Back to Journals » OncoTargets and Therapy » Volume 12
The impact of adjuvant radiotherapy on molecular prognostic markers in gliomas
Authors Harat M , Blok M, Harat A , Soszyńska K
Received 8 January 2019
Accepted for publication 21 February 2019
Published 26 March 2019 Volume 2019:12 Pages 2215—2224
DOI https://doi.org/10.2147/OTT.S200818
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Takuya Aoki
Maciej Harat,1,2 Maciej Blok,2 Aleksandra Harat,3 Krystyna Soszyńska4
1Department of Oncology and Brachytherapy, Nicolaus Copernicus University, Ludwik Rydygier Collegium Medicum, Bydgoszcz, Poland; 2Unit of Radiosurgery and Radiotherapy of CNS, Department of Radiotherapy, Franciszek Lukaszczyk Oncology Center, Bydgoszcz, Poland; 3Department of Public Health, Nicolaus Copernicus University, Ludwik Rydygier Collegium Medicum, Bydgoszcz, Poland; 4Department of Pathology, Laboratory of Clinical Genetics and Molecular Pathology, 10th Military Hospital, Bydgoszcz, Poland
Purpose: Changes in MGMT promoter methylation, IDH1 and IDH2 mutation, and 1p/19q co-deletion status in gliomas between first and subsequent resections and their associated clinical factors are poorly described. In this study, we assayed these biomarkers in the clinical setting.
Patients and methods: We used multiplex ligation-dependent probe amplification to measure MGMT promoter methylation, IDH mutation status, and 1p/19q co-deletion in 45 paired tumor samples from patients undergoing resection and subsequent re-resections for gliomas.
Results: Molecular changes were present in 20 patients (44%). At least one molecular characteristic changed over time in 89% of patients with primary grade III tumors. Gliomas with IDH wild-type and/or non-co-deleted were stable, but IDH1/2 mutation and/or co-deletion were sometimes lost at the time of recurrence. In a multivariate analysis, adjuvant radiotherapy alone was independently associated (P=0.02) with changes in molecular profile.
Conclusion: Molecular biomarkers change in gliomas during the course of the disease, most often MGMT methylation status. These changes in genetic profiles are related to adjuvant treatment with radiotherapy alone, which might be important for individualized treatment planning over the disease course.
Keywords: glioma, IDH, MGMT, 1p/19q, mutation changes, glioblastoma, anaplastic astrocytoma
Introduction
Central nervous system (CNS) tumors are characterized by certain recurrent molecular abnormalities. The WHO classification of CNS tumors1 emphasizes the importance of molecular testing, the inclusion of which is obligatory in the final histopathological report for consequent clinical decision-making. The most commonly tested biomarkers in CNS tumors are 1p/19q co-deletion, IDH1/IDH2 mutations, and methylation of the MGMT gene promoter. However, these alterations are not equivalent in terms of their impact on tumor classification, prognosis, and therapy.
Co-deletion of 1p/19q is considered a specific marker of oligodendrogliomas,2 the presence of which is closely related to isocitrate dehydrogenase (IDH) mutations.3 1p/19q co-deletion is of strong prognostic and predictive significance, being associated with a better prognosis and response to chemotherapy with alkylating drugs4 and chemoradiotherapy.5 Fluorescence in situ hybridization (FISH) is most commonly used to determine 1p/19q status.6
IDH is one of the key enzymes in the citric acid cycle. IDH mutations alter the enzyme and its product, 2-hydroxglutarate, resulting in epigenetic changes in tumor cells.7 IDH mutations are more common in WHO grade II and III tumors and in secondary gliomas after progression from a lower to higher grade, being relatively less common in de novo gliomas.8,9 IDH1/2 mutations are also prognostic, being associated with a decreased risk of aggressive disease in invasive WHO II and III gliomas10 and better responses to chemotherapy with alkylating drugs or radiotherapy.11,12
MGMT promoter methylation is a particularly important molecular event in WHO III and IV tumors. MGMT is a repair transferase that transfers O6-methylguanine to guanine. MGMT promoter methylation is associated with temozolomide responses in high-grade gliomas.13,14
However, despite the routine clinical characterization of basic mutations in CNS tumors, the full dynamics and characteristics of these mutations, for example, during disease progression, are unknown, despite possibly contributing to failure of targeted and non-targeted therapies. Even less is known about the clinical factors associated with mutations and their stability. IDH1/2 mutations are driver mutations that are expected to change only rarely; 1p/19q co-deletions are early but secondary alterations might be expected to change by clonal selection a bit more frequently (though still rarely); whereas MGMT methylation status is an epigenetic alteration that can change very easily and has no bearing on tumor classification, despite serving as a prognostic marker. Although studying these disparate alterations together cannot be regarded as a biologically driven approach, their impact on prognosis and WHO classification prompted us to examine them in a single study.
