Back to Journals » OncoTargets and Therapy » Volume 12
The LncRNA XIRP2-AS1 predicts favorable prognosis in colon cancer
Authors Zhou F, Shen F , Zheng Z , Ruan J
Received 11 May 2019
Accepted for publication 26 June 2019
Published 17 July 2019 Volume 2019:12 Pages 5767—5778
DOI https://doi.org/10.2147/OTT.S215419
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Takuya Aoki
Fengru Zhou,1 Feng Shen,1 Zhuojun Zheng,2 Jincheng Ruan1
1Department of Anus and Intestine Surgery, The Tongde Hospital of Zhejiang Province, Hangzhou, Zhejiang Province, People’s Republic of China; 2Department of Hematology, The Third Affiliated Hospital of Soochow University, Changzhou, Jiangsu Province, People’s Republic of China
Background: Colorectal cancer is a heterogeneous disease with complex genetic and epigenetic changes. LncRNA has recently been regarded as the biomarker in cancers. Novel biomarkers in colon cancer need to be identified.
Purpose: The objective of this study was to identify the differentially expressed lncRNAs between colon cancer tissue and adjacent tissue, as well as to explore its biological functions.
Patients and methods: There were 130 colon cancer patients included in this study. Of them, 6 colon cancer samples and 3 normal samples were selected for microarray profiling. Another 121 colon cancer samples with complete clinical information were used for immunohistochemical assay and survival analysis. Microarray analysis was performed to determine the differentially expressed lncRNAs between colon cancer tissue and adjacent tissue. Gain-of-function experiments was conducted in vitro and in vivo. In situ hybridization and survival analysis were applied to determine the prognostic impact on survival.
Results: LncRNA XIRP2-AS1 was significantly less expressed in colon cancer tissue. XIRP2-AS1 was remarkably downregulated in colon cancer tissues and cell lines. Functionally, XIRP2-AS1 could inhibit the proliferation and invasion ability of colon cancer cells in vitro and in vivo. Clinical sample analysis showed that XIRP2-AS1 had a favorable impact on the overall survival and progression free survival of patients with colon cancer. miR-182 was validated as the target of XIRP2-AS1 according to luciferase reporter assays, RNA immunoprecipitation and RNA pull down.
Conclusions: Our results suggested that XIRP2-AS1 may act as a favorable biomarker for patients with colon cancer.
Keywords: colon cancer, XIRP2-AS1, proliferation, invasion, survival
Background
Colorectal cancer is a heterogeneous disease with complex genetic and epigenetic changes.1 The epidemiological spectrum of colon cancer in the Chinese population is as follows:2,3 for urban men the morbidity rate is 9.74–16.71/100,000, and the mortality rate is 4.13–9.51/100,000; for urban women the morbidity rate is 8.49–13.99/100,000, and the mortality rate is 3.16–6.89/100,000; for rural men the morbidity rate is 3.86–12.66/100,000, and the mortality rate is 2.04–8.21/100,000; and for rural women the morbidity rate is 4.34–9.98/100,000, and the mortality rate is 0.95–4.68/100,000. The main clinical symptoms of colon cancer are changes in bowel habits, hemafecia, abdominal pain, intestinal obstruction and anemia. Distant metastasis occurs predominantly to the liver and lung, while right colon cancer is more prone to peritoneal and lymph node metastasis.
LncRNAs are low-fidelity transcriptional products of RNA polymerase II without a biological function,4 and they have been referred to by some scholars as “noise” in the gene transcription process. Possible sources of LncRNAs include the disrupted structure of a protein-coding gene, recombination of chromatin, reversal of noncoding genes during replication, tandem duplication of ncRNA and transposable elements inserted into a coding gene.5,6 LncRNAs can be classified into the following five subtypes: sense type, antisense type, bidirectional type, endogenous type and intergenomic type, whose main function is to regulate gene transcription. The mechanism of action of LncRNAs includes7–9 1) regulation of transcriptional interference or chromosome remodeling, 2) interaction with chromatin-modifying enzymes to modify histones and 3) interaction with transcriptional regulatory elements. In addition, LncRNAs are involved in posttranscriptional regulation via a mechanism of action that includes adsorption of small RNA precursors or endogenous competitive RNAs to small RNAs, affecting the variable cleavage of RNA. They also participate in regulation of protein translation and posttranslational modification and localization. In the present study, gene chip technology was used to screen differentially expressed LncRNAs in colon cancer and para-carcinoma tissues and to identify their functions in vivo and in vitro. Our ultimate goal was to determine their clinical significance, provide new insights into the pathogenesis of colon cancer and guide clinical treatment.
