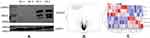Back to Journals » Breast Cancer: Targets and Therapy » Volume 14
The Involvement of Insulin-Like Growth Factor 2 Messenger Ribonucleic Acid-Binding Protein 2 in the Regulation of the Expression of Breast Cancer-Related Genes
Authors Gao C, Li L, Jin X, Song X, Li H, Xu X, Dong C , Ma B
Received 19 July 2022
Accepted for publication 22 September 2022
Published 10 October 2022 Volume 2022:14 Pages 311—322
DOI https://doi.org/10.2147/BCTT.S382566
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Pranela Rameshwar
Chao Gao,1 Li Li,2 Xixin Jin,1 Xinyu Song,1 Huiling Li,1 Xiaoli Xu,1 Chao Dong,1 Binlin Ma1
1State Key Laboratory of Pathogenes, Prevention and Treatment of High Incidence Diseases in Central Asia, Department of Breast and Thyroid Surgery, Affiliated Cancer Hospital of Xinjiang Medical University, Urumqi, People’s Republic of China; 2State Key Laboratory of Pathogenes, Prevention and Treatment of High Incidence Diseases in Central Asia, Department of Gynecology and surgery, Affiliated Cancer Hospital of Xinjiang Medical University, Urumqi, People’s Republic of China
Correspondence: Chao Dong; Binlin Ma, State Key Laboratory of Pathogenes, Prevention and Treatment of High Incidence Diseases in Central Asia, Department of Breast and Thyroid Surgery, Affiliated Cancer Hospital of Xinjiang Medical University, No. 789 of East Suzhou Street, Urumqi, 830000, People’s Republic of China, Tel +860991-7968088, Fax +860991-7968111, Email [email protected]; [email protected]
Aim: This study investigated the role and mechanism of insulin-like growth factor 2-IGF2BP2 in breast cancer.
Methods: IGF2BP2 is overexpressed in MDA-MB-231 human breast cancer cells. Thus, RNA sequencing was used to analyze the differentially expressed genes, Cell Counting Kit-8 was used to detect cell proliferation, and a Transwell assay was used to assess cell invasion. Following on from the RNA sequencing results, Interferon-induced protein with tetratricopeptide repeats 2 (IFIT2), chemokine C-C motif ligand 20 (CCL20), chemokine C-C motif ligand 5 (CCL5), and chemokine C-X-C motif ligand 10 (CXCL10) regulated by IGF2BP2 were subjected to real-time reverse transcriptase-polymerase chain reaction verification.
Results: After IGF2BP2 overexpression, 67 genes were up-regulated, and 87 genes were down-regulated. The gene with the most significant up-regulation was homeobox protein 1 (PROX1), and the gene with the most significant down-regulation was Acidic β-crystallin 4 (CRYBA4). The most enriched gene ontology (GO) terms of up-regulated differentially expressed genes are protein binding and cell membrane and of down-regulated differentially expressed genes they are ion binding, cytoplasm, and response to virus. Kyoto Encyclopedia of Genes and Genomes analysis showed that the up-regulated differential genes were mainly enriched in protein processing, the endoplasmic reticulum, and the regulation of actin cytoskeleton, while down-regulated differential genes were mainly enriched in rheumatoid arthritis, chemokine signaling pathways, toll-like receptor signaling pathways, tumor necrosis factor signaling pathways, cytokine-cytokine receptor interaction, and Notch signaling pathways. IGF2BP2 overexpression significantly promoted the proliferation and invasion of breast cancer cells (P < 0.01). Compared with the control group, the IGF2BP2 overexpression group had significantly increased expressions of IFIT2, CCL20, and CXCL10 (P < 0.05).
Conclusion: IGF2BP2 may promote the invasion and proliferation of human breast cancer cells by up-regulating breast cancer-related genes, such as IFIT2, CCL20, and CXCL10.