We therefore studied MGMT promoter methylation, IDH1/2 mutation status, and 1p/19q co-deletion in paired primary tumor and recurrence samples to examine the molecular dynamics of frequent alterations over the course of the disease in clinical practice.
Materials and methods
Patients and tissue collection
Four hundred and eighty-six consecutive glioma patients with 531 molecular diagnoses analyzed in the Department of Pathology, Laboratory of Clinical Genetics and Molecular Pathology, 10th Military Hospital between November 2015 and September 2018 were reviewed. The study was approved by the Nicolaus Copernicus University, Ludwik Rydygier Collegium Medicum institutional review board (number: KB 694/2018) and was conducted according to the principles of the Declaration of Helsinki. Patients who underwent biopsy at first and second surgeries were included. Patients provided written informed consent for every medical procedure analyzed in the study.
Tumor tissues were collected from both first and second surgeries of at least 2 months interval in 45 re-operations. In two patients, three surgeries were performed, in which case the first and last surgeries were analyzed. For each patient, the following clinical variables were defined: age, time interval between first and second surgeries (early, ≤12 months; late >12 months), tumor location, recurrences in the same lobe or distant, and treatment between diagnosis and recurrence (irradiation or irradiation plus chemotherapy).
Methylated MGMT, 1p/19q co-deletion, IDH1/IDH2 mutations, or MGMT methylation increasing at least by two categories (see MLPA section) was considered a favorable molecular profile, while a profile was considered unfavorable when none of the above was present, one favorable mutation was no longer present, or MGMT methylation decreased by least two categories.
Tumor samples and DNA extraction
Tumor specimens were formalin-fixed and paraffin-embedded. All samples were classified by histopathological examination and graded according to WHO 2016 guidelines. DNA was extracted using the Maxwell 16 FFPE Plus LEV DNA Purification Kit and Maxwell 16 Instrument (Promega Corporation, Fitchburg, WI, USA). DNA samples were purified using the DNA Clean & Concentrator Kit (Zymo Research, Irvine, CA, USA). For multiplex ligation-dependent probe amplification (MLPA), DNA was isolated from the blood of healthy volunteers for use as controls.
MLPA
MLPA and the SALSA MLPA P088-C1 kit (MRC-Holland, Amsterdam, the Netherlands) were used to detect IDH1 (R132H, R132C) and IDH2 (R172K, R172M) mutations and to detect loss of 1p and 19q. Methylation-specific MLPA (MS-MLPA) and the SALSA MS-MLPA ME011-B3 kit were used to perform promoter methylation analysis.15 Both MLPA and MS-MLPA assays were carried out by PCR according to the manufacturer’s protocol using 50 ng of normal and tumor DNA. Reference samples were included in each experiment. The PCR, DNA denaturation, and ligation steps were performed according to the manufacturer’s instructions. Amplified PCR products were separated by electrophoresis on an ABI PRISM 310 genetic analyzer (Thermo Fisher Scientific, Waltham, MA, USA) and, as an internal size standard, the LIZ-500 Genescan (Thermo Fisher Scientific) was used. Data were analyzed using the MRC-Coffalyser.Net (MRC-Holland). 1p/19q co-deletion by MLPA was defined as in Bienkowski et al,16 which clearly separates the complete co-deletion from isolated segmental deletions and normal copy numbers. Complete co-deletion was defined when most loci within each chromosomal region (subtelomeric 1p, rest of 1p, 19q) are deleted (<0.7), while isolated segmental deletion was defined when subtelomeric 1p (but not the rest of 1p) was significantly affected.