Methods
Tissue samples
Colon cancer tissue and adjacent normal colon tissue that resected in surgical procedures were collected from The Third Affiliated Hospital of Soochow University from January 2014 to April 2018. Of them, six colon cancer samples and three normal samples were selected for microarray profiling. Another 121 colon cancer samples with complete clinical information were used for immunohistochemical assay and survival analysis. Liquid nitrogen was used for tissue store at - 80°C. Each participant provided written informed consent. The use of human clinical tissues was approved by the Institutional Human Experiment and Ethics Committee of The Third Affiliated Hospital of Soochow University and The TONGDE Hospital of Zhejiang Province. All experiments were conducted under the rule of the Declaration of Helsinki.
Microarray profiling
LncRNAs with different expression in colon cancer tissues and adjacent tissue were screened by the LncRNA microarray expression profiling according to the criteria of fold change >2 and Padj <0.01. Manufacturer’s standard protocols were strictly followed. Briefly, after synthesis, label and purification of cDNA, cRNA which is labeled Cyanine-3-CTP was used to hybridize the lncRNA microarray chip. Samples were washed and then analyzed on the microarray. The results were estimated and clustered by R project.
Cell line culture
Colon cancer cell lines (SW620 and HCT116) and human colon epithelial cells (NCM460) were obtained from American Type Culture Collection (ATCC, Manassas, VA, USA). Modified RPMI-1640 medium which is supplemented with 10% FBS including 100 μg/L penicillin and 100 μg/L streptomycin was applied to maintain all cells at 5% CO2 and 37°C.
Real-time quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted by TRIzol reagent (Invitrogen, Carlsbad, CA, USA). Primers were amplified by using SYBR Green Mix (Promega). Sequences of the XIRP2-AS1 primer are listed as follows: Forward primer: GACAAGCCACAGGCAAACATT; Reverse primer: GGACTTCCATGGGACTGTGT. The primer was synthesized by Shanghai Tingzhou Biological Engineering Co., Ltd. All results were presented as 2−ΔΔCt. TaqMan MicroRNA Assays Kit (Applied Biosystems, Carlsbad, CA, USA) was applied to measure the expression level of miR-182. Endogenous control for XIRP2-AS1 and miR-182: GAPDH and U6. Each experiment repeats three times.
Lentivirus production and cell transfection
The pBLLV-CMV-IRES-ZsGreen XIRP2-AS1 cDNA lentiviral plasmid for overexpression transfection was obtained from Genelily BioTech Co., Ltd (Shanghai, People's Republic of China). Puromycin (2 μg/mL) was used to select stably transfected cells for 2 weeks. Transfection efficiency was verified by RT-qPCR. Lipofectamine 3000 (Invitrogen) was applied for plasmid transfection.
Cell counting kit-8 (CCK8) assay
Cells (2×104 cells/mL) were incubated at 5% CO2 and 37°C on 96-well plates (100 μL/well) for 24 hrs. CCK8 solution was then added to each well after 5 days of cell culture. Cell viability was estimated by a microplate reader which measure the absorbance values at a wavelength of 450 nm.
MTT assay
Transfected colon cancer cells were incubated for 24, 48, 72 and 96 hrs on a 96-well plate (1×104 cells/well). Each well was then added with MTT (10 μL of 5 mg/mL) and incubated for 4 hrs. After removal of supernatants, dimethyl sulfoxide (DMSO, Thermo Fisher Scientific, Waltham, MA, USA) (100 μL per well) was finally added. The results were estimated by a microplate reader which measure the absorbance values at a wavelength of 490 nm.
