Back to Journals » The Application of Clinical Genetics » Volume 13
The Impact of the Genetic Polymorphism in DNA Repair Pathways on Increased Risk of Glioblastoma Multiforme in the Arab Jordanian Population: A Case–Control Study
Authors Al-Khatib SM , Abdo N, Al-Eitan LN , Al-Mistarehi AHW, Zahran DJ, Al Ajlouni M, Kewan TZ
Received 9 February 2020
Accepted for publication 22 May 2020
Published 11 June 2020 Volume 2020:13 Pages 115—126
DOI https://doi.org/10.2147/TACG.S248994
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Martin Maurer
Sohaib M Al-Khatib,1 Nour Abdo,2 Laith N Al-Eitan,3 Abdel-Hameed W Al-Mistarehi,4 Deeb Jamil Zahran,5 Marwan Al Ajlouni,2 Tariq Zuheir Kewan6
1Department of Pathology and Laboratory Medicine, Faculty of Medicine, Jordan University of Science and Technology, Irbid, Jordan; 2Department of Public Health, Faculty of Medicine, Jordan University of Science and Technology, Irbid, Jordan; 3Department of Biotechnology and Genetic Engineering, Faculty of Science and Arts, Jordan University of Science and Technology, Irbid, Jordan; 4Department of Family Medicine, Faculty of Medicine, Jordan University of Science and Technology, Irbid, Jordan; 5Department of Internal Medicine, Faculty of Medicine, Jordan University of Science and Technology, Irbid, Jordan; 6Department of Internal Medicine, Cleveland Clinic Foundation, Cleveland, Ohio, USA
Correspondence: Sohaib M Al-Khatib
Department of Pathology and Laboratory Medicine, Jordan University of Science and Technology, P.O.Box 3030, Irbid 22110, Jordan
Tel +962-799-409933
Fax +962-2-72006262
Introduction: Among the Jordanian population, brain tumors are the tenth most common type of cancers in both males and females, comprising 2.8% of all newly diagnosed neoplasms. Diffuse gliomas are the most prevalent and the most aggressive primary brain tumors in adults. The incidence of diffuse gliomas varies among different populations; this variation is partially linked to genetic polymorphisms. The purpose of the study is to examine the association between (BRCA1 rs799917G>A, rs1799966T>C, EXO1 rs1047840G>A, EME1 rs12450550T>C, ERCC2 rs13181T>G, rs1799793C>T, and XRCC1 rs1799782G>A) DNA repair gene polymorphisms and glioblastoma multiforme (GBM) susceptibility, and survival in the Jordanian Arab population.
Methods: Eighty-four patients diagnosed with glioblastoma multiforme at the King Abdullah University Hospital (KAUH) between 2013 and 2018 and 225 healthy cancer-free control subjects with similar geographic and ethnic backgrounds to the patients were included in the study. Genomic DNA was extracted from the formalin-fixed paraffin-embedded tissues of the subjects. The Sequenom MassARRAY® sequencer system (iPLEX GOLD) was used. The analyses included assessments of population variability and survival.
Results: This study is the first to address the relationship between BRCA1 rs1799966 and rs799917 SNP, and the risk of GBM among the Arab Jordanian population. The findings of the study show that BRCA1 rs799917 is associated with decreased risk of GBM in the recessive model (AA vs G/G-A/G: OR, 0.46, 95% CI, 0.26– 0.82, p=0.01) and the same SNP is associated with increased risk of GBM in the overdominant model (AG vs G/G-A/A: OR, 1.72, 95% CI, 1.02– 2.89, p=0.04).