MS-MLPA is a semi-quantitative methylation profiling method. Methylation status was quantified by comparing MGMT probe relative peak area ratios in digested and undigested samples. Relative copy numbers were obtained by comparing MGMT probe relative peak area ratios with that obtained from a control sample. Methylation status was defined as absent or very low (0.00–0.25), low (0.25–0.50), moderate (0.50–0.75), or extensive (0.75).
Statistical analysis
Patient characteristics were described with mean and standard deviations or medians and ranges for continuous variables and by frequencies for categorical variables compared with Fisher’s exact test. Univariate and multivariate logistic regression was performed to evaluate the effects of clinical parameters on molecular changes. Analyses were performed using PQStat version 1.6.6.202. Two-tailed P-values <0.05 were considered significant and P-values <0.01 were considered highly significant.
Results
The pathological and molecular characteristics of the entire study cohort are summarized in Table 1, and individual tumor details are provided in Table S1. About 54% (n=264/486) of patients were male, and the average age was 47 years (range, 18–84 years); 53% (251/474) of gliomas were unmethylated or very low/low methylated, 19% (103/531) were 1p/19q co-deleted, 39% (207/531) had an IDH1 mutation, and 2% (10/531) an IDH2 mutation.
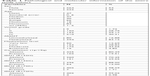  | Table 1 Pathological and molecular information of the entire study cohort |
A second surgery was performed in 9% (45/486) of cases. No WHO I, 11% (16/150) WHO II, 6% (9/152) WHO III, and 10% (20/210) WHO IV underwent re-operation during the study period.
The clinical characteristics of patients undergoing double surgery are presented in Table 2 and the molecular results in Table 3. There was no difference in the frequency of molecular alterations between the first and second surgeries (Table 3).
  | Table 2 Clinical variables of the study population undergoing surgery for recurrences |
  | Table 3 The pathological and molecular characteristics of patients undergoing double surgery |
Changes in molecular profiles
The molecular alterations in those patients undergoing second surgery are summarized in Figure 1. Overall, the molecular results changed in 44% (20/45) of cases in the second surgery. Changes in methylation level were not regarded as significant in eight cases (only one level of difference, eg, from very low to low). MGMT methylation was the most frequently observed change. Changes in both the histological and molecular characteristics occurred in 18% of cases (8/45), a change in histology alone was noted in 5% (2/45) of cases, and a change in the molecular profile alone in 26% (12/45) of cases.
Detailed directions and frequencies of changes for each biomarker are presented in Figure 2. While tumors did not change from non-co-deleted to co-deleted, 50% (4/8 patients) of cases lost co-deletion. In some cases, changes in co-deletion were equivocal but noted. There were also examples of loss of IDH1 (one case) and IDH2 mutations (one case) over the course of the disease. The first was in a man aged 39 years with fibrillary astrocytoma containing an IDH1 mutation and extensive MGMT methylation, which recurred 12 months after first surgery with transformation to anaplastic astrocytoma. MGMT methylation and IDH1 mutation were not present in the recurrent tumor. In another case, IDH2 was mutated in anaplastic oligoastrocytoma without grade transformation, with a concomitant decreased level of methylation and loss of co-deletion. Overall, 5% (2/45) of patients transitioned to a better prognostic type and 25% (11/45) of patients transitioned to a worse prognostic type.
