Back to Journals » Infection and Drug Resistance » Volume 16
The Impact of mNGS Technology in the Etiological Diagnosis of Severe Pneumonia in Children During the Epidemic of COVID-19
Authors Yu C, Guo W, Zhang Z, Ma Y, Cao X, Sun N, Cui Y, Wang Y, Cui W, Xu Y , Zhan J
Received 28 January 2023
Accepted for publication 10 April 2023
Published 21 April 2023 Volume 2023:16 Pages 2395—2402
DOI https://doi.org/10.2147/IDR.S403851
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Changjiang Yu,1,* Wei Guo,1,2,* Zhulai Zhang,1,* Yuting Ma,3,* Xiaobei Cao,1 Na Sun,1 Yingyao Cui,1 Yunshen Wang,1 Wenyu Cui,1 Yongsheng Xu,1 Jianghua Zhan2,4
1Department of Pediatric Respiratory Medicine, Tianjin Children’s Hospital, Tianjin University Children’s Hospital, Tianjin, People’s Republic of China; 2Graduate School of Tianjin Medical University, Tianjin, People’s Republic of China; 3Department of Infection, Tianjin Children’s Hospital, Tianjin University Children’s Hospital, Tianjin, People’s Republic of China; 4Department of Pediatric Surgery, Tianjin Children’s Hospital, Tianjin University Children’s Hospital, Tianjin, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Yongsheng Xu, Department of Pediatric Respiratory Medicine, Tianjin Children’s Hospital, Tianjin University Children’s Hospital, Tianjin, 300134, People’s Republic of China, Email [email protected] Jianghua Zhan, Graduate School of Tianjin Medical University, Department of Pediatric Surgery, Tianjin Children’s Hospital, Tianjin University Children’s Hospital, Tianjin, 300134, People’s Republic of China, Email [email protected]
Purpose: Metagenomic next-generation sequencing (mNGS) is an emerging technique for pathogen detection. However, most literature on the clinical application of pediatrics generally comprises case reports or small-scale cohort studies.
Patients and Methods: A total of 101 children with community-acquired severe pneumonia admitted to Tianjin Children’s Hospital from November 2021 to February 2022 were included. Pathogens in bronchoalveolar lavage fluid (BALF) specimens were detected using mNGS. The performances of mNGS and conventional tests on pulmonary infection diagnosis and pathogen identification were compared.
Results: According to our data, mNGS had a broader spectrum for pathogen detection. The mNGS results of BALF showed that the number of children with severe pneumonia hospitalized for mycoplasma pneumoniae infection was more than that for other bacterial infections during the COVID-19 epidemic. In addition, 43 cases (42.6%) had been identified with mixed infection, including 36 cases (35.6%) of Mycoplasma pneumoniae mixed with other pathogenic bacteria. Analytically, the mNGS exhibited significantly enhanced detection in the BALF as compared with the conventional laboratory pathogenic detection approaches (P < 0.05). The Pearson correlation analysis revealed positive correlation between the time of fever during hospitalization and the number of mycoplasma sequences (P < 0.05).
Conclusion: Compared with traditional methods, mNGS has a higher etiological detection rate and can comprehensively detect various pathogens of severe pneumonia. Therefore, mNGS of bronchoalveolar lavage fluid should be performed in children with severe pneumonia, which is of great significance for guiding treatment.
Keywords: mNGS, severe pneumonia, pathogen, BALF, children
Introduction
Pneumonia is a common disease in children and is the leading cause of mortality in children under 5, worldwide. It is a serious health concern that threatens children’s health and has attracted remarkable attention from clinicians around the globe.1,2 The World Health Organization reported 740180 children death under the age of 5 in 2019.3 The common pathogenic detection approaches for etiological diagnosis include traditional microbial culture, nucleic acid amplification detection, molecular immunology test, and so on. However, these diagnostic procedures are controversial because of recognized limitations such as a narrow spectrum of pathogen detection, lacking both in timeliness and sensitivity, and so on.4 Recent advances in the detection of pathogenic microorganisms have led to the emergence of rapid detection approach, metagenomic next-generation sequencing (mNGS) technology, which has changed the landscape and spectrum of the etiological diagnosis of critical and difficult cases.