Keywords: glioblastoma multiforme, DNA repair genes, SNP, overall survival, Arab population
Introduction
Cancer is a major health problem and is the leading cause of death in male and female patients in the 40–79 age range.1,2 In all cancer-related deaths, the primary central nervous tumors (PCNT), which are defined as tumors arising in the brain, brain envelops, and nerve roots account for almost 3% of all cancer-related deaths.2 The incidence rates (IR) of all PCNT are geographically variable, ranging from 17.6 per 10,000, to 18.2 per 10,000, and 22.0 per 10,000 in France,3 Australia,4 and the United States,5 respectively, with slight female predominance. Although the incidence of PCNT is relatively not high with approximately 23,770 new cases diagnosed in the US in 2016, brain tumors are considered a major health problem since they are associated with high mortality and morbidity6,7 and because of the increasing trend in incidence rates.8
Brain tumors are a heterogeneous group of neoplasms including more than 40 different entities, and brain tumors are divided into 6 major groups by the 2016 World Health Organization (WHO) Classification of Tumors of the Central Nervous System.9 Of these groups; the diffuse gliomas (DG) are the most common and the most aggressive primary brain tumors in adults.5,6,10 An important leap in the latest update to the WHO’s classification of the Central Nervous System CNS tumors reveals that the implication of molecular findings is a part of the diagnostic approach of diffuse gliomas, and is not totally relied on morphologic findings. This approach would definitely reduce the subjectivity, inconsistency, and inter-observer variability among diagnostic pathologists; and improves treatment options.11–13 The WHO depends on the presence of nuclear atypia or pleomorphism, mitotic activity or count, vascular proliferation, and necrosis to grade DG into grade II with the presence of pleomorphism, grade III with the presence of pleomorphism and high mitotic count, and grade IV tumors with the presence of pleomorphism, high mitosis and necrosis or vascular proliferation.14 Grade III and Grade IV tumors are considered high-grade gliomas (HGGs).
The diagnosis of HGGs is mostly challenging because they frequently display intratumoral morphological heterogeneity. It has recently been reported that the identification of biomarkers predictive of the outcome of patients would be a key way to improve the diagnosis of HGGs.15 Molecular markers believed to be useful tools for the management of HGG include DNA repair gene mutations. The purpose of this study is to analyze the relationship between single nucleotide polymorphisms (SNPs) in 5 DNA repair genes (BRCA1 rs799917G>A, rs1799966T>C, EXO1 rs1047840G>A, EME1 rs12450550T>C, ERCC2 rs13181T>G, rs1799793C>T, and XRCC1 rs1799782G>A) and the risk and overall survival of patients with GBM (an HGG) in the Jordanian Arab population.
Patients and Methods
Patients and Data Collection
The study population consisted of 84 patients diagnosed with GBM at King Abdullah University Hospital (KAUH) from the period of 2013 to 2018 and 225 healthy cancer-free control subjects collected over 1 year (2017–2018) with similar geographic and ethnic background to patients. All cases of GBM were independently diagnosed by a pathologist (SK) according to the 2007 WHO Classification of Tumors of the Central Nervous System. The name of the subjects and cases were coded, blinded, and confidentially was ensured. The study was given an approval from the Institutional Review Board (IRB) of (KAUH), Jordan [Institutional Review Board (IRB) code number 6/106/2017, dated 8/06/2017]. All control subjects signed a written consent form which stressed that the participation of respondents is voluntary. However, formal written informed consent was not required with a waiver by the IRB. All clinical investigations were conducted as per the ethical principles for medical research involving human subjects of the World Medical Association (WMA) Declaration of Helsinki.
DNA Extraction
Genomic DNA was extracted for the patients with GBM from formalin-fixed paraffin-embedded tissue using a commercially available kit, DNeasy Blood & Tissue Kit (Qiagen Ltd., West Sussex, UK), using the manufacturer’s protocols. Genomic DNA from control-subjects blood samples was extracted using the QIAamp® or Promega DNA Mini Kit according to the manufacture’s instruction. Agarose gel electrophoresis and ethidium bromide staining were employed to examine the quality of extracted DNA. The concentration and purity of the extracted DNA were assessed by using NanoDrop 1000® spectrophotometer. The pure DNA samples with their concentrations were sent to the Australian Genome Research Facility ([AGRF], Melbourne Node, Melbourne, Australia) for genotyping of BRCA1 rs799917G>A, rs1799966T>C, EXO1 rs1047840G>A, EME1 rs12450550T>C, ERCC2 rs13181T>G, rs1799793C>T, and XRCC1 rs1799782G>A. Genotyping with the Sequenom MassARRAY® system (iPLEX GOLD) (Sequenom, San Diego, CA, USA) was performed at the AGRF by following the manufacturer’s protocols (Sequenom, San Diego, CA, USA). Genotype distributions were compared between patients and controls. The SNPs, SNPs position, and primers sequences are shown in Table 1. Unconditional logistic regression analysis was performed to estimate the relationship between the genotype frequency and the risk of developing GBM.
