Back to Journals » International Journal of General Medicine » Volume 14
The Impacts of IL1R1 and IL1R2 Genetic Variants on Rheumatoid Arthritis Risk in the Chinese Han Population: A Case–Control Study
Authors Liu X, Peng L, Li D, He C, Xing S, Wang Y, He Y
Received 11 November 2020
Accepted for publication 16 April 2021
Published 28 May 2021 Volume 2021:14 Pages 2147—2159
DOI https://doi.org/10.2147/IJGM.S291395
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Scott Fraser
Xiaoli Liu,1,* Linna Peng,1,* Dandan Li,1 Chunjuan He,1 Shishi Xing,1 Yuhe Wang,2 Yongjun He1
1Key Laboratory of Molecular Mechanism and Intervention Research for Plateau Diseases of Tibet Autonomous Region, School of Medicine, Xizang Minzu University, Xianyang, Shaanxi, 712082, People’s Republic of China; 2Department of Clinical Laboratory, The Affiliated Hospital of Xizang Minzu University, Xianyang, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Yongjun He
Key Laboratory of Molecular Mechanism and Intervention Research for Plateau Diseases of Tibet Autonomous Region, School of Medicine, Xizang Minzu University, #6 East Wenhui Road, Xianyang, 712082, Shaanxi, People’s Republic of China
Tel +86-18691008772
Email [email protected]
Background: Rheumatoid arthritis (RA), an autoimmune systemic inflammatory disease, largely resulted from genetic factor. Our purpose was to explore the association for IL1R1 and IL1R2 genetic variants with RA susceptibility in the Chinese Han population.
Patients and Methods: A total of 508 RA patients and 494 controls were involved in this case–control study; single-nucleotide polymorphisms (SNPs) genotyping was identified by the Agena MassARRAY platform. The relationship between polymorphisms and RA susceptibility was calculated using the Pearson’s Chi-square test with odds ratios and 95% confidence intervals (CIs) in multiple genetic models. The Pearson’s Chi-square test and Student’s t-test were used for sample basic characteristic analysis. And linkage disequilibrium (LD) analysis and haplotype analysis were performed by logistic regression analysis.
Results: The result from this study showed that rs2072472 (IL1R2) was an increased risk factor of RA (adjusted OR = 1.41, p = 0.011). Stratified analysis indicated SNPs rs10490571, rs956730, rs3917318 of IL1R1, and SNPs rs4851527, rs719250, rs3218896, rs3218977, rs2072472 of IL1R2 had impacts on RA risk after stratification based on gender and average age (54 years). Finally, haplotype analysis revealed that Ars3218977Ars2072472 haplotype in IL1R2 was related to a decreased RA risk (adjusted OR = 0.79; 95% CI = 0.65– 0.94; p = 0.010). Yet, rs3917225(IL1R1) and rs11674595(IL1R2) were not significant in RA association analysis.
Conclusion: We determined SNPs (rs3917318, rs956730, rs1049057) of IL1R1 and SNPs (rs3218977, rs719250, rs4851527, rs3218896, rs2072472) of IL1R2 were correlated with the RA susceptibility in the Chinese Han population.