Clinical variables and molecular changes
Molecular instability over time was not related to patient age (P=0.55) or sex (P=0.08), but was associated with tumor grade (P=0.0015). The highest percentage of changes in molecular characteristics was in grade III tumors, occurring in 89% (8/9) of cases (Table 4 Section A). Grade IV tumors were generally stable (80%, 16/20), while 44% (7/16) of grade II tumors underwent significant molecular changes, a quarter of which had significant prognostic influence. Malignant transformation was observed in 15% of cases (7/45). There was no correlation between molecular instability and grade transformation. The highest percentage of molecular changes (67%, 6/9 cases) without grade transformation was in WHO III group (Table 4 Section A). MGMT methylation level changed significantly more often in grade III tumors than other grades (P=0.0018, Table 4 Section A).
  | Table 4 Changes in molecular results of paired primary and recurrent tumors according to clinical variables |
There were no differences in general molecular stability between early and late recurrences. However, IDH1 mutations were typical of late recurrences (P=0.008, Table 4 Section B). There were no differences in molecular profile associated with age (Table 4 Section C) or primary tumor location and recurrence site (local or distant) (Table 4 Section D). However, these data are limited due to the presence of only five distant recurrences (stable profile in two cases and unstable in three cases).
Adjuvant radiotherapy alone but not radiochemotherapy was associated with molecular instability: almost 80% (11/14) of cases treated with radiotherapy had profile changes. Radiotherapy was associated with changes to a worse prognostic molecular profile (P=0.0058, Table 4 Section E). Patients with grade II gliomas showed a trend toward significant changes after irradiation to a better prognostic profile (P=0.0481).
In multivariate logistic regression analyses including all parameters (age, tumor grade, time between first and second surgeries, recurrence location, radiotherapy, and chemotherapy), only radiotherapy was independently associated with changes in molecular profile between surgeries (OR 334 [95% CI 2.2–50,098], P=0.023; Table 5).
  | Table 5 Multivariate logistic regression in the double surgery cohort |
Discussion
1p/19q co-deletion, MGMT promoter methylation, and IDH1/2 mutation testing have gained importance in routine clinical decision-making in patients with CNS tumors.17 While the value of these molecular markers at first diagnosis has been demonstrated,5,9,14,18 only a few studies have examined these biomarkers at the time of recurrence. Therefore, to our knowledge, this is one of the largest cohorts of gliomas analyzed with regard to changes in these biomarkers over the course of the disease. Consistent with data from whole-exome sequencing studies of astrocytic tumors,19–22 the mutational profiles at recurrence often changed.
The MLPA assay used in our study is a relatively new and cost-effective method for the detection of multiple genetic alterations in tumor tissue in a single procedure.16 MLPA has been shown to be reproducible and concordant with FISH or loss of heterozygosity assays for the presence or absence of the 1p/19q deletion.23,24 In a consecutive series of 165 diffuse gliomas tested in the clinical setting, MLPA analysis was shown to be accurate for the assessment of the most important molecular markers for diagnostic tumor typing and was recommended for routine diagnostic work-up.16
Identification of the 1p/19q allelic status in gliomas, primarily those with a major oligodendroglial component, has become an excellent molecular complement to tumor histology to identify tumors sensitive to chemotherapy. Although 1p/19q status is indicated in the routine clinical setting for prognostic assessment and aids in making therapeutic decisions, repeat 1p/19q testing of tumor recurrences is not recommended, as the deletion typically constitutes an early genetic event.25 Our results, however, suggest that changes may occur over the course of the disease that might be clinically relevant, and the frequency of losing the 1p/19q co-deletion status has not been commonly reported.19 Although 1p/19q co-deletion is an early tumorigenic event, IDH1/2 mutations are thought to precede them.6,26
Over 70% of anaplastic gliomas carry IDH mutations, with higher frequencies in oligodendroglial tumors.9 IDH1 alterations in high-grade tumors are known to be derived from earlier lesions.9,20 In most current published series, IDH1 mutations were never lost during progression and remained clonal in all progressed tumors.20–22 In one paper, IDH mutation status has been reported to change in a small subset of gliomas, most commonly due to the deletion of the mutant allele on chromosome 2 and, less commonly, amplification of the mutant allele in recurrent or post-therapy specimens.27 Although rare, the mechanism for the deletion appears to be selective pressure on the tumor to no longer be dependent on the mutant IDH1/2 protein once additional alterations provide a growth advantage.