 |
Table 1 The SNPs, SNPs Positions and Primers Sequences for BRCA1, EXO1, EME1, ERCC2, XRCC1 Genes |
Statistical Analysis
Overall survival (OS) was calculated from the date of diagnosis to the date of death or the last visit for those who were alive at the time of final data collection and analysis. All statistical analyses were performed using the Statistical Package for the Social Sciences (SPSS), version 25.0 (SPSS, Inc., Chicago, IL, USA). The clinical characteristics and response rate of the patients were compared using chi-square tests. The Hardy–Weinberg equilibrium (HWE) test was estimated by a goodness-of-fit χ2 test. The Kaplan–Meier method was used to construct survival curves, and the results were compared using a log-rank/Wilcoxon (Gehan) statistic. The association between polymorphism and the risk for GBM was calculated using unconditional logistic regression. Actuarial life table survival analysis was conducted to obtain the overall survival probabilities. The survival curves were displayed using Graph Pad Prism 6 software. All significant variables (p < 0.05) were entered into a multivariate model to adjust for possible confounders.
Ethical Approval
The study was approved by the Institutional Review Board of KAUH, Jordan. All the control subjects were voluntarily involved and signed a written informed consent form. All the clinical investigations were conducted according to the principles given in the Declaration of Helsinki.
Results
Demographic and Clinical Data
In this study, 84 patients with GBM and 225 ethnically and geographically matched healthy controls were enrolled, as shown in Table 2. According to the study findings, 60% of the patients were male while 60% of the controls were female. The mean age for cases was 45.4 and the controls were 30.6 years. The findings of the study revealed that the right side of the brain was involved more commonly than the left side (45% of the cases). By the end of the study, 46% of the patients were dead while 54% of them were alive with a median survival of 35 months. By definition, an HGG is infiltrative and involving more than one lobe, the most commonly involved lobe was the parietal lobe in (57% of the cases). All the SNPs were in the HWE and normally distributed with p-value >.05. Table 3 shows the (HWE) p-values for the cases and the controls.
 |
Table 2 Demographic and Clinical Data of 84 GBM Patients of Jordanian Arab Descent |
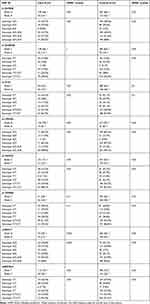 |
Table 3 The Frequency of Allele and Genotype and the (HWE) p-values for the Cases and the Controls for All SNPs Alive and Dead Patients |
Association Between BRCA1, EXO1, EME1, ERCC2, and XRCC1 Genes Polymorphisms and the Risk of GBM
The distribution of both genotypic frequencies and allelic frequencies of the seven SNPs was performed and shown in Table 4. Unconditional logistic regression analysis was used to estimate the association between the genotype frequency and the risk of developing GBM. None of the BRCA1 rs799917G>A, rs1799966T>C, EXO1 rs1047840G>A, EME1 rs12450550T>C, ERCC2 rs13181T>G, rs1799793C>T, and XRCC1 rs1799782G>A showed a clear association with the overall susceptibility for GBM in any allelic model (p= 0.10, 0.50, 0.73, 0.53, 0.77, 0.13, and 0.79, respectively).
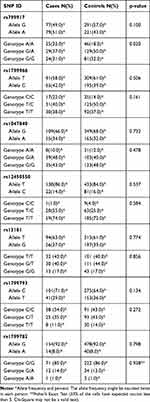 |
Table 4 The Frequency of Allele and Genotype for DNA Repair Gene SNPs in GBM Patients and Controls |
Additional analysis based on four genetic models (codominant, dominant, recessive, and overdominant) was performed and showed that BRCA1 rs799917 was associated with decreased risk of GBM in the recessive model (AA vs G/G-A/G: OR, 0.46, 95% CI, 0.26–0.82, p=0.01) and the same SNP was associated with increased risk of GBM in the overdominant model (AG vs G/G-A/A: OR, 1.72, 95% CI, 1.02–2.89, p=0.04). The results are shown in Table 5.