Flow cytometry
The flow cytometry assays were performed as previously described.10 Annexin V-FITC early apoptosis kit was applied for cell apoptosis analysis. Colon cancer cells with overexpressed XIRP2-AS1 and negative control cells were analyzed on the flow cytometer (FACScan; BD Biosciences). The results were calculated by CellQuest software (BD Biosciences).
Transwell assay
Cell migration and invasion ability were evaluated by Transwell chambers (8-μm pore size; Corning Costar, Cambridge, MA, USA). Cells were seeded into the upper chamber. Lower chamber was supplemented by 20% serum which was considered as a chemoattractant. The filters were fixed and stained with methanol and 0.1% crystal violet later after 48-hr incubation. Gently abrasion could be observed in the upper faces of the filters. In the lower faces of filters, cells were counted and photographed under the microscope. Each experiment repeats three times.
Wound-healing assay
Equal numbers of SW620 and HCT116 cells with transfection of XIRP2-AS1 overexpressed plasmid and NC plasmid were plated into 6-well plates. Then, the cell monolayers were wounded with a pipette tip to draw a gap on the plates. Colon cancer cells that migrated into the cleared section were observed under microscope at the specific time points.
In situ hybridization (ISH)
The ISH assays were performed as previously described.11 Digoxigenin antibody (Roche, 11,093,274, 1:1000) was used to label a locked nucleic acid probe which contains complementary sequences to a section of XIRP2-AS1 (custom LNA detection probe, Exiqon). Then, the probe was synthesized. There were two independent pathologists evaluating the intensity and the extent of staining. The pathologists were blinded to the experiment.
Luciferase reporter assays
XIRP2-AS1 cDNA with miR-182 predictive binding site and point mutations of the miR-182 seed region binding site were cloned into pmirGLO Dual-Luciferase miRNA Target Expression Vector (Promega) to form pmirGLO-XIRP2-AS1-wt reporter vector and pmirGLO-XIRP2-AS1-Mut reporter vector. These vectors were then co-transfected with miR-182 and miR-NC into HEK-293FT cells by using Lipofectamine 3000 (Invitrogen). After 48 hrs of transfection, the Dual-Luciferase Reporter Assay System (Promega) was performed. The manufacturer’s instructions were strictly followed.
RNA immunoprecipitation (RIP)
Manufacturer’s protocol of the EZMagna RIP Kit (Millipore) was strictly followed. Colon cancer cells were lysed by complete RIP lysis buffer. The co-incubations of cell extract and anti-argonaute 2 or control anti-IgG antibodies conjugated with magnetic beads were conducted at 4°C for 6 h. After removal of proteins of the beads, RT-qPCR was performed for purified RNA measurement.
RNA pull-down assay
Briefly, the 3ʹend biotinylated miR-182 or miR-182-mut was transfected into SW620 cells for 24 hrs at 20 nmol/L. Streptavidin-coated magnetic beads (Ambion, Life Technologies) were used for cell incubation. Pull-down assay was performed in biotin-coupled RNA complex. The abundance of XIRP2-AS1 in bound fractions was calculated according to the results of RT-qPCR.
Animal studies
BALB/c-nude mice (4–5 weeks of age) were purchased from Shanghai SLAC Laboratory Animal Co., Ltd, People's Republic of China. The animal experiments were approved by the Institutional Animal Care and Use Committee of both institutions. Also, NIH principles of laboratory animal care were followed for the welfare of the animals. The growth of tumor was monitored and recorded every 5 days. The mice were sacrificed 1 month later. Tumors were removed from the body and weighed.
In abdominal metastasis model, the mice were anesthetized. Then, operation was performed by a right lateral flank incision. About 100 μL SW620-Luc-vector and SW620-Luc-XIRP2-AS1 cells (2×107) contained Hank’s balanced salt solution were injected into the right abdominal cavity. Bioluminescence images were collected by the Interactive Video Information Systembioluminescence imaging system (Caliper Life Sciences) 4 weeks later.