We observed significant changes in IDH during the course of disease in two cases, which were mutated to wild-type but not vice versa. A multiple stereotactic biopsy study of grade II and III gliomas revealed intratumoral homogeneity of IDH1,28 suggesting that the differences observed here are unlikely to represent sampling of other parts of the same, incompletely resected tumor. However, other multiregional sampling analyses also identified a close correlation between regional heterogeneity and the history of clonal evolution that might account for our result.22
Grade III tumors were most often genetically unstable, and, when changes did occur, these were of adverse prognostic impact. Decreased MGMT promoter methylation was most frequent, IDH1 and IDH2 mutations were lost, and co-deletion 1p/19q decreased or was lost in 50% of cases at the time of recurrence. Systemic therapy may, therefore, be better administered at the time of primary diagnosis when the tumors are genetically favorable. It remains uncertain and of clinical trial interest of when to start treatment with alkylating agents.29 Our results raise concerns about the “treat-the-recurrence” strategy due to the possible change to a favorable profile over time. This hypothesis is in line with EORTC trial results supporting the use of early adjuvant PCV chemotherapy in patients with co-deleted tumors rather than at progression.4 Furthermore, these kinetics were not restricted to grade III tumors, since negative prognostic changes with respect to 1p/19 co-deletion were also found in grade II tumors. Grade IV tumors were relatively stable in terms of genetic alterations (80%), with methylated to no or low methylated status changes occurring in 15% of grade IV lesions.
We observed other molecular dynamics in low-grade gliomas with respect to MGMT methylation. The molecular phenotype changed from no or low methylated to high or very high methylated status, in line with the hypothesis that irradiation produces favorable genetic changes in grade II tumors. Therefore, our results support a strategy of irradiating unmethylated WHO grade II gliomas not only for therapy but also due to the favorable impact on genetic profile at recurrence that would introduce the possibility of effective salvage with temozolomide.
The potential influence of treatment on genetic stability has previously been discussed.20,30 Radiation exposure induces epigenetic effects in human cell lines,31,32 and treatment of breast cancer cells with ionizing radiation results in DNA methylation changes.33 Alterations in DNA methylation may alter gene expression.33 Our data provide supportive clinical evidence that tumor genetic profiles are changed by radiation, since radiotherapy was significantly associated with molecular alterations at second surgery in multivariate analysis. However, radiation was mostly given to patients with grade III tumors, and what factor has a stronger impact on molecular instability remains to be elucidated.
Genetic stability was not obviously correlated with early recurrence. This stable genetic profile has been noted elsewhere.34 The direct influence of treatment on MGMT promoter methylation status and protein expression has been examined in glioblastoma U343 cells;35 radiation, steroids, and temozolomide had no effect on MGMT promoter status. Another hypothesis is the existence of frequent spatial errors in radiation dose delivery that promote cellular clones are not altered by radiation. As highly infiltrative masses, glioblastomas are particularly susceptible to spatial errors during irradiation, which are common in MRI-based radiation therapy.36,37 In general, the worst molecular profiles were noted in grade III recurrences, suggesting that other molecular events may be important in these tumors, that is, no spatial error of target selection, but selective pressure of more aggressive cells epigenetically altered in response to radiation. In grade IV lesions, typical early recurrence of genetically the same tumors supports the concept of progression of the primary tumor due to ineffective treatment rather than true recurrence. Therefore, recurrent cells not changed by treatment could be recognized by genetic signature.
It is unclear whether all three alterations occurred in the same cell population or as a result of clonal expansion after treatment. Intratumoral heterogeneity, with distinct clones arising separately in different tumor areas and expansion of one clone due to alterations promoting survival or resistance to therapy, is often presented as the main reason for genetic changes over time.6 Parkinson et al38 studied intratumoral and between-treatment MGMT promoter methylation in ten glioblastoma multiforme patients and showed that differences between first and second surgeries occurred irrespective of primary tumor homogeneity. Conversely, another study observed only genetic discordance in cases with a primary GBM and heterogeneous MGMT methylation status, with possible subsequent subclonal expansion in the recurrence.39 A recent retrospective study of MGMT methylation in GBM showed stability in 75% of cases.34 However, while these results do not explain the nature of changes between GBM pairs, they are consistent with our findings that genetic alterations in WHO IV tumors are generally uncommon but when they do occur are unfavorable.