Keywords: rheumatoid arthritis, RA, IL1R1/IL1R2, single-nucleotide polymorphism, SNP, susceptibility
Introduction
Rheumatoid arthritis (RA) is a common autoimmune systemic inflammatory disease that has complex multifactorial pathogenesis finally leading to the development of synovitis, joint damage, and structural bone damage1 and causing severe disability and reduced health-related quality of life of patients.2 The RA population minimum prevalence is 1.16% in women and 0.44% in men.3 RA is influenced by both genetic and environmental factors.4 The genetic factors may explain up to 60% of the susceptibility to RA,5 and genetic variants affect both the development and severity of rheumatoid arthritis.6 Recent studies have expanded the number of RA susceptibility variants.7–9 Besides, many studies have also confirmed that IL1R1 and IL1R2 polymorphisms were associated with the risk of certain inflammation-related diseases, such as inflammatory bowel disease, Alzheimer’s disease.10,11 However, the association of IL1R1 and IL1R2 gene polymorphisms with RA risk has not been clarified. As we all know, there were several cytokines involved in the pathogenesis of RA,12,13 which was an inflammatory disease. Among these reported cytokines, the interleukin-1 (IL-1) family as pro-inflammatory cytokine has been illustrated to play crucial roles in the pathogenesis of RA, promoting the bone and cartilage inflammation and degradation by affecting cell proliferation and differentiation.14 The cytokine IL-1 mediates its biological functions exactly by binding to the signaling receptor IL-1R1. Contrarily, the other receptor IL-1R2 which was known as the decoy receptor can seclude the active forms of IL-1 or obstruct the signaling complex assembly.15 Thus, IL-1R1 and IL-1R2 played crucial roles in the process of RA development. Genetically, IL1R1 and IL1R2, encoding interleukin 1 receptor type 1 (IL1-R1) and interleukin 1 receptor type 2 (IL1-R2), respectively.16,17 IL-1α (IL-1A) and IL-1β (IL-1B) are two subtypes of IL-1, and it has been demonstrated that untreated RA patients had significant upregulation of both IL-1B and IL1R genes compared with healthy individuals,18 which indicated the complex roles of IL1R1 and IL1R2 in the pathogenesis of RA. Genetic polymorphisms could affect the gene function, furthermore affecting the occurrence and development of disease. Published studies reported that IL1R1 and IL1R2 gene polymorphisms had an impact on the risk of osteonecrosis of the femoral head (ONFH) and aggressive periodontitis,19,20 indicating the potential importance of IL1R1 and IL1R2 genetic polymorphisms in the inflammatory disease. Thus, here we try to explore the relationship between IL1R1 and IL1R2 gene polymorphisms and the susceptibility of RA in the Chinese Han population to elucidate the IL1R1 and IL1R2 risk variants may be an important factor in determining the progression in early RA.
The impacts of IL1R1 (rs10490571, rs956730, rs3917225, rs3917318), IL1R2 (rs11674595, rs4851527, rs719250, rs3218896, rs3218977, rs2072472) on RA risk among the Chinese Han population were investigated in this case–control study, which may provide more theoretical basis for the function of IL1R1 and IL1R2 in RA pathogenesis in the Chinese Han population.
Patients and Methods
Study Subjects
In total, 1002 Chinese subjects including 508 RA patients and 494 geographically ethnicity-matched healthy subjects who were from the Shanxi provincial Yanliang 630 Hospital (Xi’an City, Shaanxi, China) were enrolled in this study. All subjects were individuals without related relationship from Shaanxi province in northern China and at least three generations of Han ancestors. Before beginning the research, all participants have provided written informed consent. All experiments were carried out adhering to the protocol of Helsinki’s Declaration. And the study protocol was approved by the ethics committee of the Affiliated Hospital of Xizang Minzu University (20,190–5). RA patients were diagnosed by routine biochemical blood analysis (including C‐reaction protein [CRP], rheumatoid factor, erythrocyte sedimentation rate [ESR], anti‐cyclic citrulline antibody [CCP]), anti-keratin auto antibody [AKA] and X‐rays of small joints, and the patients were firstly diagnosed as RA according to current criteria for classifying rheumatoid arthritis.21 All control subjects were healthy individuals who had a routine physical examination in the same period. We excluded the healthy controls with immune diseases or more aggressive metabolic and endocrinological disease. The clinical information of each participants were collected by consulting medical records.
SNP Selection and Genotyping
Firstly, a preliminary selection of SNPs with minor allele frequencies >5% was performed in the 1000 Genome Projects (http://www.internationalgenome.org/) based on the CHB (Chinese Han in Beijing) data in order to ensure the successful genotyping and valid statistical analysis in our study population. Secondly, the linkage disequilibrium (LD) haplotype analysis was observed using the haploview software (version 4.2) analysis. Then, the functional prediction of the tagSNPs which constructed the haplotypes was performed in dbSNP database (https://www.ncbi.nlm.nih.gov/snp/), which indicated these tagSNPs were introns affecting the activity of splice sites. Finally, four SNPs (rs3917318, rs956730, rs10490571 and rs3917225) in IL1R1 and six SNPs (rs3218977, rs719250, rs4851527, rs11674595, rs3218896 and rs2072472) in IL1R2 were selected for further research.