We have done mNGS studies of bronchoalveolar lavage fluid with community-acquired pneumonia, and found that the positive coincidence rate of the mNGS method for pathogens was much higher than that of the CT method (culture and targeted polymerase chain reaction tests).5 However, further studies are needed for severe pneumonia. This study aimed to investigate the significance of mNGS examination of alveolar lavage fluid in children with severe pneumonia.
Materials and Methods
Patients and Definitions
One hundred and one children with community-acquired severe pneumonia treated in Tianjin Children’s Hospital were selected from November 2021 to February 2022. All children were excluded from COVID-19 by nasopharyngeal PCR. Among the 101 children, 44 were males and 57 were females. The age of the children ranged from 1 to 16 years old. Inclusion criteria were as follows: (1) All children met the WHO diagnostic criteria for severe pneumonia,6 and fiberoptic bronchoscopy was performed.7 (2) The clinical data of children were complete. (3) Informed consent was signed for bronchoscopy and mNGS examination. Exclusion criteria were as follows: (1) Hospitalization for other reasons. (2) Basic diseases such as immune deficiency disease. General demographic data, clinical data, and imaging information were collected material. The clinical characteristics of the children are shown in Table 1.
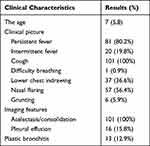 |
Table 1 Clinical Characteristics of Severe Pneumonia in Children |
mNGS Detection
mNGS detection was performed by a medical laboratory. Its microbial database covers bacteria, fungi, atypical pathogens (Mycoplasma, Chlamydia, Bartonella, etc.), viruses, and parasites. NGS technology extracts total nucleic acid from clinical samples, removes human nucleic acid through dehostization, randomly interrupts PCR-Free library construction, and performs quantitative quality inspection of Qubit library. Bioinformatics analysis identifies specific microbial species and then excludes the microbial community background that often occurs in different sampling sites and specimen types to find possible pathogenic microorganisms in the detected clinical specimens. The genome sequence was obtained from the National Center of Biotechnology Information (fp:/fip.acbi alm nhgov/genomes/). The pathogen detection index provided by mNGS technology includes the number of specific nucleic acid sequence reads identified by pathogen. The larger the number of reads, the higher the relative content of the pathogen detected.
Statistical Analysis
SPSS 22.0 software was used for data processing and statistical analysis. The measurement data that conformed to the normal distribution were expressed as ±s, and the t-test was used for the comparison between groups; the measurement data that was not normally distributed was expressed as M (Q1, Q3), and the comparison between groups was used Mann–Whitney rank sum test. The correlation between continuous variables was assessed using Pearson’s correlation test. The enumeration data were expressed as cases (%), the detection rate of each group was compared by χ²-test, and P<0.05 was considered statistically significant.
Result
Basic Situation
The patients underwent the first fiberoptic bronchoscopy 1–3 days after admission. The results revealed 58 cases (57.4%) of single infection, including 51 cases (50.5%) of Mycoplasma pneumoniae (MP), 5 cases (4.9%) of Streptococcus pneumoniae (Sp), 1 case (1%) each for Haemophilus influenzae (Hi) and human herpesvirus 6B. In addition, 43 cases (42.6%) were found with mixed infection, including 36 cases (35.6%) of Mycoplasma pneumoniae with other pathogenic bacteria and 7 cases (6.9%) of bacteria with bacteria/virus mixed infection. The detailed results are shown in Table 2.
 |
Table 2 Detection of Pathogens in Bronchoalveolar Lavage Fluid with mNGS |
Mycoplasma was the most important pathogen in this study, accounting for 86.1% of the total cases. The infection detection rates of male and female children were 93.2% and 80.7%, respectively, and exhibited no difference in the detection rates of different genders (P= 0.07). The sample size in early childhood (1–3 years old) was relatively small, with 4 cases, of which 3 cases of Mycoplasma infection. In 31 cases of preschool age (3–6 years old) and 66 cases of school age (6–12 years old), the detection rates of mycoplasma were 61.3% and 98.5%, respectively. The variation was found statistically significant (P< 0.001), indicating that school-age children are more susceptible to Mycoplasma pneumonia.