This study has some limitations. First, our methods are relatively simple and cannot provide deep knowledge about gliomagenesis from the whole genome perspective. Nevertheless, we analyzed and presented clinical data that are commonly used in practice. Second, the level of methylated MGMT and co-deletion may have been affected by the percentage of normal (unmethylated) tissue present in the sample. Moreover, whether very low MGMT methylation should be treated as methylated or unmethylated is currently unknown. However, we did not focus on the prognostic or predictive significance of the results of the assay, rather changes in the profile. To avoid observer bias, all measurements were blinded and verified in each case by the same medical geneticist and preselected by a pathologist to exclude inflammation or necrosis. While there are several assays for determining IDH1/2 mutations, 1p/19q co-deletion, and MGMT promoter methylation, there is currently no accepted gold standard. The 1p/19q MLPA assessment may result in misclassification and overestimation of the rate of 1p/19q co-deleted tumors; however, the same method and threshold were used in both the examinations. In addition, the nomenclature of changing to either better or worse molecular profiles in the recurrent specimen may be misleading, since whether the molecular alterations at the time of recurrence are of prognostic significance is currently unknown. Our findings regarding the influence of adjuvant radiotherapy alone on molecular profiles were limited by the small number of low-grade gliomas and grade III gliomas treated with radiochemotherapy. We expect to see more patients with grade III tumors treated with radiochemotherapy in the future, which might shed further light on this observation. Finally, while the study size was relatively small, this represents the largest paired cohort published to date.
Conclusion
Here we examined the results of the most common biomarkers tested in primary and recurrent gliomas in clinical practice. The status of the biomarkers changed during the course of the disease, most often MGMT methylation status. These changes in genetic profiles of gliomas were related to adjuvant treatment with radiotherapy alone, which might be important for individualized treatment planning over the disease course.
Acknowledgment
The authors received no specific funding for this work.
Disclosure
The authors report no conflicts of interest in this work.
References
Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 2016;131(6):803–820. doi:10.1007/s00401-016-1545-1 | ||
Burger PC, Minn AY, Smith JS, et al. Losses of chromosomal arms 1p and 19q in the diagnosis of oligodendroglioma. A study of paraffin-embedded sections. Mod Pathol. 2001;14(9):842–853. doi:10.1038/modpathol.3880400 | ||
Labussiere M, Idbaih A, Wang XW, et al. All the 1p19q codeleted gliomas are mutated on IDH1 or IDH2. Neurology. 2010;74(23):1886–1890. doi:10.1212/WNL.0b013e3181e1cf3a | ||
van Den Bent MJ, Brandes AA, Taphoorn MJ, et al. Adjuvant procarbazine, lomustine, and vincristine chemotherapy in newly diagnosed anaplastic oligodendroglioma: long-term follow-up of EORTC brain tumor group study 26951. J Clin Oncol. 2013;31(3):344–350. doi:10.1200/JCO.2012.43.2229 | ||
Cairncross G, Wang M, Shaw E, et al. Phase III trial of chemoradiotherapy for anaplastic oligodendroglioma: long-term results of RTOG 9402. J Clin Oncol. 2013;31(3):337–343. doi:10.1200/JCO.2012.43.2674 | ||
Horbinski C, Miller CR, Perry A. Gone FISHing: clinical lessons learned in brain tumor molecular diagnostics over the last decade. Brain Pathol. 2011;21(1):57–73. doi:10.1111/j.1750-3639.2010.00453.x | ||