We isolated the genomic DNA from the blood samples using the GoldMag-Mini Whole Blood Genomic DNA Purification Kit (GoldMag. Co. Ltd., Xi’an, China) as described by the manufacturer’s instructions. The extracted DNA was labeled and stored at −20°C until used. The genes of interest were amplified using appropriate primers which was designed using the Agena Bioscience Assay Design Suite V2.0 software (https://agenacx.com/online-tools/). The information of primers used in this study are displayed in Supplementary Table 1. Then, SNP genotyping was carried out using Agena MassARRAY platform with iPLEX gold chemistry (Agena Bioscience, San Diego, CA, USA), and data analysis was performed using Agena Bioscience TYPER, Version 4.0.
Statistical Analysis
We compared the difference of gender distribution among two groups (RA patients and controls) using Pearson’s Chi-square tests, and assessed the distribution difference of age using Student’s t-test, both of them were performed on the SPSS 19.0 (SPSS, Chicago, IL, USA) and Microsoft Excel. In the control group, Hardy-Weinberg equilibrium (HWE) of the genotype frequencies was assessed by the Fisher’s exact test. The allele frequencies of two groups were also evaluated with the Pearson’s Chi-square test. The correlation of IL1R1 and IL1R2 polymorphisms with RA susceptibility was assessed using logistic regression analysis by calculating odds ratios (ORs) and 95% confidence intervals (CIs) with adjustment for age and gender based on the multiple genetic model analysis. Besides, the gender and average age (45 years old) stratification analyses between IL1R1 (rs10490571, rs956730, rs3917225, rs3917318), IL1R2 (rs11674595, rs4851527, rs719250, rs3218896, rs3218977, rs2072472) and susceptibility of RA were also conducted. In addition, false discovery rate (FDR) test was performed in this study. All p-values were two-tailed, and p < 0.05 means statistical significance.
Results
Characteristics of Study Subjects
There were 508 patients (134 males and 374 females) with RA and 494 healthy individuals (124 males and 370 females) in this case–control study. The basic characteristics of all participants are shown in Table 1. The age at diagnosis of the RA patient group and the control group was 54.34 ± 12.01 years and 54.03 ± 8.83 years, respectively. The age and gender between RA cases and controls (p > 0.05) was well distributed while the age distribution between RA cases and controls was significantly different (p < 0.001).
 |
Table 1 Basic Characteristics of Both Cases and Controls |
Association Between IL1R1 and IL1R2 Variants and Rheumatoid Arthritis Risk
Table 2 displays the basic information about ten polymorphisms of IL1R1 and IL1R2. All ten SNPs retained for their genotype distribution were in accordance with HWE (p > 0.05) in the control group. We found no difference for allele frequency distribution between RA patients and controls. And we explored the correlations between IL1R1 and IL1R2 genetic variants and RA risk under the multiple genetic models (Table 3 and Supplemental Table 2). We noted that among these ten SNPs, only rs2072472 (IL1R2) was a risk factor of RA susceptibility in codominant (adjusted OR = 1.41, 95% CI: 1.08–1.84, p = 0.011) and dominant model (adjusted OR = 1.35, 95% CI: 1.05–1.75, p = 0.021). However, no statistical significance was found between other models of rs2072472 (IL1R2) and RA risk. And we found no significant difference for genetic models of other candidate nine loci in IL1R1 and IL1R2 with RA risk, either. However, after FDR analysis, we got a negative result for the association between IL1R1 and IL1R2 variants and RA risk in total.