Keywords: breast cancer, RNA-binding protein, IGF2BP2, bioinformatics
Introduction
Breast cancer is one of the most common female malignant tumors in the world, which seriously affects the life and health of women. The latest statistics show that there are about 2.26 million new cases of breast cancer in the world in 2020, accounting for about 11. 5% of all new cancer cases in the world. 7%, with about 680,000 deaths, and breast cancer has replaced lung cancer as the most common malignant tumor in the world for the first time.1 However, at present, the early diagnosis of breast cancer is hampered by the lack of biomarkers with high sensitivity and specificity. Insulin-like growth factor 2-IGF2BP2 also known as p62/IMP2 is a breast cancer susceptibility protein,2 which is overexpressed in the tumor tissues of some breast cancer patients, and some patients have elevated IGF2BP2 autoantibody levels in their serum.3 In human breast cancer cell lines, overexpression of IGF2BP2 can increase cell migration and reduce cell adhesion to extracellular matrix proteins.4 As an RNA-binding protein, IGF2BP regulates RNA stability5 and mRNA localization6 and translation,7 and it can also regulate transcription by influencing transcription factors.8 The latest report shows that IGF2BP2 enhances the stability of B3GNT6mRNA through M6A methylation, thus promoting the progression of pancreatic cancer.9 However, the mechanism of IGF2BP2 as a susceptibility gene and a regulator of cancer cell migration in breast cancer is less studied.
This study explored the effects and underlying mechanisms of IGF2BP2 in breast cancer. IGF2BP2 is overexpressed in MDA-MB-231 human cells, so cell proliferation and invasion were evaluated. The down-stream genes of IGF2BP2 were analyzed using bioinformatics and verified using real-time reverse transcriptase-polymerase chain reaction (RT-qPCR). The findings provide experimental evidence for understanding the pathogenesis of breast cancer.
Methods
Cell Culture and Transfection
MDA-MB-231 cells were purchased from Procell Life Science & Technology Co. Ltd. They were cultured in Dulbecco’s Modified Eagle Medium (DMEM) medium, containing 10% fetal bovine serum and antibiotics, at 37℃, with 5% CO2. For transfection, the cells were divided into three groups: the control group, the IGF2BP2 overexpression (Ov) group, and the IGF2BP2 IGF2BP2 Ov negative control (NC) group. The cells were plated in 6-well plates until cell confluence reached 90%, after which cell transfection was performed using Lipofectamine 2000. At 48 h after transfection, the cells were collected for analysis.
RNA Sequencing
The total RNA was extracted from the cells with TRIZOL. The RNA-seq library was prepared with a VAHTS Stranded mRNA-seq Library Prep Kit using 1μg of total RNA. Polyadenylated mRNAs were converted into double-stranded cDNA using VAHTS mRNA trapping beads and fragmented mRNAs. After end repair and A tailing, the DNA was ligated to a diluted VAHTS RNA Adapter. The products of 300–500 bps were amplified, purified, quantified, and stored at −80℃ before sequencing. The second cDNA strand labeled with 2’- Deoxyuridine-5’- Triphosphate called dUTP was not amplified, thereby achieving strand-specific sequencing. High-throughput sequencing was performed using the Illumina HiSeq X Ten system.
The Screening of Differential Genes and the Bioinformatics Analysis
The differentially expressed genes were screened according to the criteria of Fold change ≥2 and false discovery rate <0.05. They were subjected to GO and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis using DAVID online software.
The Cell Counting Kit-8 Assay
At 48 h after transfection, the cells were seeded into a 100 μL, 2000–5000 cells/well 96-well plate. After culture for 24 h, 10 μL of Cell Counting Kit-8 (CCK-8) solution was added to each well, and the cells were incubated for 4 h. The absorbance at 450 nm was measured using a microplate reader.
The Transwell Invasion Assay
The upper chamber of the Transwell room is pre-painted with Matrigel from Becton Dickinson. At 37℃ for 4 h. The cells were prepared in cell suspension after 48 h of transfection, with 60 µL of cell suspension being added to the upper chamber of the Transwell chamber, and incubated at 37℃. Complete DMEM medium was added to the lower chamber. After 24 h, the upper chamber was removed, and the non-invaded cells were wiped with a cotton swab. The cells were washed twice with PBS, fixed with paraformaldehyde for 20 min, and stained with crystal violet in the dark for 20 min. After being washed with PBS three times, the cells were observed under a 400× microscope. Three fields of view were randomly selected, and the invaded cells were counted.