Comparison of Traditional Detection Methods and mNGS in Different Types of Pathogens
The comparison between mNGS and traditional methods is shown in Table 3. The mNGS portend significantly higher etiological detection rate compared with traditional detection methods: bacteria (12.87% vs 2.97%), mycoplasma (86.13% vs 44.55%), and the extent of variation was statistically significant (P<0.05). Tuberculosis, fungal and viral infections were not detected by traditional methods in this study. One case of tuberculosis infection, 3 cases of fungal infection, and 24 cases of viral infection were found in NGS examination.
 |
Table 3 Comparison of Traditional Methods and mNGS in Different Kinds of Pathogens |
Correlation Analysis of the Number of Sequences Detected by Mycoplasma in NGS
The Pearson correlation analysis revealed a positive correlation between the time of fever and the number of mycoplasma sequences in bronchoalveolar lavage fluid during hospitalization (P=0.049). However, the count number of white blood cells, CRP, PCT, LDH, and FER exhibit no significant correlation (P> 0.05), as shown in Table 4. The above-mentioned findings indicated that the number of mycoplasma sequences are associated with disease recovery. The higher the number of mycoplasma sequences, the longer the time span for a child’s body temperature to return to normal during hospitalization.
 |
Table 4 Correlation Analysis of the Number of Mycoplasma Sequences |
The Difference Between Pure Mycoplasma Infection and Mycoplasma Mixed Infection
Two groups of patients were included: single Mycoplasma pneumoniae infection group (51 cases) and a mixed infection group of Mycoplasma pneumoniae with pathogenic bacteria (36 cases). Both groups of patients were compared and analyzed for the number of white blood cells, CRP, PCT, LDH, FER, and other indicators, as shown in Table 5. The results revealed the absence of significant differences between the two groups, making it difficult to distinguish simple mycoplasma infection from mixed infection through blood tests. mNGS of BALF are of great significance for children with mixed infections.
 |
Table 5 Comparison of Single Infection and Mixed Infection Groups |
Clinical Impact of mNGS Result
The clinical impact and role of mNGS results are shown in Figure 1. A total of 51 children had their treatment changed as a result of mNGS results (both positive and negative), representing approximately 50% of the total number of patients. Forty-two children were treated with first-line macrolide antibiotics after positively diagnosing mycoplasma pneumonia. In addition, 3 children with Staphylococcus aureus infection were treated with linezolid, 3 children with fungal infection were administered antifungal therapy, and 1 patient was diagnosed with tuberculosis infection and received active anti-tuberculosis treatment. Another mixed-infected patient diagnosed with adenovirus pneumonia was treated with human immunoglobulin. One patient discontinued antibiotics regimen after confirming only human herpesvirus 6B infection.
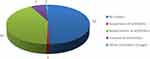 |
Figure 1 Clinical impact of mNGS result. |
Discussion
The COVID-19 outbreak fetched significant concerns to the global health and medical environment. China devised a strong prevention and management strategy after being affected badly by pandemic, resulting in the elevated use of broad-spectrum antibiotics. Antimicrobial resistance (AMR) will likely become an important challenge in the post-pandemic era for the foreseeable future.8 Therefore, higher requirements are put forward for the detection method of pneumonia etiology. Recently, mNGS has achieved remarkable results with high sensitivity in diagnosing clinical infectious diseases and discovering new pathogens. Using high-throughput sequencing technology to analyze patient samples’ nucleic acid content, the diagnosis spans a broad spectrum of infections and identifies many potential pathogens than any routine tests.9 Additionally, it outwitted some of the limitations of routine testing and helped achieve clinically actionable results within a reasonable time frame. Considering the potential of some sequencing platforms to provide same-day results, mNGS could revolutionize routine diagnosis.10 To our knowledge, this article is currently the largest mNGS study based on pediatric cases.
Our study found that among 101 patients with severe pneumonia, only 18 cases were infected with Streptococcus pneumoniae, and 87 were infected with mycoplasma. The number of mycoplasma infections was about 4.8 times that of streptococcus pneumoniae infections, higher than the results of other studies.11–13 This may be related to the strengthening of prevention of respiratory diseases in children during the COVID-19 epidemic, resulting in changes in the types of pathogenic bacteria susceptible to children, which requires further long-term observation. Although there are regional differences in the pathogen spectrum of children with respiratory tract infections, it can be seen from the results of this study and literature report that in recent years, MP was the most commonly detected bacteria among children aged ≥5 years hospitalized with CAP.14 In children 3–15 years of age, Mycoplasma pneumoniae pneumonia (MPP) accounts for 7–20% of community-acquired pneumonia.15,16 In recent years, reports of severe Mycoplasma pneumoniae pneumonia (SMPP) and refractory Mycoplasma pneumoniae pneumonia (RMPP) have gradually increased.17,18 Mixed infection is relatively common in SMPP or RMPP.17,19 Therefore, when the initial macrolide treatment effect is not good, it should be noted that it is a factor of drug-resistant strains or mixed infections. Mixed infection should be considered for children with RMPP combined with typical signs of bacterial infection, such as lung abscess, empyema, and sharply elevated inflammatory markers. Empirical antimicrobial therapy should be administered in addition to basic treatment. However, it is necessary to actively search for other pathogenic evidence for cases without typical manifestations. The application of NGS detection from BALF portends an opportunity for researchers to effectively solve this problem. The composition analysis of the pathogen spectrum not only provides the diagnostic basis for clinical empirical treatment, but also prevents downstream complications in advance, shortens the treatment time, improves prognosis, and relieves the pain in children.