 |
Table 5 Genotype Distribution of the BRCA1, EXO1, EME1, ERCC2, and XRCC1 SNPs in GBM Patients |
Association Between BRCA1, EXO1, EME1, ERCC2, and XRCC1 Genes Polymorphisms and the Survival Rate of GBM
The Kaplan–Meier curve and Log-rank test were performed for the survival analysis for the all patients with GBM Figure 1. None of the BRCA1 rs799917G>A, rs1799966T>C, EXO1 rs1047840G>A, EME1 rs12450550T>C, ERCC2 rs13181T>G, rs1799793C>T, and XRCC1 rs1799782G>A SNP showed significant survival results with a p value of 0.22, 0.57, 0.37, 0.68, 0.4, 0.23, and 0.47, respectively. Figure 1
Discussion
The study successfully genotyped 7 SNPs in (BRCA1 rs799917G>A, rs1799966T>C, EXO1 rs1047840G>A, EME1 rs12450550T>C, ERCC2 rs13181T>G, rs1799793C>T, and XRCC1 rs1799782G>A) and found some association at rs799917G>A SNP and GBM risk.
The DNA in our cells is continually vulnerable to injury by exposure to damaging agents. Reactive oxygen species (ROS), chemicals, ultraviolet light, radiotherapy, chemotherapeutic agents, and dietary carcinogens are DNA-damaging agents. DNA damage may affect the ability of the cell to divide and survive secondarily to structural change in the molecular sequence. The inability to repair the damaged DNA will ultimately lead to tumor formation.16
The group of processes taken by the cell to recognize the effect of these harmful agents on the genome and fix them is known as DNA repair.17 More than 130 of human DNA repair genes functioning in four pathways have been identified; three of them are base excision repair (BER) pathways and one is DNA double-strand breaks (DSB) repair pathway.18–21 The excision-based repair pathways act by removing the damaged base, they include the BER pathway, the nucleotide excision repair (NER) pathway and the mismatch repair (MMR) pathway.
The major enzymes in the BER pathway are DNA glycosylases where the damaged oxidized, reduced, or methylated base is removed by the corresponding base-specific DNA glycosylase enzyme. The abased site generated by the glycosylase is then repaired by DNA synthesis and ligation.18,22
The NER pathway is a complex multistep pathway reserved for huge DNA damages caused by environmental carcinogens. Bulky fragments of damaged DNA are removed as oligonucleotides.19 The MMR pathway is responsible for fixing of the DNA polymerases errors when they incorrectly copy microsatellites.
The microsatellites are certain DNA base pairs or motifs that are repeated tens of times.20,23 MLH1, MSH2, PMS2, and MSH6 are among the common genes involved in the MMR pathway.
The double-stranded breaks in the chromosome are produced either spontaneously during DNA replication or by ionizing radiation. A large number of genes are connected to (DSB) repair pathway, most notably tumor suppressor genes including BRCA1, BRCA2, P53 and XRCC3 gene products. Repairing broken DNA strands can be a homologous or non-homologous repair process. The homologous recombination process is based on resection of the broken ends, strand exchange, and ligation. In the non-homologous repair pathway, the broken ends are directly re-joined.21,24
Essential meiotic endonuclease 1 (EME1) gene in human is mapped to chromosome 17 (17q21.3).25 In the NER pathway, EME1 endonuclease is essential to overcome any blocks in DNA replication by processing the stalled replication forks. By forming a heterodimer stable complex with its partner protein methyl methanesulfonate-sensitive UV-sensitive 81 (Mus81), the Mus81±Eme1 complex is able to cleave branched DNA structures including replication forks and 3-prime flaps.25,26
XRCC1 encodes X-ray repair complementing defective repair in Chinese hamster cells 1 that is involved in repair of DNA single-strand breaks, BER, and NER by interaction with DNA ligase III.27 XRCC1 gene has 17 exons and spans approximately of 31.9 kb and mapped to chromosome 19 (19q13.3).28
The ERCC2/XPD protein consists of 760 amino acids and contains 23 exons and 21 introns.29 It is mapped to chromosome 19 (19q13.32). ERCC2/XPD is a subunit of human transcriptional initiation factor TFIIH/repair factor and participates in NER by an ATP-dependent DNA helicase activity.30 EXO1 enzyme or HEX1 for human exonuclease 1, is encoded by EXO1 Gene containing 14 exons and 13 introns; and maps to the chromosome 1 (1q42-q43). EXO1 gene is part of RAD2 family that is involved in the NER pathway.31 EXO1 exhibits a 5′→3′ exonuclease activity and participates in DNA MMR pathway by interacting with MSH2.32