 |
Table 2 The Result of Distribution Analysis of Allele Frequencies of IL1R1/IL1R2 SNPs in Case and Control Groups |
 |
Table 3 Association Analysis Result Between IL1R2 Rs2072472 Genetic Variants and Rheumatoid Arthritis Risk |
The Correlation of IL1R1 and IL1R2 Variants with RA Risk Based on the Gender Stratification
Furthermore, we stratified our study subjects based on gender after excluding the age factor to investigate the association of the candidate SNPs in this study with RA susceptibility. As shown in Table 4, the frequency of rs3917318-A (IL1R1) confirmed a significantly higher probability of RA susceptibility than the G allele in males (adjusted OR = 1.44, 95% CI = 1.02–2.04, p = 0.040). Similarly, rs3917318 (IL1R1) was also associated with the elevated RA risk in codominant model (adjusted OR=1.97, 95% CI = 1.01–3.83, p = 0.045 for “A/A” genotype) and dominant model (adjusted OR = 1.84, 95% CI = 1.04–3.27, p = 0.037 for “G/A-A/A” genotype) in males but non-significant in females. Also, SNP rs10490571 (IL1R1) was a factor of elevated RA risk based on the dominant model (adjusted OR = 1.68, 95% CI = 1.01–2.81, p = 0.047) in males. However, rs956730 (IL1R1) was related to the decreased RA susceptibility in recessive model (adjusted OR = 0.48, 95% CI = 0.24–0.97, p = 0.042) in females. For SNPs in IL1R2, rs719250-T showed a distinctly less likelihood of RA susceptibility than the C allele in males (adjusted OR = 0.66, 95% CI = 0.45–0.97, p = 0.034). Similarly, it was also correlated with the reduced chances of RA susceptibility in codominant model (adjusted OR = 0.33, 95% CI = 0.12–0.91, p = 0.032), recessive model (adjusted OR = 0.37, 95% CI = 0.13–0.99, p = 0.049), and log-additive model (adjusted OR = 0.66, 95% CI = 0.44–0.97, p = 0.036) in males but non-significant in females. In addition, in males, rs3218977 was also (adjusted OR = 0.40, 95% CI = 0.18–0.88, p = 0.023) a protective factor in RA risk. Conversely, in females, rs2072472 (IL1R2) was a factor of elevated RA risk in codominant model (adjusted OR = 1.41, 95% CI = 1.03–1.92, p = 0.031). And no significant difference was found for rs3917225 of IL1R1 and rs11674595, rs4851527, rs3218896 of IL1R2 with RA risk in gender stratification analysis (Supplemental Table 3). However, after FDR analysis, we got a negative result for the association between IL1R1 and IL1R2 variants and RA risk in gender stratified analyses.
 |
Table 4 Association Analysis Result Between IL1R1/ IL1R2 Genetic Variants and Rheumatoid Arthritis Risk Based on the Gender Stratification |
The Correlation of IL1R1 and IL1R2 Variants with RA Risk Based on the Age Stratification
Significant differences between candidate SNPs and RA risk were also observed in the average age (54 years) stratification analysis after excluding the gender factor. As shown in Table 5, rs956730 (IL1R1) was a protective factor of RA risk in recessive model (adjusted OR = 0.37, 95% CI = 0.14–0.97, p = 0.043 for “A/A” genotype) in patients aged older than 54 years. In the codominant model of rs4851527 (IL1R2), both of the G/A and A/A genotypes were associated with a significantly protective effect for RA in patients aged younger than 54 years(adjusted OR = 0.63, 95% CI = 0.43–0.93, p = 0.023 for “G/A” genotype) and patients aged older than 54 years (adjusted OR = 0.47, 95% CI = 0.23–0.96, p = 0.039 for “A/A” genotype), respectively. Besides, rs4851527(IL1R2) was also associated with a significantly protective effect for RA in patients aged younger than 54 years based on dominant model (adjusted OR = 0.68, 95% CI = 0.47–0.97, p = 0.035) and patients aged older than 54 years based on recessive model (adjusted OR = 0.41, 95% CI = 0.21–0.82, p = 0.011). In addition, the recessive model of rs3218977 (IL1R2) was a protective factor of RA in patients (>54 years) (adjusted OR = 0.40, 95% CI = 0.18–0.88, p = 0.023), contrarily, rs3218977 (IL1R2) was an elevated risk factor of RA in the log-additive model. Also, both of rs719250 (IL1R2) and rs3218896 (IL1R2) had impacts on the RA risk in patients aged younger than 54 years. In patients aged older than 54 years, rs2072472 (IL1R2) was an increased risk factor of RA in A/G genotype of codominant model (adjusted OR = 1.67, 95% CI = 1.14–2.45, p = 0.009) and dominant model (adjusted OR = 1.47, 95% CI = 1.02–2.12, p = 0.039). And no significant difference was found for rs10490571, rs3917225 and rs3917318 of IL1R1 and rs11674595 of IL1R2 with RA risk in average age (54 years) stratification analyses (Supplemental Table 4). However, no significant association between IL1R1 and IL1R2 variants and susceptibility to RA was observed after a FDR adjustment in the average age (54 years) stratified analysis.