mNGS is distinguished from the traditional test group in term of the positive rate of samples. By obtaining the sequence information of all nucleic acid fragments in the sample, mNGS can detect all types of microorganisms in a relatively short period by reading biological information with higher sensitivity and specificity.20 However, some studies reported a lower positive rate of blood NGS with low sensitivity,21 indicating that NGS is not suitable for all types of pathogenic examination. Although sputum culture isolation is a time-consuming approach with poor sensitivity and relatively difficult to obtain evidence of etiology, it is still hailed as the core method for detecting pathogenic microorganisms.22 The advantage is that, on the one hand, the cultured isolates are live microorganisms, not dead bacteria, which helps to identify pathogenic bacteria. On the other hand, culture can execute strain identification and drug susceptibility testing of positive pathogenic microorganisms and provide information such as drug susceptibility/resistance and minimum inhibitory concentration, which is convenient for guiding clinical anti-infection treatment. The diagnosis of M. pneumoniae primarily relies on culture, serology, and nucleic acid amplification. However, the limited experimental site and detection time of the above-mentioned conventional detection method is the leading cause of using serum-specific IgM antibody as primary method for the diagnosis of Mycoplasma pneumoniae.23 But it has a time window and individual differences in antibody production, an indication for false negatives.24 MP-IgM generally begins to increase within 1 week of initial infection, peaks at 3rd week and then declines gradually.25 The persistence period is up to several years, which is not affected by antibiotic treatment. Therefore, mNGS, in combination with traditional methods, can draw more accurate conclusions.
mNGS results have significant clinical implications in diagnosis and treatment. Although all diagnoses required a combination of clinical manifestations, mNGS was used for the final diagnosis in 53 patients (52.5%). A retrospective study in Shanghai, China, found that a significant proportion of infections were modified based on mNGS (61.4%).26 It is changing the clinician’s way of thinking about the treatment of undiagnosed patients, shifting the focus from traditional medicine to genome-based diagnosis.27 mNGS results also directly led to changes in treatment. Because of the rapid etiologic results, antibiotics can be escalated or adjusted as quickly as possible. mNGS can also rule out bacterial infections, thus reducing the use of antibiotics. Based on mNGS results, treatment regimens can be optimized, and drug resistance caused by empirical medication can be reduced, reducing medical costs.
In children with severe community-acquired pneumonia, mNGS in bronchoalveolar lavage fluid exhibit a higher detection rate than traditional methods, especially in the detection of bacteria, mycoplasma, and co-infections, and aids in timely and accurate clinical interventions to provide a basis for clinical diagnosis and treatment. In 2018, the US Food and Drug Administration (FDA) released a set of guidelines to promote the efficient development of mNGS technology and guide the management and treatment method.28 In 2020, Chinese experts proposed the “Expert Consensus on the Clinical Application of Metagenomics Sequencing Technology in Moderate and Severe Infections (1st Edition)”.29 mNGS has been the center of attention for many experts and scholars. However, we also found that the cost of mNGS as a new detection method is still high,30 especially for developing countries. In this study, only DNA was tested, and RNA was not sequenced, mainly due to economic factors. Additional information needed to be obtained from mNGS data in terms of detection of virulence factors, identification and typing of antimicrobial resistance markers, and minimum inhibitory concentrations (MICs), but these are still in their experimental stages.10 With the continuous development of technology, mNGS will show a broader application prospect.
Data Sharing Statement
The raw sequence data reported in this paper have been deposited in the Genome Sequence Archive (Genomics, Proteomics & Bioinformatics 2021) in National Genomics Data Center (Nucleic Acids Res 2022), China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences (GSA: CRA008160) that are publicly accessible at https://ngdc.cncb.ac.cn/gsa.