BRCA1 gene is one of the caretaker tumor suppressor genes which is mapped to chromosome 17 (17q21.31), it includes 22 exons and spans 110 kb of DNA.33,34 BRCA1 gene encodes breast cancer type 1 susceptibility protein which involves in repair of double-strand DNA breaks (DSB).34,35 This role in tumor suppression and DNA repair is emphasized by the fact that the risk of many malignancies including hematologic such as mantle cell lymphoma, acute myeloid leukemia, acute lymphocytic leukemia, and chronic lymphocytic leukemia; and non-hematologic like breast and ovarian is increased by inactivation of any component within the BRCA pathway.35
The incidence of central nervous system tumors is highly variable among different ethnic groups worldwide. A descriptive epidemiologic study by EJ Maile et al (2016) depicted differences in tumor incidence by ethnicity for gliomas, pituitary tumors, meningiomas, and cranial and paraspinal nerve tumors. Their study revealed that the incidence of gliomas is significantly lower among blacks, Chinese, and south Asians in comparison to whites (p<.01). However, the incidence of meningioma is much higher among blacks in comparison to others (p<.01).36 That variation in gliomas incidence by ethnicity cannot be explained by environmental factors and may be partially attributed to gene polymorphism among different ethnic groups.37
DNA repair pathway is one of the most important pathways that play a crucial role in carcinogenesis.38 This study analyzed five genes and seven polymorphisms involved in DNA repair pathway. The selection of the study was based on both original and review articles examining the relationship between DNA repair SNPs and the risk of gliomas.39,40 Existing studies show conflicting inconsistent results among different DNA repair genes polymorphisms and the risk of glioma that may be caused by the insufficient size of populations of interest in each separate study.
Therefore, a meta-analysis and systemic review of existing studies should increase the statistical power. In their meta-analysis, M Adel Fahmideh et al (2014) reviewed 27 studies examining 105 SNPs in 42 DNA repair genes. Their findings depict that all of their models, except the recessive model, reveal significant relationship with the rs13181 polymorphism. They found positive association between the risk of glioma and ERCC2 rs13181 polymorphism (OR=1.18 (1.06–1.31), p=.002), but no statistically significant association between ERCC2 rs1799793 and XRCC1 rs1799782 polymorphisms and the risk to develop gliomas.41 However, this study shows no statistically significant relationship between any of ERCC2 rs13181, ERCC2 rs1799793 and XRCC1 rs1799782 gene polymorphisms and the susceptibility to GBM (p=.77, 0.13, and.79, respectively).
SNPs in BRCA1 gene alter its function and stability by modifying microRNA (miRNA) binding sites.42 Accordingly, the BRCA1 mRNA has a role to play in the overall risk and response to treatment of different types of tumors.43–45
Many studies show that specific haplotypes of BRCA1 gene are predictive of clinical benefit to chemotherapeutic agents and survival of cancer patients.46,47 For example, the presence of at least one AAAG allele is associated with better overall survival vs patients with no AAAG allele.48
The association between BRCA1 gene polymorphism and cancer risk is not constant. Although the presence of rs799917 T allele in comparison with rs799917 C allele is associated with weaker reduction (increased expression) of BRCA1 protein and lower risk of non-breast cancers, the BRCA1 CC genotype is associated with lower risk of breast cancer.49,50 The apparent inconsistency between BRCA1 gene polymorphisms and the risk of different cancers may be attributed to parallel variation in the expression of BRCA1 messenger RNA during different stages of tumor development.
The relationship between BRCA1 gene polymorphisms and the risk of glioma was reviewed by a study of GP Xu et al (2018) who examined the strength of the association between BRCA1 gene polymorphisms and cancer risk. They performed a meta-analysis to examine the association between BRCA1 polymorphisms (rs799917, rs1799950, rs1799966, or rs16941) and cancer risk. In their meta-analysis, they evaluated 35 studies contained 19 articles involving 28,094 cases and 50,657 controls. Odds ratios with 95% confidence intervals were calculated to evaluate the strength of the relationship.
