumor grade, are linked with expression of the cluster members. Therefore, we believe that the miR-503 cluster as a whole functions as a tumor suppressor complex in human endometrial cancers. We also believe that this tumor suppressor function is likely disrupted in a coordinated manner in endometrial cancers by hyper-methylation. The list of experimentally validated miR-503 cluster targets reinforces this tumor suppressor role in which up-regulation of many of the targets by methylation inhibition of miRNA expression would be beneficial to establishing, maintaining, and expanding an endometrial cancer. This makes the entire miR-503 cluster an attractive subject for further research and an attractive therapeutic target in endometrial adenocarcinomas for adjuvant use of cytidine analogs along with conventional chemotherapies.
IRB approval
Tumor and control tissues were obtained under informed consent from patients in the University of Iowa Hospitals and Clinics Department of Obstetrics and Gynecology (IRB#200209010) and archived along with complete medical record information in the WHTR (IRB #200910784).
Acknowledgments
This work was supported, in part, by NIH R01CA99908 and R01CA184101 to Kimberly K. Leslie and the University of Iowa Carver College of Medicine Department of Obstetrics and Gynecology Research Development Fund. We are grateful to the Department of Obstetrics and Gynecology WHTR and Gynecologic Malignancy Repository (Dr Donna Santillan, Director) and for the continued invaluable assistance of the University of Iowa Institute of Human Genetics Genomics Facility, in particular, Mary Boes and Garry Hauser.
Author contributions
EJD and JGB conceived, designed, and managed the project under the mentorship of KKL. EC, AW, and MDM carried out all the nucleic acid purifications, cell culture, in vitro cell treatments, and miR-specific qPCR assays. EJD and JGB performed all data analyses. All authors contributed to writing the manuscript and approved the submission. All authors contributed toward data analysis, drafting and revising the paper and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
Ferlay J, Soerjomataram I, Bray FInternational Agency for Research on Cancer. GLOBOCAN 2012 v1.1, Cancer Incidence and Mortality Worldwide: IARC CancerBase No. 11 [Internet]. Lyon: France; 2014. Available from: http://globocan.iarc.fr. | ||
SEER Cancer Stat Facts: Endometrial Cancer. Bethesda, MD: National Cancer Institute. Available from: http://seer.cancer.gov/statfacts/html/corp.html. | ||
Sheikh MA, Althouse AD, Freese KE, et al. USA endometrial cancer projections to 2030: should we be concerned? Future Oncol. 2014;10(16):2561–2568. | ||
Cote ML, Ruterbusch JJ, Olson SH, Lu K, Ali-Fehmi R. The growing burden of endometrial cancer: a major racial disparity affecting Black women. Cancer Epidemiol Biomark Prev. 2015;24(9):1407–1415. | ||
Devor EJ, Leslie KK. The oncoplacental gene placenta-specific protein 1 is highly expressed in endometrial tumors and cell lines. Obstet Gynecol Int. 2013;2013:807849. | ||
Devor EJ, Reyes HD, Santillan DA, et al. Placenta-specific protein 1: a potential key to many oncofetal-placental OB/GYN research questions. Obstet Gynecol Int. 2014;2014:678984. | ||
Devor EJ, Reyes HD, Gonzalez-Bosquet J, et al. Placenta-specific protein 1 expression in human papillomavirus 16/18-positive cervical cancers is associated with tumor histology. Int J Gynecol Cancer. 2017;27(4):784–790. | ||
Devor EJ, Gonzalez-Bosquet J, Warrier A, et al. p53 mutation status is a primary determinant of placenta-specific protein 1 expression in serous ovarian cancers. Int J Oncol. 2017;50(5):1721–1728. | ||