Horbinski C. What do we know about IDH1/2 mutations so far, and how do we use it? Acta Neuropathol. 2013;125(5):621–636. doi:10.1007/s00401-013-1106-9 | ||
Hartmann C, Meyer J, Balss J, et al. Type and frequency of IDH1 and IDH2 mutations are related to astrocytic and oligodendroglial differentiation and age: a study of 1,010 diffuse gliomas. Acta Neuropathol. 2009;118(4):469–474. doi:10.1007/s00401-009-0561-9 | ||
Yan H, Parsons DW, Jin G, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360(8):765–773. doi:10.1056/NEJMoa0808710 | ||
Eckel-Passow JE, Lachance DH, Molinaro AM, et al. Glioma groups based on 1p/19q, IDH, and TERT promoter mutations in tumors. N Engl J Med. 2015;372(26):2499–2508. doi:10.1056/NEJMoa1407279 | ||
Hartmann C, Hentschel B, Wick W, et al. Patients with IDH1 wild type anaplastic astrocytomas exhibit worse prognosis than IDH1-mutated glioblastomas, and IDH1 mutation status accounts for the unfavorable prognostic effect of higher age: implications for classification of gliomas. Acta Neuropathol. 2010;120(6):707–718. doi:10.1007/s00401-010-0781-z | ||
Houillier C, Wang X, Kaloshi G, et al. IDH1 or IDH2 mutations predict longer survival and response to temozolomide in low-grade gliomas. Neurology. 2010;75(17):1560–1566. doi:10.1212/WNL.0b013e3181f96282 | ||
Esteller M, Garcia-Foncillas J, Andion E, et al. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med. 2000;343(19):1350–1354. doi:10.1056/NEJM200011093431901 | ||
Hegi ME, Diserens AC, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352(10):997–1003. doi:10.1056/NEJMoa043331 | ||
Jeuken JW, Cornelissen SJ, Vriezen M, et al. MS-MLPA: an attractive alternative laboratory assay for robust, reliable, and semiquantitative detection of MGMT promoter hypermethylation in gliomas. Lab Invest. 2007;87(10):1055–1065. doi:10.1038/labinvest.3700664 | ||
Bienkowski M, Wohrer A, Moser P, et al. Molecular diagnostic testing of diffuse gliomas in the real-life setting: A practical approach. Clin Neuropathol. 2018;37(4):166–177. doi:10.5414/NP301110 | ||
Weller M, Stupp R, Hegi ME, et al. Personalized care in neuro-oncology coming of age: why we need MGMT and 1p/19q testing for malignant glioma patients in clinical practice. Neuro Oncol. 2012;14(Suppl 4):iv100–iv108. doi:10.1093/neuonc/nos206 | ||
Jenkins RB, Blair H, Ballman KV, et al. A t(1;19)(q10;p10) mediates the combined deletions of 1p and 19q and predicts a better prognosis of patients with oligodendroglioma. Cancer Res. 2006;66(20):9852–9861. doi:10.1158/0008-5472.CAN-06-1796 | ||
Aihara K, Mukasa A, Nagae G, et al. Genetic and epigenetic stability of oligodendrogliomas at recurrence. Acta Neuropathol Commun. 2017;5(1):18. doi:10.1186/s40478-017-0422-z | ||
Bai H, Harmanci AS, Erson-Omay EZ, et al. Integrated genomic characterization of IDH1-mutant glioma malignant progression. Nat Genet. 2016;48(1):59–66. doi:10.1038/ng.3457 | ||
Johnson BE, Mazor T, Hong C, et al. Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science. 2014;343(6167):189–193. doi:10.1126/science.1239947 | ||
Suzuki H, Aoki K, Chiba K, et al. Mutational landscape and clonal architecture in grade II and III gliomas. Nat Genet. 2015;47(5):458–468. doi:10.1038/ng.3273 | ||
Franco-Hernandez C, Martinez-Glez V, de Campos JM, et al. Allelic status of 1p and 19q in oligodendrogliomas and glioblastomas: multiplex ligation-dependent probe amplification versus loss of heterozygosity. Cancer Genet Cytogenet. 2009;190(2):93–96. doi:10.1016/j.cancergencyto.2008.09.017 | ||