Statistical analysis
Statistical analysis was calculated by SPSS 22.0 statistical software and GraphPad Prism 7.0. Numerical data are expressed as mean±SD. The Student’s t-test and one-way ANOVA were used to compare differences among groups. Overall survival (OS) was calculated from the time at diagnosis to the time of death of any cause. Progression-free survival (PFS) was calculated from the time that patients receive treatment to the time of disease progression. Kaplan–Meier (Log-rank test) and Cox’s regression model were used for univariate and multivariate survival analysis, respectively. P<0.05 was considered as statistically significant.
Results
LncRNA XIRP2-AS1 was significantly down-regulated in colon cancer tissues
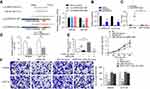
A total of 60 differentially expressed lncRNAs (Log2 [fold change] >1, Padj <0.01) were screened as shown in Figure 1A and B, of which, XIRP2-AS1 significantly decreased in colon cancer tissues (Figure 1A). LncRNA XIRP2-AS1, an antisense transcript of XIRP2 gene, is localized in Chromosome 2q24.3 with 2 exons. At present, its biological effects on tumors remain unclear.
The upregulation of XIRP2-AS1 expression suppressed the viability and invasion of colon cancer cells
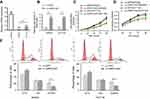
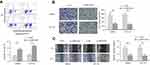
The results from RT-qPCR showed that the XIRP2-AS1 expression in colon cancer cells was significantly downregulated compared to that in the normal human colon epithelial cells (NCM460) (Figure 2A; P<0.05). After transfecting the overexpressed XIRP2-AS1 plasmid, the expressions of XIRP2-AS1 in SW620 and HCT116 cells were obviously upregulated (Figure 2B; P<0.05). Further CCK8 and MTT assays showed that the cell proliferation and viability in the XIRP2-AS1-overexpressed group were lower than those in the empty vector group (Figure 2C and D; P<0.05). The overexpression of XIRP2-AS1 markedly inhibited cell cycle (Figure 2E; P<0.05) but promoted cell apoptosis (Figure 3A; P<0.05). Moreover, the overexpression of XIRP2-AS1 also significantly weakened the invasive capacity of cancer cells according to the Transwell and wound-healing assays (Figure 3B and C ; P<0.05).
The in vivo verification of effects of XIRP2-AS1
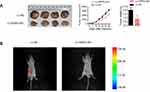
To further verify the effects of XIRP2-AS1 in vivo, BALB/c-nude mice were subcutaneously injected with the XIRP2-AS1-overexpressed SW620 cells and corresponding negative control cells, respectively. As shown in Figure 4A, the overexpression of XIRP2-AS1 significantly decreased the volume and weight of tumors (P<0.05). To validate the effects of XIRP2-AS1 on the metastasis of cancer cells, we established an abdominal metastasis model. In brief, approximately 2×107 XIRP2-AS1-overexpressed and control SW620 cells were injected into the right abdominal cavity of BALB/c-nude mice. One month later, the bioluminescence imaging was used to evaluate the invasion of cancer cells in vivo. Our results demonstrated that the XIRP2-AS1 overexpression significantly inhibited the metastasis of cancer cells (Figure 4B; P<0.05).
XIRP2-AS1 had a favorable effect on the survival of patients with colon cancer
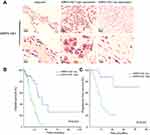
A total of 121 colon cancer tissues with complete clinical information were collected for the survival analysis. The results from ISH manifested that the XIRP2-AS1 expression was lower in tumor tissues than that in adjacent tissues (Figure 5A). Subsequently, according to the staining intensity of XIRP2-AS1 in ISH, the intensity scored 0–2 was classified as low expression and the intensity scored 3–4 was classified as high expression. The baseline clinical characteristics of all patients are listed in Table 1. The median OSs were 40 and 16 months in the high- and low-expressed XIRP2-AS1 groups, respectively (Figure 5B). The median PFS in the high-expressed XIRP2-AS1 group was not reached and was 11 months in the low-expressed XIRP2-AS1 group (Figure 5C). These abovementioned differences were statistically significant according to the univariate Log-rank test (P<0.001). In addition, further multivariate analysis confirmed that expression of XIRP2-AS1 was an independent risk factor for the OS of the patients with colon cancer (HR: 3.254, 95% CI: 1.647–6.426, P<0.001) (Table 2). Meanwhile, the low expression of XIRP2-AS1 was also identified as an independent risk factor for PFS according to the multivariate analysis (HR: 8.790, 95% CI: 2.644–29.224, P<0.001) (Table 3). Taken together, our results indicated that the XIRP2-AS1 had favorable effects on the OS and PFS of the patients with colon cancer.