RT-qPCR
Total RNA was extracted using TRIZOL reagent and reverse transcribed into cDNA at 25℃ for 5 min, 42℃ for 60 min, and 70℃ for 5 min. The SYBRGreen PCR kit was used for PCR amplification. The primer sequences are listed in Table 1. β-actin was used as an internal reference. The polymerase chain reaction system includes 10 μL of SYBR ®Select Master Mix, Upstream and downstream primers 0.5 μL respectively, 2 μL cDNA, and 7 μL ddH2O. RT-qPCR was conducted using a fluorescence quantitative PCR instrument of Hangzhou Longgene Scientific Instruments Co., Ltd. China. The PCR procedure involved pre-denaturation at 95℃ for 5 min and 45 cycles each at 95℃ for 5 s and 60℃ for 35s. The 2−ΔΔCT method was used to calculate the relative expression level of each gene.
 |
Table 1 Primer Sequences |
Statistical Methods
GraphPad Prism 7.00 was used for data analysis. F-test was used for multi-comparison of the three groups, and an independent sample t-test was used for a comparison of two of the groups. P < 0.05 indicated that the difference was statistically significant and was marked as *P < 0.05, **P < 0.01, and ***P < 0.001.
Results
The Screening of the Differentially Expressed Genes
The confirmatory data showing the expression profile of IGF2BP2 is shown in Figure 1A. The differential gene expression level is shown in Figure 1B volcano map and Figure 1C heat map. The results show that compared with the control group, 67 genes were up-regulated and 87 genes were down-regulated in the IGF2BP2 Ov group. The gene with the most significant up-regulation was PROX1, and the gene with the most significant down-regulation was CRYBA4. The top 10 differentially expressed genes up-regulated and down-regulated in breast cancer cells overexpressing IGF2BP2 are shown in Table 2. The gene expression levels in this study and the findings of previous studies10–13 suggest that the genes of IFIT2, CCL20, CCL5, and CXCL10 may play a role in breast cancer.
 |
Table 2 The Top 10 Differentially Expressed Genes (Up-Regulated and Down-Regulated) in Breast Cancer Cells Overexpressing IGF2BP2 |
GO Enrichment Analysis of the Differentially Expressed Genes
GO enrichment analysis was performed to analyze the functions of the differentially expressed genes, and the top 10 GO terms are displayed. The enriched GO term in the molecular function of up-regulated differentially expressed genes was mainly protein binding, and, in the cellular component, it was cell membrane as shown in Figure 2A. There was no enriched term with respect to the biological process. The molecular functions of down-regulated differentially expressed genes are mainly enriched in ion binding, cellular components are mainly enriched in cytoplasm, and biological processes are mainly enriched in response to viruses as shown in Figure 2B.
KEGG Pathway Enrichment Analysis of the Differentially Expressed Genes
KEGG pathway enrichment analysis showed that the differentially expressed genes participated in 68 classical signaling pathways, 17 of which were statistically significant. The up-regulated differential genes are mainly concentrated in the protein processing of endoplasmic reticulum, including HSPA2, TXNDC5 and actin cytoskeleton regulation, including APC2, ITGA7 and other genes as shown in Table 3 and Figure 3. The down-regulated differential genes were mainly enriched in rheumatoid arthritis, including ATP6V1B1, CCL20, CCL3L3, and CCL5, chemokine signaling pathways, including CCL20, CXCL10, CCL3L3, CCL5, and other genes, toll-like receptor signaling pathways, including CXCL10, CCL3L3, CCL5, and other genes, TNF signaling pathways, including CCL20, CXCL10, CCL5, and other genes, cytokine-cytokine receptor interaction, including CCL20, CXCL10, CCL3L3, CCL5, and other genes, and Notch signaling pathways, including DLL1, DLL3, and other genes as shown in Table 3 and Figure 3.
 |
Table 3 The Top 5 Enriched Signaling Pathways of Up-Regulated and Down-Regulated Differential Genes |
Relationship Between Differential Genes and Immune Cell Infiltration and Kaplan-Meier Plotter Survival Analysis
As shown in Figure 4A, the expression of IGF2BP2, IFIT2, CCL20, CCL5, and CXCL10 was positively correlated with the degree of infiltration of B Cell, CD8+ T Cell, CD4+ T Cell, Macrophage, Neutrophil, Dendritic Cell, partila.cor > 0, P < 0 05. The Kaplan-Meier Plotter survival analysis of downstream (Figure 4B) differentially expressed genes IFIT2, CCL20, CCL5, and CXCL10 showed that the high expression of CCL20 could improve the 5-year survival rate of breast cancer patients. The high expression of IFIT2, CCL5 and CXCL10 decreased the 5-year survival rate of patients.
Figure 4 Continued.
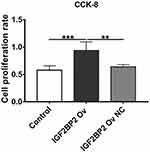
The Proliferation of MDA-MB-231 Cells
The results of the CCK-8 assay are shown in Figure 5. The cell proliferation in the IGF2BP2 Ov group was significantly higher than that in the control group, P < 0.01. This indicates that overexpression of IGF2BP2 could promote the proliferation of MDA-MB-231 cells.