Ethics Approval and Informed Consent
This study was conducted complying with the Declaration of Helsinki. This study was reviewed and approved by the Ethics Committee of Tianjin Children’s Hospital. All the patients’ parents gave their Parental written consent for this study. All methods were carried out in relevant regulations and guidelines.
Acknowledgments
We thank all participants and staff of this study and the physicians at the Tianjin University Children’s Hospital. Changjiang Yu, Wei Guo, Zhulai Zhang and Yuting Ma are co-first authors for this study.
Funding
This work was supported by Tianjin Science and Technology Bureau (21JCYBJC00460) and Tianjin Children’s Hospital (Y2020013).
Disclosure
The authors declare no conflicts of interest in this work.
References
1. de Benedictis FM, Kerem E, Chang AB, et al. Complicated pneumonia in children. Lancet. 2020;396(10253):786–798. doi:10.1016/S0140-6736(20)31550-6
2. Alamneh YM, Adane F. Magnitude and predictors of pneumonia among under-five children in Ethiopia: a systematic review and meta-analysis. J Environ Public Health. 2020;2020:1606783. doi:10.1155/2020/1606783
3. World Health Organization (WHO). Pneumonia. [Updated November 11, 2021].Available from: https://www.who.int/news-room/fact-sheets/detail/pneumonia.
4. Qian YY, Wang HY, Zhou Y, et al. Improving pulmonary infection diagnosis with metagenomic next generation sequencing. Front Cell Infect Microbiol. 2020;10:567615. doi:10.3389/fcimb.2020.567615
5. Guo W, Cui X, Wang Q, et al. Clinical evaluation of metagenomic next-generation sequencing for detecting pathogens in bronchoalveolar lavage fluid collected from children with community-acquired pneumonia. Front Med. 2022;9:952636. doi:10.3389/fmed.2022.952636
6. Bradley JS, Byington CL, Shah SS, et al. The management of community-acquired pneumonia in infants and children older than 3 months of age: clinical practice guidelines by the pediatric infectious diseases society and the infectious diseases society of America. Clin Infect Dis. 2011;53:e25–76. doi:10.1093/cid/cir531
7. Faro A, Wood RE, Schechter MS, et al. Official American Thoracic Society technical standards: flexible airway endoscopy in children. Am J Respir Crit Care Med. 2015;191:1066–1080. doi:10.1164/rccm.201503-0474ST
8. Haqqi A, Awan UA, Ahmed H, Afzal MS. Antimicrobial resistance vs COVID-19: a bigger challenge in the post-pandemic era. J Formos Med Assoc. 2021;120(1537–1538):1537–1538. doi:10.1016/j.jfma.2021.03.003
9. Miller S, Chiu C. The role of metagenomics and next-generation sequencing in infectious disease diagnosis. Clin Chem. 2021;68:115–124. doi:10.1093/clinchem/hvab173
10. Schuele L, Cassidy H, Peker N, Rossen JWA, Couto N. Future potential of metagenomics in microbiology laboratories. Expert Rev Mol Diagn. 2021;21:1273–1285. doi:10.1080/14737159.2021.2001329
11. Su DQ, Huang HL, Zhuo ZQ. Pathogen distribution and bacterial resistance in children with severe pneumonia: a single-center retrospective study. Medicine. 2021;100:e27128. doi:10.1097/MD.0000000000027128
12. Gareca Perales J, Soleto Ortiz L, Loayza Mafayle R, et al. Diagnosis of community-acquired pneumonia in hospitalized children: a multicenter experience in Bolivia. Pediatr Infect Dis J. 2021;40:32–38. doi:10.1097/INF.0000000000002909
13. Wang H, Gu J, Li X, et al. Broad range detection of viral and bacterial pathogens in bronchoalveolar lavage fluid of children to identify the cause of lower respiratory tract infections. BMC Infect Dis. 2021;21:152. doi:10.1186/s12879-021-05834-0
14. Kutty PK, Jain S, Taylor TH, et al. Mycoplasma pneumoniae Among children hospitalized with community-acquired pneumonia. Clinical infectious diseases: an official publication of the infectious diseases society of America. Clin Infect Dis. 2019;68:5–12. doi:10.1093/cid/ciy419