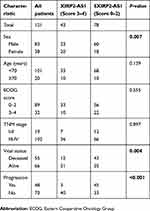 |
Table 1 Clinical characteristics of patients with colon cancer |
 |
Table 2 Univariate and multivariate analysis of colon cancer patients on overall survival |
 |
Table 3 Univariate and multivariate analysis of colon cancer patients on progress-free survival |
XIRP2-AS1 targeted and regulated miR-182
In our study, the targeted miRNA that reverses complemented recognition sequence of XIRP2-AS1 was screened by bioinformatics (miRcode http://www.mircode.org/), and then the miR-182 was identified. Next, luciferase reporter assay was performed, and the pmirGLO-XIRP2-AS1-wt reporter vector with miR-182 and miR-NC was co-transfected into the SW620 cells. The results showed that compared to miR-NC group, the luciferase activity in miR-182 group significantly decreased. Whereas after transfecting with pmirGLO-XIRP2-AS1-mut reporter vector, there was no significant change in the luciferase activity in miR-182 group (Figure 6A). For further verification, anti-AGO2 was used for the RIP assay of SW620 extract. The results demonstrated that compared to anti-IgG immunoprecipitates, XIRP2-AS1 and miR-182 were enriched in AGO2 as a matter of priority (Figure 6B). RNA pull-down assay indicated that XIRP2-AS1 was more enriched in the miR-182-wt than that in the miR-182-mut that consists broken binding site of XIRP2-AS1 (Figure 6C). RT-qPCR showed that the overexpression of XIRP2-AS1 reduced the expression of miR-182 in colon cancer cells (Figure 6D). Moreover, the miR-182 mimic-induced upregulations of the proliferation (Figure 6E) and invasion (Figure 6F) of cancer cells could be largely reversed by the overexpression of XIRP2-AS1 in the rescue experiments.
Discussion
Previous research has demonstrated that a variety of LncRNAs are involved in the development of colon cancer. The first exon of the H19 gene produces a highly conserved miRNA 675, and high expression of miRNA675 promotes the growth of human colorectal cancer cells.12 P53 is a recognized tumor suppressor gene, and miRNA675 inhibits P53 and P53-dependent protein expression.13 In addition, miRNA675 can affect a range of cellular transcriptional processes, such as direct downregulation of Igf1r,14 Smad1, Smad5, cdc6,15 cell adhesion factor 11 (Cadherin-11),16 Cadherin-13,17 Rb,18 transforming growth factor beta-inducible protein,19 calneuron 1 (CALN1)20 and microphthalmia-associated transcription factor21 H19 is overexpressed in colorectal cancer tissues and cells. In a mouse model of colon cancer, Yoshimizu et al found that downregulating H19 expression significantly increased the number of colon polyps. These results suggest that H19 may have a dual role in oncogenes and tumor suppressor genes in colorectal cancer. The expression level of CCAT1 in colorectal cancer tissues is more than 200 times that in normal intestinal mucosa tissues, and the specificity and sensitivity of colorectal cancer tissues to CCAT1 expression are higher than controls.22 MaCleland et al23 reported that the expression of CCAT1 is related to the degree of differentiation and the clinical stage of colorectal cancer, suggesting that CCAT1 can be used as a biological indicator to evaluate the prognosis of patients. CCAT1-l is a subtype of CCAT1 that is found mainly in the nucleus and spatially close to the enhancer. It is a colorectal cancer-specific LncRNA with a full length of 5200 nt that is transcribed 515 kb upstream of the MYC gene. Xiang et al24 found that knockout of CCAT1-L in colorectal cancer cell lines significantly reduced MYC gene transcription, suggesting that CCAT1-L can play a role in the progression of colorectal cancer by increasing the expression of MYC, which may play a reinforcing role in MYC gene expression. Lin et al25 found via ISH that MALAT-1 is expressed in more than 50% of colorectal cancer, breast cancer and pancreatic cancer tissues, while expression levels are lower in normal control tissues. Studies have