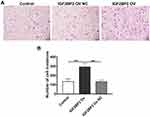
The Invasion Ability of MDA-MB-231 Cells
The Transwell assay detected cell invasion ability, and the results are shown in Figure 6. Compared with the control group and the IGF2BP2 Ov NC group, the IGF2BP2 Ov group had significantly more cells passing through the basement membrane of the chamber, P < 0.01. This indicates that overexpression of IGF2BP2 can enhance the invasion ability of MDA-MB-231 cells.
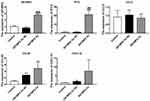
RT-qPCR Verification of IFIT2, CCL20, CCL5, and CXCL10
So as to further verify the relationship between IGF2BP2 and its downstream target genes, RT-qPCR was performed to evaluate the breast cancer-related genes of IGF2BP2, IFIT2, CCL20, CCL5, and CXCL10. The F-test results showed that the levels of the IGF2BP2, IFIT2, and CLCL10 genes were significantly higher in the IGF2BP2 Ov group than in the control group or the IGF2BP2 Ov NC group as shown in Figure 7, P < 0.05. The CCL20 gene was significantly increased in the IGF2BP2 Ov group compared to the control group, P < 0.05. However, there was no significant change in the CCL5 gene, P > 0.05.
Discussion
IGF2BP2 is a member of the IGF2BP protein family, which is composed of IGF2BP1–3, and it is also known as IMP-2 or VICKZ2. IGF2BP2 is an RNA-binding protein, which can bind to the 5’ untranslated region of IGF2 mRNA and regulate its translation. IGF2BP2 plays an important role in metabolism.14 It has been reported that the up-regulation of IGF2BP2 expression can lead to poor prognosis of patients with pancreatic ductal adenocarcinoma.15 In colorectal cancer, LncRNA 91H can promote IGF2 expression, by interacting with IGF2BP2, and promote tumor invasion and metastasis.16 In liver cancer, RHPN1-AS1 can promote the expression of IGF2BP2 and accelerate the proliferation and metastasis of liver cancer cells.17 The above studies indicate that IGF2BP2 is involved in various cancers, and, as an oncogene, it may be a molecular target for cancer treatment. However, the mechanism of IGF2BP2 in breast cancer has been less clear.
In this study, in breast cancer cells with IGF2BP2 overexpression, RNA sequencing identified 67 up-regulated differential genes and 87 down-regulated differential genes, of which PROX1 was the most significantly up-regulated and CRYBA4 was the most significantly down-regulated. In addition, it has been reported that the expression of Prox1 in breast cancer is decreased, and it is related to lymph node metastasis, TNM stage and differentiation.18 It is of great significance for clinical evaluation of patients’ condition, and it is also a new direction of targeted drug therapy research and development. It has been reported that CRYBA4 can be used as a prognostic marker in renal clear cell carcinoma, but no related studies have been found in breast cancer.19 Through GO functional enrichment of these differentially expressed genes, it was found that they were mainly enriched in protein binding, ion binding, the cell membrane, and cytoplasm. KEGG enrichment analysis showed that differentially expressed genes were involved in 68 signaling pathways, of which 17 signaling pathways were statistically significant, including protein processing in the endoplasmic reticulum, actin cytoskeleton regulation, the rheumatoid arthritis signaling pathway, the chemokine signaling pathway, the Toll-like receptor signaling pathway, the TNF signaling pathway, and the Notch signaling pathway. The other 51 signal pathways were not statistically significant. The Notch signaling pathway is already known to play an important role in breast cancer. Recently, Zhu et al reported20 that after silencing Linc-OIP5, the expression of Notch signaling ligand Jagged1 was down-regulated, thus reducing Notch signaling, and promoting the apoptosis of breast cancer MDA-MB-231 cells. A Toll-like receptor is a type of pattern recognition receptor and when stimulated by corresponding ligands, these receptors can activate downstream signaling pathways and release a large number of cytokines. They have anti-pathogen infection and anti-tumor effects, and they can also change the tumor microenvironment and promote the angiogenesis of tumor tissues and the proliferation and metastasis of tumor cells.21
There have been many studies concerning breast cancer-related genes. One study has shown that the flavonoid compound baicalein can inhibit radiotherapy and chemotherapy resistance in breast cancer by up-regulating IFIT2. It is known that CCL20 regulates cell migration and MMP-9 expression by activating PKC-α through Src kinase, thereby activating downstream the Akt, JNK, and NF-κB pathways.22 It can also promote the transcription of c-fos and c-myc and participate in the proliferation of breast cancer cells. The chemokine CXCL10 and its receptor CXCR3 play an important role in breast cancer bone metastasis and osteoclast activation and can mediate the NF-κB signaling pathway.23 CCL5 is an inflammatory mediator with carcinogenic activity in breast cancer.24 In this study, IGF2BP2 was overexpressed in breast cancer MDA-MB-231 cells. The CCK-8 and Transwell experiments showed that IGF2BP2 significantly promoted cell proliferation and invasion. The genes of IFIT2, CCL20, CCL5, and CXCL10 were verified by RT-qPCR, and the results suggested that the expression of IFIT2, CCL20, and CXCL10 in the IGF2BP2 Ov group was significantly higher. However, the CCL5 gene was not significantly changed.