They found no significant relationship between overall cancer risk and BRCA1 rs1799966, and rs799917 polymorphisms in any genetic models. However, when dealing with cancers as subgroups, they found that among Asian populations, the rs799917 polymorphism is associated with decreased risk of esophageal squamous carcinoma, gastric carcinoma, cervical cancer, and non-Hodgkin lymphoma.51
This study is the first to address the relationship between BRCA1 rs1799966 and rs799917 SNP, and the risk of GBM among the Arab Jordanian population. The findings of the study show that BRCA1 rs799917 is associated with decreased risk of GBM in the recessive model (AA vs G/G-A/G: OR, 0.46, 95% CI, 0.26–0.82, p=0.01) and the same SNP is associated with increased risk of GBM in the overdominant model (AG vs G/G-A/A: OR, 1.72, 95% CI, 1.02–2.89, p=0.04).
Conclusion
BRCA1 rs799917 is associated with decreased risk of GBM in the recessive model (AA vs G/G-A/G: OR, 0.46, 95% CI, 0.26–0.82, p=0.01) and the same SNP is associated with increased risk of GBM in the overdominant model (AG vs G/G-A/A: OR, 1.72, 95% CI, 1.02–2.89, p=0.04) in the Jordanian Arab population.
Acknowledgment
The study was supported by the Jordan University of Science and Technology Grant number 20170196.
Disclosure
The authors declare that they have no conflict of interest.
References
1. Curado M-P, Edwards B, Shin HR, et al. Cancer Incidence in Five Continents. Vol. IX. IARC Press, International Agency for Research on Cancer; 2007.
2. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. doi:10.3322/caac.21332
3. Baldi I, Gruber A, Alioum A, et al. Descriptive epidemiology of CNS tumors in France: results from the gironde registry for the period 2000–2007. Neuro-Oncology. 2011;13(12):1370–1378. doi:10.1093/neuonc/nor120
4. Wöhrer A, Waldhör T, Heinzl H, et al. The Austrian brain tumour registry: a cooperative way to establish a population-based brain tumour registry. J Neurooncol. 2009;95(3):401. doi:10.1007/s11060-009-9938-9
5. Ostrom QT, Gittleman H, Fulop J, et al. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2008–2012. Neuro-Oncology. 2015;17(suppl4):iv1–iv62. doi:10.1093/neuonc/nov189
6. Davis FG, Freels S, Grutsch J, Barlas S, Brem S. Survival rates in patients with primary malignant brain tumors stratified by patient age and tumor histological type: an analysis based on surveillance, epidemiology, and end results (SEER) data, 1973–1991. J Neurosurg. 1998;88(1):1–10. doi:10.3171/jns.1998.88.1.0001
7. DeAngelis LM. Brain tumors. N Eng J Med. 2001;344(2):114–123. doi:10.1056/NEJM200101113440207
8. Miranda-Filho A, Piñeros M, Soerjomataram I, Deltour I, Bray F. Cancers of the brain and CNS: global patterns and trends in incidence. Neuro-Oncology. 2016;now166. doi:10.1093/neuonc/now166
9. Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–820. doi:10.1007/s00401-016-1545-1
10. Tabouret E, Nguyen AT, Dehais C, et al. Prognostic impact of the 2016 WHO classification of diffuse gliomas in the French POLA cohort. Acta Neuropathol. 2016;132(4):625–634. doi:10.1007/s00401-016-1611-8
11. Dubbink HJ, Atmodimedjo PN, Kros JM, et al. Molecular classification of anaplastic oligodendroglioma using next-generation sequencing: a report of the prospective randomized EORTC Brain Tumor Group 26951 Phase III trial. Neuro-Oncology. 2016;18(3):388–400. doi:10.1093/neuonc/nov182
12. Labussiere M, Di Stefano A, Gleize V, et al. TERT promoter mutations in gliomas, genetic associations and clinico-pathological correlations. Br J Cancer. 2014;111(10):2024–2032. doi:10.1038/bjc.2014.538
13. Network CGAR. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med. 2015;2015(372):2481–2498.
14. Louis DN, Ohgaki H, Wiestler OD, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114(2):97–109. doi:10.1007/s00401-007-0243-4
15. De Tayrac M, Saikali S, Aubry M, et al. Prognostic significance of EDN/RB, HJURP, p60/CAF-1 and PDLI4, four new markers in high-grade gliomas. PLoS One. 2013;8(9):e73332. doi:10.1371/journal.pone.0073332
16. Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. Molecular biology of the cell 4th edn (New York: garland science). Ann Bot. 2002;91:401.