15. Shim JY. Current perspectives on atypical pneumonia in children. Clin Exp Pediatr. 2020;63:469–476. doi:10.3345/cep.2019.00360
16. Krafft C, Christy C. Mycoplasma pneumonia in children and adolescents. Pediatr Rev. 2020;41:12–19. doi:10.1542/pir.2018-0016
17. Zhou Y, Shan Y, Cui Y, et al. Characteristics and outcome of severe mycoplasma pneumoniae pneumonia admitted to PICU in Shanghai: a retrospective cohort study. Crit Care Explor. 2021;3:e0366. doi:10.1097/CCE.0000000000000366
18. Gong H, Sun B, Chen Y, Chen H. The risk factors of children acquiring refractory mycoplasma pneumoniae pneumonia: a meta-analysis. Medicine. 2021;100:e24894. doi:10.1097/MD.0000000000024894
19. Zhang X, Chen Z, Gu W, et al. Viral and bacterial co-infection in hospitalised children with refractory Mycoplasma pneumoniae pneumonia. Epidemiol Infect. 2018;146:1384–1388. doi:10.1017/S0950268818000778
20. Gu W, Deng X, Lee M, et al. Rapid pathogen detection by metagenomic next-generation sequencing of infected body fluids. Nat Med. 2021;27:115–124. doi:10.1038/s41591-020-1105-z
21. Hogan CA, Yang S, Garner OB, et al. Clinical impact of metagenomic next-generation sequencing of plasma cell-free DNA for the diagnosis of infectious diseases: a multicenter retrospective cohort study. Clin Infect Dis. 2021;72:239–245. doi:10.1093/cid/ciaa035
22. van Belkum A, Durand G, Peyret M, et al. Rapid clinical bacteriology and its future impact. Ann Lab Med. 2013;33:14–27. doi:10.3343/alm.2013.33.1.14
23. Kumar S, Garg IB, Sethi GR. Serological and molecular detection of Mycoplasma pneumoniae in children with community-acquired lower respiratory tract infections. Diagn Microbiol Infect Dis. 2019;95:5–9. doi:10.1016/j.diagmicrobio.2019.03.010
24. Copete AR, Vera C, Herrera M, et al. Mycoplasma pneumoniae in children with and without community-acquired pneumonia. What do PCR and serology say? Pediatr Infect Dis J. 2020;39:e104–e108. doi:10.1097/INF.0000000000002636
25. Lin LJ, Chang FC, Chi H, et al. The diagnostic value of serological studies in pediatric patients with acute Mycoplasma pneumoniae infection. J Microbiol Immunol Infect. 2020;53:351–356. doi:10.1016/j.jmii.2018.09.001
26. Miao Q, Ma Y, Wang Q, et al. Microbiological diagnostic performance of metagenomic next-generation sequencing when applied to clinical practice. Clin Infect Dis. 2018;67:S231–s240. doi:10.1093/cid/ciy693
27. Han D, Li Z, Li R, et al. mNGS in clinical microbiology laboratories: on the road to maturity. Crit Rev Microbiol. 2019;45:668–685. doi:10.1080/1040841X.2019.1681933
28. U.S. Food and Drug Administration (FDA). Considerations for Design, Development, and Analytical Validation of next Generation Sequencing (Ngs)–based in vitro Diagnostics (IVDs) Intended to Aid in the Diagnosis of Suspected Germline Diseases. [Updated April 13, 2018]. Available from: https://www.fda.gov/downloads/MedicalDevices/DeviceRegulationandGuidance/GuidanceDocuments/UCM509838.pdf.
29. Consensus Group Of Experts On Application Of Metagenomic Next Generation Sequencing In The Pathogen Diagnosis In Clinical Moderate And Severe Infections, Professional Committee Of Sepsis And Shock Chinese Research Hospital Association, Professional Committee Of Microbial Toxins Chinese Society For Microbiology et al. 宏基因组学测序技术在中重症感染中的临床应用专家共识(第一版)[Expert consensus for the application of metagenomic next generation sequencing in the pathogen diagnosis in clinical moderate and severe infections (first edition)]. Zhonghua Wei Zhong Bing Ji Jiu Yi Xue. 2020;32:531–536. Chinese. doi:10.3760/cma.j.cn121430-20200228-00095
30. Greninger AL. The challenge of diagnostic metagenomics. Expert Rev Mol Diagn. 2018;18:605–615. doi:10.1080/14737159.2018.1487292
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