shown that the expression level of MALAT-1 is positively correlated with the degree of colorectal cancer metastasis.18 Ji et al reported that26 resveratrol treatment of MALAT-1 can reduce nuclear localization of β-catenin to attenuate cytokines and β-catenin signaling, thereby inhibiting the invasion and metastasis of colon cancer cells. Therefore, MALAT-1 may be able to enhance the invasion and metastasis of colorectal cancer cells. Kogo et al27 found that HOTAIR increases expression in stage IV colorectal cancer and is associated with poor prognosis and liver metastasis, indicating that HOTAIR can be used to evaluate the prognosis of patients with colorectal cancer. Fer1L4 was first discovered in gastric cancer tissues,27 and its expression in gastric cancer is downregulated compared with adjacent normal tissues. In recent years, it has been shown that the expression of Fer1L4 is also downregulated in colorectal cancer tissues, and the expression of miR-106a-5p is negatively correlated with the expression of Fer1L4 in colorectal cancer tissues and cells. Yue et al28 showed that Fer1L4 inhibits the development of colon cancer by downregulating the expression of miR-106a-5p, and miR-106a-5p may act as a tumor suppressor gene in colon cancer.
In the present study, it was found that the expression of LncRNA XIRP2-AS1 in colon cancer tissues was lower than that in adjacent tissues, which was confirmed by the in vitro experiments. In the SW620 and HCT116 cell lines, XIRP2-AS1 expression significantly increased, and the high-expressed XIRP2-AS1 significantly reduced cell proliferation, blocked cell cycle but increased cell apoptosis. It was also found that the exogenously increased XIRP2-AS1 inhibited the invasion and migration of colon cancer cells. In the in vivo experiments, tumor imaging systems showed that the overexpressing of XIRP2-AS1 significantly decreased the invasion of colon cancer cells. Moreover, XIRP2-AS1 levels were also significantly lower than those in control mice that were injected with conventional colon cancer cells. To further study the effect of XIRP2-AS1 on the prognosis of clinical patients, 121 tumor tissues were collected for the retrospective analysis of colon cancer patients diagnosed in our hospital. All patients received routine treatment and regular follow-up according to the in situ hybridization score (0–4 points). The patients with a score of 0–2 points were classified into the low-expressed XIRP2-AS1 group, while those with 3–4 points were classified into the high-expressed XIRP2-AS1 group. According to multivariate analysis, the expression of XIRP2-AS1 was an independent risk factor for OS and PFS, and it was not affected by any other factors. The results of median OS and PFS suggested that XIRP2-AS1 had a positive effect on the clinical treatment of colon cancer. To further explore the mechanism of XIRP2-AS1 regulation, miR-182 was screened and identified as a target via luciferase reporter assay, RIP and RNA pull-down assays. The miR-182, a key factor affecting XIRP2-AS1, could negatively regulate the activity of miR-182, which was coincided with the previous studies29–32 on colorectal cancers. Considering that the expression of XIRP2-AS1 in normal colon cancer cells was relatively low, we did not perform loss-of-function assay in this study. And the further downregulating of XIRP2-AS1 might not lead to significant changes in cell proliferation, cell apoptosis and cell invasion. Instead, we performed rescue experiment, and the results showed that cell proliferation and invasion did not significantly increase after transfecting with miR-182 mimic alone. Meanwhile, the downregulating of XIRP2-AS1 in colon cancer cells might not largely affect the biological function of cells. The classic signal pathways of XIRP2-AS1 regulation as well as the downstream target genes of miR-182 also need to be further explored, which could be the main future direction of our research.