17. Friedberg EC, Walker GC, Siede W, Wood RD. DNA Repair and Mutagenesis. American Society for Microbiology Press; 2005.
18. Lu A-L, Li X, Gu Y, Wright PM, Chang D-Y. Repair of oxidative DNA damage. Cell Biochem Biophys. 2001;35(2):141–170. doi:10.1385/CBB:35:2:141
19. Friedberg EC. How nucleotide excision repair protects against cancer. Nat Rev Cancer. 2001;1(1):22. doi:10.1038/35094000
20. Aquilina G, Bignami M. Mismatch repair in correction of replication errors and processing of DNA damage. J Cell Physiol. 2001;187(2):145–154. doi:10.1002/jcp.1067
21. Khanna KK, Jackson SP. DNA double-strand breaks: signaling, repair and the cancer connection. Nat Genet. 2001;27(3):247. doi:10.1038/85798
22. David SS, O’Shea VL, Kundu S. Base-excision repair of oxidative DNA damage. Nature. 2007;447(7147):941. doi:10.1038/nature05978
23. Richard G-F, Kerrest A, Dujon B. Comparative genomics and molecular dynamics of DNA repeats in eukaryotes. Microbiol Mol Biol Rev. 2008;72(4):686–727.
24. Haber JE. Partners and pathways: repairing a double-strand break. Trends Genet. 2000;16(6):259–264. doi:10.1016/S0168-9525(00)02022-9
25. Ciccia A, Constantinou A, West SC. Identification and characterization of the human mus81-eme1 endonuclease. J Biol Chem. 2003;278(27):25172–25178. doi:10.1074/jbc.M302882200
26. Boddy MN, Gaillard P-HL, McDonald WH, Shanahan P, Yates JR, Russell P. Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell. 2001;107(4):537–548. doi:10.1016/S0092-8674(01)00536-0
27. Taylor RM, Thistlethwaite A, Caldecott KW. Central role for the XRCC1 BRCT I domain in mammalian DNA single-strand break repair. Mol Cell Biol. 2002;22(8):2556–2563. doi:10.1128/MCB.22.8.2556-2563.2002
28. Siciliano M, Bachinski L, Dolf G, Carrano A, Thompson L Chromosomal assignments of human DNA-repair genes that complement chinese-hamster ovary (cho) cell mutants. Paper presented at: CYTOGENETICS AND CELL GENETICS1987.
29. Frederick GD, Amirkhan RH, Schultz RA, Friedberg EC. Structural and mutational analysis of the xeroderma pigmentosum group D (XPD) gene. Hum Mol Genet. 1994;3(10):1783–1788. doi:10.1093/hmg/3.10.1783
30. Coin F, Marinoni J-C, Rodolfo C, Fribourg S, Pedrini AM, Egly J-M. Mutations in the XPD helicase gene result in XP and TTD phenotypes, preventing interaction between XPD and the p44 subunit of TFIIH. Nat Genet. 1998;20(2):184. doi:10.1038/2491
31. Wilson DM, Coleman MA, Adamson AW, Christensen M, Lamerdin JE, Carney JP. Hex1: a new human Rad2 nuclease family member with homology to yeast exonuclease 1. Nucleic Acids Res. 1998;26(16):3762–3768. doi:10.1093/nar/26.16.3762
32. Schmutte C, Marinescu RC, Sadoff MM, Guerrette S, Overhauser J, Fishel R. Human exonuclease I interacts with the mismatch repair protein hMSH2. Cancer Res. 1998;58(20):4537–4542.
33. Miki Y, Swensen J, Shattuck-Eidens D, et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266(5182):66–71. doi:10.1126/science.7545954
34. Wang Y, Cortez D, Yazdi P, Neff N, Elledge SJ, Qin J. BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes Dev. 2000;14(8):927–939.