 |
Table 5 Association Analysis Result Between IL1R1/ IL1R2 Genetic Variants and Rheumatoid Arthritis Risk Based on the Age Stratification |
LD and Haplotype Analysis
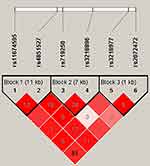
We performed the LD haplotype analysis among above ten SNPs in IL1R1 and IL1R2 genes at last (Figure 1). LD haplotypes in IL1R1 and IL1R2 genes on chromosome 2 were formed by rs3218977 (IL1R2) and rs2072472 (IL1R2), and a haplotype Ars3218977Ars2072472 which was significant with RA risk was found (adjusted OR = 0.79; 95% CI = 0.65–0.94; p = 0.010). In addition, the False discovery rate (FDR) test also illustrated that the Ars3218977Ars2072472 haplotype was associated with the risk of RA. Table 6 shows the frequency distribution of haplotypes in RA patients and control groups. The other two haplotypes Ars3218977Grs2072472 and Grs3218977Ars2072472 were also obtained; nevertheless, no significant linkage was found (p > 0.05) between them and RA risk.
 |
Table 6 The Haplotype Analysis Result of IL1R1/ IL1R2 Polymorphisms and Their Associations with Rheumatoid Arthritis Risk |
Discussion
We explored the association between the ten SNPs on IL1R1 and IL1R2 genes and the RA susceptibility in the Chinese Han population in this study. We concluded that rs2072472, rs719250, rs4851527, and rs3218977 in IL1R2, and rs10490571, rs3917318, and rs956730 in IL1R1 were associated with the RA risk.
The IL1R1 and IL1R2 genes play a pathogenic role in inflammation and tissue destruction of RA for encoding cytokine receptors (IL-1R1, IL-1R2) for IL-1 which plays the crucial role in the immune pathogenesis of many diseases including RA.22 More and more studies reported that IL1R1 and IL1R2 gene polymorphisms had impacts on inflammation-related diseases.10,11 IL1R1 (on chromosome 2q12) is an important mediator involved in many cytokine‐induced immune and inflammatory responses,23 An et al reported that the rs10490571 and rs3917225 in IL1R1 gene were associated with an increased risk of osteonecrosis of the femoral head in the allele model, codominant model, dominant model, and log‐additive model, respectively.19 Kouhia et al indicated that SNPs (rs956730, rs3917225) in the IL1R1 gene provided evidence for association with hand osteoarthritis.24 Another study involving the association between five SNPs polymorphisms in IL1R1 (rs10490571, rs956730, rs3917225 and rs3917318) and osteoarthritis risk, the result found that rs3917225 in IL1R1 was associated with increasing the risk of knee OA.25,26 In our study, rs10490571 (IL1R1) was proved to be related to the increased RA risk in males. Besides, we also demonstrated that rs956730 (IL1R1) was also associated with the decreased RA susceptibility in females. For rs3917318 (IL1R1), it was related to the elevated risk of RA in males, which indicated the unfavorable effects of genetic factors in IL1R1 on RA development. Besides, we did not find the significant association between rs3917225 genetic polymorphisms and RA risk. Contrary to our results, Chang et al confirmed that rs3917225(IL1R1) was a protective factor of age-related hearing impairment, in which Chronic inflammation is an important factor in Taiwan.27 The inconsistency may result from the geographical difference.