In conclusion, the in vivo and in vitro results demonstrated that the expression of LncRNA XIRP2-AS1 was downregulated in colon cancer, and it involved the inhibitions of the proliferation and invasion of tumor cells. According to the clinical retrospective studies, LncRNA XIRP2-AS1 had clinical significance for the treatment of colon cancer. miR-182, a famous oncogene, was the target for XIRP2-AS1, and the XIRP2-AS1/miR-182 pathway might be a novel biomarker and potential therapeutic target for colon cancer. At present, there are few studies on LncRNA XIRP2-AS1, and its regulatory mechanism or signal transduction pathway remain unclear. Therefore, further research could be needed, and its prognostic significance should be further confirmed via large-scale clinical studies.
Availability of data and materials
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
Author contributions
All authors contributed to data analysis, drafting or revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Hinoue T, Weisenberger DJ, Lange CPE, et al. Genome-scale analysis of aberrant DNA methylation in colorectal cancer. Genome Res. 2012;22:271–282. doi:10.1101/gr.117523.110
2. Lu Y, Kweon -S-S, Tanikawa C, et al. Large-scale genome-wide association study of East Asians identifies loci associated with risk for colorectal cancer. Gastroenterology. 2019;156:1455–1466. doi:10.1053/j.gastro.2018.11.066
3. Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi:10.3322/caac.21338
4. Spizzo R, Almeida MI, Colombatti A, Calin GA. Long non-coding RNAs and cancer: a new frontier of translational research? Oncogene. 2012;31:4577–4587. doi:10.1038/onc.2011.621
5. Kapranov P, Cheng J, Dike S, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi:10.1126/science.1138341
6. Liu C, Fu H, Liu X, et al. LINC00470 coordinates the epigenetic regulation of ELFN2 to distract GBM cell autophagy. Mol Ther. 2018;26:2267–2281. doi:10.1016/j.ymthe.2018.06.019
7. Zhang Z-K, Li J, Guan D, et al. Long noncoding RNA lncMUMA reverses established skeletal muscle atrophy following mechanical unloading. Mol Ther. 2018;26:2669–2680. doi:10.1016/j.ymthe.2018.09.014
8. Ma M, Zhang Y, Weng M, et al. lncRNA GCAWKR promotes gastric cancer development by scaffolding the chromatin modification factors WDR5 and KAT2A. Mol Ther. 2018;26:2658–2668. doi:10.1016/j.ymthe.2018.09.002
9. Zhu KP, Ma XL, Zhang CL. LncRNA ODRUL contributes to osteosarcoma progression through the miR-3182/MMP2 Axis. Mol Ther. 2017;25:2383–2393. doi:10.1016/j.ymthe.2017.06.027
10. Koblizek M, Lebedeva A, Fiser K. flowIO: flow cytometry standard conformance testing, editing, and export tool. Cytometry A. 2018. doi:10.1002/cyto.a.23563
11. Dong J, Wang R, Ren G, et al. HMGA2-FOXL2 axis regulates metastases and epithelial-to-mesenchymal transition of chemoresistant gastric cancer. Clin Cancer Res. 2017;23:3461–3473. doi:10.1158/1078-0432.CCR-16-2180
12. Raveh E, Matouk IJ, Gilon M, et al. The H19 Long non-coding RNA in cancer initiation, progression and metastasis - a proposed unifying theory. Mol Cancer. 2015;14:184. doi:10.1186/s12943-015-0458-2
13. Liu C, Chen Z, Fang J, Xu A, Zhang W, Wang Z. H19-derived miR-675 contributes to bladder cancer cell proliferation by regulating p53 activation. Tumour Biol. 2016;37:263–270. doi:10.1007/s13277-015-3779-2
14. Keniry A, Oxley D, Monnier P, et al. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nat Cell Biol. 2012;14:659–665. doi:10.1038/ncb2521