35. Friedenson B. The BRCA1/2 pathway prevents hematologic cancers in addition to breast and ovarian cancers. BMC Cancer. 2007;7(1):152. doi:10.1186/1471-2407-7-152
36. Maile EJ, Barnes I, Finlayson AE, Sayeed S, Nervous System AR. Intracranial tumour incidence by Ethnicity in England, 2001–2007: a descriptive epidemiological study. PLoS One. 2016;11(5):e0154347. doi:10.1371/journal.pone.0154347
37. Jacobs DI, Walsh KM, Wrensch M, et al. Leveraging ethnic group incidence variation to investigate genetic susceptibility to glioma: a novel candidate SNP approach. Front Genet. 2012;3:203. doi:10.3389/fgene.2012.00203
38. Goode EL, Ulrich CM, Potter JD. Polymorphisms in DNA repair genes and associations with cancer risk. Cancer Epidemiol Prev Biomarkers. 2002;11(12):1513–1530.
39. Wood RD, Mitchell M, Sgouros J, Lindahl T. Human DNA repair genes. Science. 2001;291(5507):1284–1289. doi:10.1126/science.1056154
40. Chang JS, Yeh R-F, Wiencke JK, et al. Pathway analysis of single-nucleotide polymorphisms potentially associated with glioblastoma multiforme susceptibility using random forests. Cancer Epidemiol Prev Biomarkers. 2008;17(6):1368–1373. doi:10.1158/1055-9965.EPI-07-2830
41. Adel Fahmideh M, Schwartzbaum J, Frumento P, Feychting M. Association between DNA repair gene polymorphisms and risk of glioma: a systematic review and meta-analysis. Neuro-Oncology. 2014;16(6):807–814. doi:10.1093/neuonc/nou003
42. Nicoloso MS, Sun H, Spizzo R, et al. Single-nucleotide polymorphisms inside microRNA target sites influence tumor susceptibility. Cancer Res. 2010;70(7):2789–2798. doi:10.1158/0008-5472.CAN-09-3541
43. Taron M, Rosell R, Felip E, et al. BRCA1 mRNA expression levels as an indicator of chemoresistance in lung cancer. Hum Mol Genet. 2004;13(20):2443–2449. doi:10.1093/hmg/ddh260
44. Quinn JE, James CR, Stewart GE, et al. BRCA1 mRNA expression levels predict for overall survival in ovarian cancer after chemotherapy. Clin Cancer Res. 2007;13(24):7413–7420. doi:10.1158/1078-0432.CCR-07-1083
45. Thrall M, Gallion H, Kryscio R, Kapali M, Armstrong DK, Deloia JA. BRCA1 expression in a large series of sporadic ovarian carcinomas: a gynecologic oncology group study. Int J Gynecol Cancer. 2006;16(Suppl 1):166–171. doi:10.1136/ijgc-00009577-200602001-00026
46. Kim H-T, Lee J-E, Shin E-S, et al. Effect of BRCA1 haplotype on survival of non–small-cell lung cancer patients treated with platinum-based chemotherapy. Journal of Clinical Oncology. 2008;26(36):5972–5979. doi:10.1200/JCO.2008.16.6496
47. Shim HJ, Yun JY, Hwang JE, et al. BRCA1 and XRCC1 polymorphisms associated with survival in advanced gastric cancer treated with taxane and cisplatin. Cancer Sci. 2010;101(5):1247–1254. doi:10.1111/j.1349-7006.2010.01514.x
48. Laroche-Clary A, Chaire V, Le Morvan V, et al. BRCA1 haplotype and clinical benefit of trabectedin in soft-tissue sarcoma patients. Br J Cancer. 2015;112(4):688–692. doi:10.1038/bjc.2014.624
49. Zhang X, Wei J, Zhou L, et al. A functional BRCA1 coding sequence genetic variant contributes to risk of esophageal squamous cell carcinoma. Carcinogenesis. 2013;34(10):2309–2313. doi:10.1093/carcin/bgt213
50. Alyahri N, Abdi S, Khan W, et al. Novel associations between brca1 variants C. 181 T> G (Rs28897672) and ovarian cancer risk in saudi females. J Med Biochem. 2019;38(1):13–21. doi:10.2478/jomb-2018-0037
51. Xu G-P, Zhao Q, Wang D, et al. The association between BRCA1 gene polymorphism and cancer risk: a meta-analysis. Oncotarget. 2018;9(9):8681. doi:10.18632/oncotarget.24064
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.