Unlike the function of IL1R1, IL1R2 (on chromosome 2q11.2) played an inability to signal after IL-1 binding for lacking an intracellular TIR domain,28 thus, IL1R2 suppressing an inflammatory response. Arend et al proved that increased concentrations of soluble forms (sIL-1R2) are found in the synovial fluid.29 In addition, Xie et al have provided a result that IL1R2 was a protective factor in IgA nephropathy.30 And another suggested that IL1R2 was an important regulator of arthritis by acting specifically on macrophages as a decoy receptor for IL‐1.31 However, only several studies based on the IL1R2 gene polymorphisms were provided. An et al concluded that the rs11674595 in IL1R2 showed an increased risk of osteonecrosis of the femoral head in the Chinese Han population living in central China (Henan Province of China), yet, they got a negative result in the study of other five SNPs (rs4851527, rs719250, rs3218896, rs3218977, rs2072472).19 Xia et al also observed a negative result for the impacts of rs4851527(IL1R2) and rs2072472 (IL1R2) genetic polymorphisms on ankylosing spondylitis risk in Northern Chinese Han population.32 Nevertheless, for the rs11674595, we got a negative result in the RA risk, also, significant differences were obtained for rs719250, rs3218977, rs2072472, rs3218896 and rs4851527 in RA risk (see in Result part). Lastly, the result of haplotype analysis also concluded that haplotype Ars3218977Ars2072472 which was significant with RA risk, which was also proved by False discovery rate (FDR) test. Hence, IL1R2 gene may play a crucial function in affecting RA. The above inconsistent result may result from the difference of geographical location, population difference lifestyle, and environmental exposure in specific Chinese population. All of candidate SNPs in this study are located in the intron sequence according to the retrieval in the dbSNP database (https://www.ncbi.nlm.nih.gov/snp/), several studies have proved that the intronic SNPs conferred susceptibilities by affecting gene expression,33 so we suppose that these SNPs might have potential function to IL1R1 and IL1R2 expression level.
Several limitations of this investigation should be acknowledged. Firstly, the limited number of subjects with RA is a limitation of this study. Secondly, the relationship between variables such as clinical indicators and RA risk was not performed because the relevant data is incomplete. Thirdly, all the samples were collected from hospital, type false-positive error for association study may be generated. Fourthly, RA is a heterogeneous disease with many other risk factors, we did not investigate the gene–gene and gene–environment interactions because of the limited data. Last but not least, functional and molecular mechanism research were warrant to do in future. Despite the limitations mentioned above, our present results provided scientific evidence of IL1R1 and IL1R2 genes with RA in the future studies.
Conclusions
In conclusion, our study proved that SNPs rs3917318, rs956730, rs10490571 of IL1R1 and rs3218977, rs719250, rs4851527, rs3218896 and rs2072472 of IL1R2 were associated with the RA susceptibility in the Chinese Han population, which has not previously been reported and provided new genetic markers for RA susceptibility assessment. We believe that the IL1R1 and IL1R2 genes may be a new insight into the treatment of RA. Larger well‐designed epidemiological studies with more diverse populations and functional evaluations should be conducted.
Data Sharing Statement
All relevant data are within the manuscript.
Ethical Approval and Consent to Participate
This study was performed in accordance with the World Medical Association Declaration of Helsinki and was approved by the ethics committee of the Affiliated Hospital of Xizang Minzu University (20,190-5). Written informed consent was provided from all of the subjects before participating.