15. Dey BK, Pfeifer K, Dutta A. The H19 long noncoding RNA gives rise to microRNAs miR-675-3p and miR-675-5p to promote skeletal muscle differentiation and regeneration. Genes Dev. 2014;28:491–501. doi:10.1101/gad.234419.113
16. Kim NH, Choi SH, Lee TR, Lee CH, Lee AY. Cadherin 11, a miR-675 target, induces N-cadherin expression and epithelial-mesenchymal transition in melasma. J Invest Dermatol. 2014;134:2967–2976. doi:10.1038/jid.2014.257
17. Shi Y, Wang Y, Luan W, et al. Long non-coding RNA H19 promotes glioma cell invasion by deriving miR-675. PLoS One. 2014;9:e86295. doi:10.1371/journal.pone.0086295
18. Xu C, Yang M, Tian J, Wang X, Li Z. MALAT-1: a long non-coding RNA and its important 3ʹ end functional motif in colorectal cancer metastasis. Int J Oncol. 2011;39:169–175. doi:10.3892/ijo.2011.1007
19. Zhu M, Chen Q, Liu X, et al. lncRNA H19/miR-675 axis represses prostate cancer metastasis by targeting TGFBI. Febs J. 2014;281:3766–3775. doi:10.1111/febs.12902
20. Li H, Yu B, Li J, et al. Overexpression of lncRNA H19 enhances carcinogenesis and metastasis of gastric cancer. Oncotarget. 2014;5:2318–2329. doi:10.18632/oncotarget.1913
21. Kim NH, Choi S-H, Kim C-H, Lee CH, Lee TR, Lee A-Y. Reduced MiR-675 in exosome in H19 RNA-related melanogenesis via MITF as a direct target. J Invest Dermatol. 2014;134:1075–1082. doi:10.1038/jid.2013.478
22. Nissan, A., Stojadinovic A, Mitrani-Rosenbaum S et al. Colon cancer associated transcript-1: a novel RNA expressed in malignant and pre-malignant human tissues. Int J Cancer. 2012;130:1598–1606. doi:10.1002/ijc.26170
23. McCleland ML, Mesh K, Lorenzana E, et al. CCAT1 is an enhancer-templated RNA that predicts BET sensitivity in colorectal cancer. J Clin Invest. 2016;126:639–652. doi:10.1172/JCI83265
24. Xiang JF, Yin Q-F, Chen T, et al. Human colorectal cancer-specific CCAT1-L lncRNA regulates long-range chromatin interactions at the MYC locus. Cell Res. 2014;24:513–531. doi:10.1038/cr.2014.35
25. Lin R, Maeda S, Liu C, Karin M, Edgington TS. A large noncoding RNA is a marker for murine hepatocellular carcinomas and a spectrum of human carcinomas. Oncogene. 2007;26:851–858. doi:10.1038/sj.onc.1209846
26. Ji Q, Liu X, Fu X, et al. Resveratrol inhibits invasion and metastasis of colorectal cancer cells via MALAT1 mediated Wnt/beta-catenin signal pathway. PLoS One. 2013;8:e78700. doi:10.1371/journal.pone.0078700
27. Kogo R, Shimamura T, Mimori K, et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011;71:6320–6326. doi:10.1158/0008-5472.CAN-11-1021
28. Yue B, Sun B, Liu C, et al. Long non-coding RNA Fer-1-like protein 4 suppresses oncogenesis and exhibits prognostic value by associating with miR-106a-5p in colon cancer. Cancer Sci. 2015;106:1323–1332. doi:10.1111/cas.12759
29. Jin Y, Zhang ZL, Huang Y, Zhang KN, Xiong B. MiR-182-5p inhibited proliferation and metastasis of colorectal cancer by targeting MTDH. Eur Rev Med Pharmacol Sci. 2019;23:1494–1501. doi:10.26355/eurrev_201902_17107
30. Liu X, Xu T, Hu X, et al. Elevated circulating miR-182 acts as a diagnostic biomarker for early colorectal cancer. Cancer Manag Res. 2018;10:857–865. doi:10.2147/CMAR.S158016
31. Jia L, Luo S, Ren X, et al. miR-182 and miR-135b mediate the tumorigenesis and invasiveness of colorectal cancer cells via targeting ST6GALNAC2 and PI3K/AKT pathway. Dig Dis Sci. 2017;62:3447–3459. doi:10.1007/s10620-017-4755-z
32. Zhang Y, Wang X, Wang Z, Tang H, Fan H, Guo Q. miR-182 promotes cell growth and invasion by targeting forkhead box F2 transcription factor in colorectal cancer. Oncol Rep. 2015;33:2592–2598. doi:10.3892/or.2015.3833
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.