Conclusion
In summary, this study has found that IGF2BP2 may promote the invasion and proliferation of human breast cancer cells by up-regulating breast cancer-related genes, such as IFIT2, CCL20, and CXCL10. The results provide experimental evidence explaining the pathogenesis of breast cancer, which could lead to the development of treatment targets for this disease.
Abbreviation
IGF2BP2, insulin-like growth factor 2 mRNA binding protein 2; IFIT2, tetratricopeptide repeats 2; CCL20, chemokine C-C motif ligand 20; CCL5, chemokine C-C motif ligand 5; CXCL10, chemokine C-X-C motif ligand 10; PROX1, Homeobox protein 1; CRYBA4, Acidic β-crystallin 4; GO, gene ontology; RT-qPCR, real-time reverse transcriptase-polymerase chain reaction; Ov, overexpression; NC, negative control; DMEM, Dulbecco’s Modified Eagle Medium; cDNA, complementary deoxyribonucleic acid; KEGG, Kyoto Encyclopedia of Genes and Genomes; CCK-8, Cell Counting Kit-8; PBS, phosphate-buffered saline; F-test, A one-way analysis of variance; TNF, tumor necrosis factor.
Funding
Natural Science Foundation of Xinjiang Uygur Autonomous Region (No. 2017D01C376). State Key Laboratory of Pathogenes,Prevention,Treatment of Central Asia High Incidence Diseases Fund (SKL-HIDCA-2022-JZ2).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. doi:10.3322/caac.21660
2. Liu G, Zhu T, Cui Y, et al. Correlation between IGF2BP2 gene polymorphism and the risk of breast cancer in Chinese Han women. Biomed Pharmacother. 2015;69:297–300. doi:10.1016/j.biopha.2014.12.017
3. Yang L, Giulio F, Jian-Ying. Z. p62/IMP2 stimulates cell migration and reduces cell adhesion in breast cancer. Oncotarget. 2015;6(32):32656.
4. McMullen Emily R, Gonzalez Maria E, Skala Stephanie L, et al. CCN6 regulates IGF2BP2 and HMGA2 signaling in metaplastic carcinomas of the breast. Breast Cancer Res Treat. 2018;172(3):577–586. doi:10.1007/s10549-018-4960-2
5. Pan X, Huang B, Ma Q, et al. Circular RNA circ-TNPO3 inhibits clear cell renal cell carcinoma metastasis by binding to IGF2BP2 and destabilizing SERPINH1 mRNA. Clin Transl Med. 2022;12(7). doi:10.1002/ctm2.994
6. Jie L, Xinya G, Zhanqiang Z, et al. CircCD44 plays oncogenic roles in triple-negative breast cancer by modulating the miR-502–5p/KRAS and IGF2BP2/Myc axes. Mol Cancer. 2021;20(1). doi:10.1186/s12943-021-01444-1
7. Valerie S, Hansen KM, Morris DR, LeBoeuf Renée C, Abrass CK. RNA-binding protein IGF2BP2/IMP2 is required for laminin-β2 mRNA translation and is modulated by glucose concentration. Ren Physiol. 2012;303(1):F75–F82. doi:10.1152/ajprenal.00185.2012
8. Xiaoge H, Wan-Xin P, Huaixiang Z, et al. IGF2BP2 regulates DANCR by serving as an N6-methyladenosine reader. Cell Death Differ. 2020;27(6):1782–1794. doi:10.1038/s41418-019-0461-z
9. Pei C, Yufan W, Ding S, et al. IGF2BP2 promotes pancreatic carcinoma progression by enhancing the stability of B3GNT6 mRNA via m6A methylation. Cancer Med. 2022. doi:10.1002/cam4.5096
10. Koh SY, Moon JY, Unno T, Cho SK. Baicalein suppresses stem cell-like characteristics in radio- and chemoresistant MDA-MB-231 human breast cancer cells through up-regulation of IFIT2. Nutrients. 2019;11(3):624. doi:10.3390/nu11030624
11. Brett E, Sauter M, Machens HG, Duscher D. Abstract 90: an unrecognized role of Ccl5 in triple negative breast tumor construction. Plast Reconstr Surg Glob Open. 2020;8(4 Suppl):59.