Acknowledgment
This study was carried out with the assistance of blood samples and data collection from the Shanxi provincial Yanliang 630 Hospital. We express our sincere thanks and gratitude to participants who were involved in this study. Xiaoli Liu and Linna Peng are co-first authors for this study.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
We acknowledge financial support from the Natural Science Foundation of Tibet Autonomous Region (Grant No. xz2019zrg-28 (z)) and the National Natural Science Foundation of China (Grant No. 81960291).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Scott DL, Wolfe F, Huizinga TW. Rheumatoid arthritis. Lancet. 2010;376(9746):1094–1108. doi:10.1016/S0140-6736(10)60826-4
2. Lillegraven S, Kvien TK. Measuring disability and quality of life in established rheumatoid arthritis. Best Pract Res Clin Rheumatol. 2007;21(5):827–840. doi:10.1016/j.berh.2007.05.004
3. Ngian GS. Rheumatoid arthritis. Aust Fam Physician. 2010;39(9):626–628.
4. Elemam NM, Hannawi S, Maghazachi AA. Role of chemokines and chemokine receptors in rheumatoid arthritis. ImmunoTargets Ther. 2020;9:43–56. doi:10.2147/ITT.S243636
5. Perricone C, Ceccarelli F, Valesini G. An overview on the genetic of rheumatoid arthritis: a never-ending story. Autoimmun Rev. 2011;10(10):599–608. doi:10.1016/j.autrev.2011.04.021
6. Deane KD, Demoruelle MK, Kelmenson LB, Kuhn KA, Norris JM, Holers VM. Genetic and environmental risk factors for rheumatoid arthritis. Best Pract Res Clin Rheumatol. 2017;31(1):3–18. doi:10.1016/j.berh.2017.08.003
7. Okada Y, Wu D, Trynka G, et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature. 2014;506(7488):376–381. doi:10.1038/nature12873
8. Leng RX, Di DS, Ni J, et al. Identification of new susceptibility loci associated with rheumatoid arthritis. Ann Rheum Dis. 2020;79(12):1565–1571. doi:10.1136/annrheumdis-2020-217351
9. Raslan HM, Attia HR, Hamed Ibrahim M, et al. Association of anti-cyclic citrullinated peptide antibodies and rheumatoid factor isotypes with HLA-DRB1 shared epitope alleles in Egyptian rheumatoid arthritis patients. Int J Rheum Dis. 2020;23(5):647–653. doi:10.1111/1756-185X.13819
10. Latiano A, Palmieri O, Pastorelli L, et al. Associations between genetic polymorphisms in IL-33, IL1R1 and risk for inflammatory bowel disease. PLoS One. 2013;8(4):e62144. doi:10.1371/journal.pone.0062144
11. Chen K, Tang Y, Zhao X, Hou C, Li G, Zhang B. Association of IL1R2 rs34043159 with sporadic alzheimer’s disease in southern Han Chinese. Eur J Neurol. 2020;27(10):1844–1847. doi:10.1111/ene.14319
12. Xu WD, Su LC, He CS, Huang AF. Plasma interleukin-38 in patients with rheumatoid arthritis. Int Immunopharmacol. 2018;65:1–7. doi:10.1016/j.intimp.2018.09.028
13. Kragstrup TW, Andersen T, Heftdal LD, et al. The IL-20 cytokine family in rheumatoid arthritis and spondyloarthritis. Front Immunol. 2018;9:2226. doi:10.3389/fimmu.2018.02226
14. Mateen S, Zafar A, Moin S, Khan AQ, Zubair S. Understanding the role of cytokines in the pathogenesis of rheumatoid arthritis. Clin Chim Acta. 2016;455:161–171. doi:10.1016/j.cca.2016.02.010
15. Schlüter T, Schelmbauer C, Karram K, Mufazalov IA. Regulation of IL-1 signaling by the decoy receptor IL-1R2. J Mol Med. 2018;96(10):983–992. doi:10.1007/s00109-018-1684-z
16. Akdis M, Burgler S, Crameri R, et al. Interleukins, from 1 to 37, and interferon-γ: receptors, functions, and roles in diseases. J Allergy Clin Immunol. 2011;127(3):
17. Li G, Cui S, Du J, et al. Association of GALC, ZNF184, IL1R2 and ELOVL7 with parkinson’s disease in Southern Chinese. Front Aging Neurosci. 2018;10:402. doi:10.3389/fnagi.2018.00402
18. Addobbati C, da Cruz HLA, Adelino JE, et al. Polymorphisms and expression of inflammasome genes are associated with the development and severity of rheumatoid arthritis in Brazilian patients. Inflamm Res. 2018;67(3):255–264. doi:10.1007/s00011-017-1119-2
19. An F, Wang J, Gao H, et al. Impact of IL1R1 and IL1R2 gene polymorphisms on risk of osteonecrosis of the femoral head from a case-control study. Mol Genet Genomic Med. 2019;7(3):e00557. doi:10.1002/mgg3.557
20. Kamei H, Ishihara Y, Fuma D, et al. Interleukin-1 receptor gene variants are associated with aggressive periodontitis in the Japanese. Arch Oral Biol. 2014;59(7):756–763. doi:10.1016/j.archoralbio.2014.04.006
21. Aletaha D, Neogi T, Silman AJ, et al. 2010 rheumatoid arthritis classification criteria: an American College of Rheumatology/European League Against Rheumatism collaborative initiative. Ann Rheum Dis. 2010;69(9):1580–1588. doi:10.1136/ard.2010.138461