Muys BR, Lorenzi JC, Zanette DL, et al. Placenta-enriched LincRNAs MIR503HG and LINC00629 decrease migration and invasion potential of JEG-3 cell line. PLoS One. 2016;11(3):e0151560. | ||
Mogilyansky E, Rigoutsos I. The miR-17/92 cluster: a comprehensive update on its genomics, genetics, functions and increasingly important and numerous roles in health and disease. Cell Death Differ. 2013;20(12):1603–1614. | ||
Santillan MK, Leslie KK, Hamilton WS, et al. “Collection of a lifetime: a practical approach to developing a longitudinal collection of women’s healthcare biological samples”. Eur J Obstet Gynecol Reprod Biol. 2014;179:94–99. | ||
Schroeder A, Mueller O, Stocker S, et al. The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol Biol. 2006;7:3. | ||
Albitar L, Pickett G, Morgan M, Davies S, Leslie KK. Models representing type I and type II human endometrial cancers: Ishikawa H and Hec50co cells. Gynecol Oncol. 2007;106(1):52–64. | ||
Korch C, Spillman MA, Jackson TA, et al. DNA profiling analysis of endometrial and ovarian cell lines reveals misidentification, redundancy and contamination. Gynecol Oncol. 2012;127(1):241–248. | ||
Nishida M, Kasahara K, Oki A, Satoh T, Arai Y, Kubo T. Establishment of eighteen clones of Ishikawa cells. Hum Cell. 1996;9(2):109–116. | ||
Nishida M. The Ishikawa cells from birth to the present. Hum Cell. 2002;15(3):104–117. | ||
Kandoth C, Schultz N. Cancer Genome Atlas Research Network, et al. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497(7447):67–73. | ||
Murali R, Soslow RA. Weigelt B: classification of endometrial cancer: more than two types. Lancet Oncol. 2014;15(7):e268–e278. | ||
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402–408. | ||
Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3(6):1101–1108. | ||
Snedecor GW. Cochran WG: Statistical Methods. 8th ed. Ames, IA: Iowa State University Press; 1989. | ||
Chou CH, Chang NW, Shrestha S, et al. miRTarBase 2016: updates to the experimentally validated miRNA-target interactions database. Nucleic Acids Res. 2016;44(D1):D239–D247. | ||
Yin ZL, Wang YL, Ge SF, et al. Reduced expression of miR-503 is associated with poor prognosis in cervical cancer. Eur Rev Med Pharmacol Sci. 2015;19(21):4081–4085. | ||
Xu YY, Wu HJ, Ma HD, Xu LP, Huo Y, Yin LR. MicroRNA-503 suppresses proliferation and cell-cycle progression of endometrioid endometrial cancer by negatively regulating cyclin D1. FEBS J. 2013;280(16):3768–3779. | ||
Li Q, Qiu XM, Li QH, et al. MicroRNA-424 may function as a tumor suppressor in endometrial carcinoma cells by targeting E2F7. Oncol Rep. 2015;33(5):2354–2360. | ||
Jurcevic S, Olsson B, Klinga-Levan K. MicroRNA expression in human endometrial adenocarcinoma. Cancer Cell Int. 2014;14(1):88. | ||
Tochigi H, Kajihara T, Mizuno Y, et al. Loss of miR-542-3p enhances IGFBP-1 expression in decidualizing human endometrial stromal cells. Sci Rep. 2017;7:40001. | ||
Zhao Z, Li R, Sha S, Wang Q, Mao W, Liu T. Targeting HER3 with miR-450b-3p suppresses breast cancer cells proliferation. Cancer Biol Ther. 2014;15(10):1404–1412. | ||
Liu F, Yu X, Huang H, et al. Upregulation of microRNA-450 inhibits the progression of lung cancer in vitro and in vivo by targeting interferon regulatory factor 2. Int J Mol Med. 2016;38(1):283–290. | ||
Jin C, Li M, Ouyang Y, Tan Z, Jiang Y. MiR-424 functions as a tumor suppressor in glioma cells and is down-regulated by DNA methylation. J Neurooncol. 2017;133(2):247–255. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.