12. Jin WJ, Kim B, Kim H, Kim J, Lee ZH. NF-kappa B signaling regulates cell autonomous regulation of CXCL10 in breast cancer 4T1 cells. Mol Biol Cell. 2016;27:e295.
13. Liangping L, Gang F, Tao C, Lijun Z. Circ_0000514 promotes breast cancer progression by regulating the miR-296-5p/CXCL10 axis. J Biochem. 2021;170(6):753–761.
14. Mingyu L, Xiaozhi R, Ling L, et al. IGF-2 mRNA binding protein 2 regulates primordial germ cell development in zebrafish. Gen Comp Endocrinol. 2021;313. doi:10.1016/j.ygcen.2021.113875
15. Shan H, Zheng W, Yunyun C, Wenzhen W, Linlin H. Insulin-like growth factor 2 mRNA binding protein 2 promotes aerobic glycolysis and cell proliferation in pancreatic ductal adenocarcinoma via stabilizing GLUT1 mRNA. Acta Biochim Biophys Sin. 2019;51(7):743–752. doi:10.1093/abbs/gmz048
16. Tianyi G, Xiangxiang L, Bangshun H, Yuqin P, Shukui W. Long non-coding RNA 91H regulates IGF2 expression by interacting with IGF2BP2 and promotes tumorigenesis in colorectal cancer. Artif Cells Nanomed Biotechnol. 2020;48(1):664–671. doi:10.1080/21691401.2020.1727491
17. Hu F, Zheng H, Wei W, et al. RHPN1-AS1 drives the progression of hepatocellular carcinoma via regulating miR-596/IGF2BP2 axis. Curr Pharm Des. 2020;25(43):4630–4640. doi:10.2174/1381612825666191105104549
18. Xianfu S, Tao H, Chengjuan Z, et al. Long non-coding RNA LINC00968 reduces cell proliferation and migration and angiogenesis in breast cancer through up-regulation of PROX1 by reducing hsa-miR-423-5p. Cell Cycle. 2019;18(16):1908–1924. doi:10.1080/15384101.2019.1632641
19. Ming-Hui Z, Jian-Guo Q, Yong X, Xue-Hua Z. IDUA, NDST1, SAP30L, CRYBA4, and SI as novel prognostic signatures clear cell renal cell carcinoma. J Cell Physiol. 2019;234(9):16320–16327.
20. Q Zhu, Yongsheng L, Xiangmei D, Yue Y, Hongyan W, Sufen G. Linc-OIP5 loss regulates migration and invasion in MDA-MB-231 breast cancer cells by inhibiting YAP1/JAG1 signaling. Oncol Lett. 2020;19(1):103–112. doi:10.3892/ol.2019.11071
21. Negin N, Mahdyieh NA, Homa D. The role of toll-like receptors in breast cancer. J Qazvin Univ Med Sci. 2019;23(3):262–277.
22. Boafo KL, Shujing W, Youjing S, Qiang W. Multifaceted roles of CCL20 (C-C motif chemokine ligand 20): mechanisms and communication networks in breast cancer progression. Bioengineered. 2021;12(1):6923–6934.
23. Emily T, Jeremiah S, Peterson EA, Schroeder JA, Suwon K. Cxcl10 chemokine induces migration of ING4-deficient breast cancer cells via a novel crosstalk mechanism between the Cxcr3 and Egfr receptors. Mol Cell Biol. 2021;42(2). doi:10.1128/MCB.00382-21
24. Yukie F, Natsuko I, Koji M, et al. Significant association between high serum CCL5 levels and better disease-free survival of patients with early breast cancer. Cancer Sci. 2020;111(1):209–218. doi:10.1111/cas.14234
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.