22. Stříž IIL. 1 family cytokines in chronic inflammatory disorders. Vnitr Lek. 2019;65(2):81–85.
23. Dinarello CA. Interleukin-1, interleukin-1 receptors and interleukin-1 receptor antagonist. Int Rev Immunol. 1998;16(5–6):457–499. doi:10.3109/08830189809043005
24. Näkki A, Kouhia ST, Saarela J, et al. Allelic variants of IL1R1 gene associate with severe hand osteoarthritis. BMC Med Genet. 2010;11:50. doi:10.1186/1471-2350-11-50
25. Na Y, Bai R, Zhao Z, et al. IL1R1 gene polymorphisms are associated with knee osteoarthritis risk in the Chinese Han population. Oncotarget. 2017;8(3):4228–4233. doi:10.18632/oncotarget.13935
26. Smith AJ, Keen LJ, Billingham MJ, et al. Extended haplotypes and linkage disequilibrium in the IL1R1-IL1A-IL1B-IL1RN gene cluster: association with knee osteoarthritis. Genes Immun. 2004;5(6):451–460. doi:10.1038/sj.gene.6364107
27. Chang NC, Yang HL, Dai CY, et al. The association of genetic polymorphisms in interleukin-1 receptors type 1 and type 2 with age-related hearing impairment in a Taiwanese population: a case control study. J Otolaryngol Head Neck Surg. 2020;49(1):16.
28. McMahan CJ, Slack JL, Mosley B, et al. A novel IL-1 receptor, cloned from B cells by mammalian expression, is expressed in many cell types. EMBO J. 1991;10(10):2821–2832. doi:10.1002/j.1460-2075.1991.tb07831.x
29. Peters VA, Joesting JJ, Freund GG. IL-1 receptor 2 (IL-1R2) and its role in immune regulation. Brain Behav Immun. 2013;32:1–8. doi:10.1016/j.bbi.2012.11.006
30. Xie M, Zhang D, Zhang Y, et al. Association of genetic polymorphisms in IL-1R1 and IL-1R2 genes with IgA nephropathy in the Han Chinese population. Oncotarget. 2017;8(31):50673–50679. doi:10.18632/oncotarget.16929
31. Shimizu K, Nakajima A, Sudo K, et al. IL-1 receptor type 2 suppresses collagen-induced arthritis by inhibiting IL-1 signal on macrophages. J Immunol. 2015;194(7):3156–3168. doi:10.4049/jimmunol.1402155
32. Xia Y, Liu YQ, Chen K, Wang LC, Ma CY, Zhao YR. Association of IL-1R2 genetic polymorphisms with the susceptibility of ankylosing spondylitis in Northern Chinese Han population. Mod Rheumatol. 2015;25(6):908–912. doi:10.3109/14397595.2015.1024302
33. Zhao H, Yang W, Qiu R, et al. An intronic variant associated with systemic lupus erythematosus changes the binding affinity of Yinyang1 to downregulate WDFY4. Genes Immun. 2012;13(7):536–542. doi:10.1038/gene.2012.33
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.