Natte R, van Eijk R, Eilers P, et al. Multiplex ligation-dependent probe amplification for the detection of 1p and 19q chromosomal loss in oligodendroglial tumors. Brain Pathol. 2005;15(3):192–197. | ||
Woehrer A, Hainfellner JA. Molecular diagnostics: techniques and recommendations for 1p/19q assessment. CNS Oncol. 2015;4(5):295–306. doi:10.2217/cns.15.28 | ||
Watanabe T, Nobusawa S, Kleihues P, Ohgaki H. IDH1 mutations are early events in the development of astrocytomas and oligodendrogliomas. Am J Pathol. 2009;174(4):1149–1153. doi:10.2353/ajpath.2009.080958 | ||
Mazor T, Chesnelong C, Pankov A, et al. Clonal expansion and epigenetic reprogramming following deletion or amplification of mutant IDH1. Proc Natl Acad Sci U S A. 2017;114(40):10743–10748. doi:10.1073/pnas.1708914114 | ||
Kunz M, Thon N, Eigenbrod S, et al. Hot spots in dynamic (18)FET-PET delineate malignant tumor parts within suspected WHO grade II gliomas. Neuro Oncol. 2011;13(3):307–316. doi:10.1093/neuonc/noq196 | ||
Wick W, Platten M, Meisner C, et al; NOA-08 Study Group of Neuro-oncology Working Group (NOA) of German Cancer Society. Temozolomide chemotherapy alone versus radiotherapy alone for malignant astrocytoma in the elderly: the NOA-08 randomised, phase 3 trial. Lancet Oncol. 2012;13(7):707–715. doi:10.1016/S1470-2045(12)70164-X | ||
Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. doi:10.1038/nature07385 | ||
Kovalchuk O, Burke P, Besplug J, Slovack M, Filkowski J, Pogribny I. Methylation changes in muscle and liver tissues of male and female mice exposed to acute and chronic low-dose X-ray-irradiation. Mutat Res. 2004;548(1–2):75–84. doi:10.1016/j.mrfmmm.2003.12.016 | ||
Pogribny I, Koturbash I, Tryndyak V, et al. Fractionated low-dose radiation exposure leads to accumulation of DNA damage and profound alterations in DNA and histone methylation in the murine thymus. Mol Cancer Res. 2005;3(10):553–561. doi:10.1158/1541-7786.MCR-05-0074 | ||
Antwih DA, Gabbara KM, Lancaster WD, Ruden DM, Zielske SP. Radiation-induced epigenetic DNA methylation modification of radiation-response pathways. Epigenetics. 2013;8(8):839–848. doi:10.4161/epi.25498 | ||
Brandes AA, Franceschi E, Tosoni A, et al. O(6)-methylguanine DNA-methyltransferase methylation status can change between first surgery for newly diagnosed glioblastoma and second surgery for recurrence: clinical implications. Neuro Oncol. 2010;12(3):283–288. doi:10.1093/neuonc/nop050 | ||
Jung TY, Jung S, Moon KS, et al. Changes of the O6-methylguanine-DNA methyltransferase promoter methylation and MGMT protein expression after adjuvant treatment in glioblastoma. Oncol Rep. 2010;23(5):1269–1276. | ||
Harat M, Malkowski B, Makarewicz R. Pre-irradiation tumour volumes defined by MRI and dual time-point FET-PET for the prediction of glioblastoma multiforme recurrence: A prospective study. Radiother Oncol. 2016;120(2):241–247. doi:10.1016/j.radonc.2016.06.004 | ||
Munck Af Rosenschold P, Costa J, Engelholm SA, et al. Impact of [18F]-fluoro-ethyl-tyrosine PET imaging on target definition for radiation therapy of high-grade glioma. Neuro Oncol. 2015;17(5):757–763. doi:10.1093/neuonc/nou316 | ||
Parkinson JF, Wheeler HR, Clarkson A, et al. Variation of O(6)-methylguanine-DNA methyltransferase (MGMT) promoter methylation in serial samples in glioblastoma. J Neurooncol. 2008;87(1):71–78. doi:10.1007/s11060-007-9486-0 | ||
Barresi V, Caffo M, De Luca G, Giuffrè G. O-6-methylguanine-DNA methyltransferase promoter methylation can change in glioblastoma recurrence due to intratumor heterogeneity. Glioma. 2018;1(6):208–213. doi:10.4103/glioma.glioma_38_18 |
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.


