Back to Journals » OncoTargets and Therapy » Volume 8
The functional role of microRNA in acute lymphoblastic leukemia: relevance for diagnosis, differential diagnosis, prognosis, and therapy
Authors Luan C , Yang Z, Chen B
Received 15 July 2015
Accepted for publication 9 September 2015
Published 13 October 2015 Volume 2015:8 Pages 2903—2914
DOI https://doi.org/10.2147/OTT.S92470
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Dr Faris Farassati
Chengxin Luan,1,* Zixue Yang,2,* Baoan Chen1
1Department of Hematology and Oncology, Zhongda Hospital, School of Medicine, Southeast University, 2State Key Laboratory of Bioelectronics, School of Biological Science and Medical Engineering, Southeast University, Nanjing, People’s Republic of China
*These authors contributed equally to this work
Abstract: MicroRNAs (miRNAs), a new class of noncoding RNAs, which can hybridize to target messenger RNAs and regulate their expression posttranscriptionally, express differentially in distinct stages of lymphopoiesis and influence the direction of lymphoid precursor maturation. Hence, there is aberrant expression of miRNAs involved in malignant lymphopoiesis, and these aberrations can be used as signatures of acute lymphoblastic leukemia (ALL) with different subtypes. In addition, changes in the expression of several miRNAs may have functional relevance with leukemogenesis or drug resistance. As a result, the reversal of the expression of these miRNAs may alleviate the disease to some extent and improve clinical outcomes. However, among the studies of miRNAs, there are still some problems that need to be solved to understand the function of miRNAs in ALL more thoroughly.
Keywords: ALL, microRNA, lymphopoiesis, molecular diagnosis, lymphoid malignant, molecular therapy
Introduction
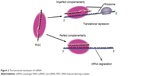
MicroRNAs (miRNAs) are small RNA sequences of approximately 18 to 25 nucleotides. They come from 70 to 100 nucleotide hairpin precursors cleaved by a complex protein system including the RNase III Drosha and Dicer1–4 shown in Figure 1. miRNAs sequences distribute throughout the whole genome and are classified as intergenic or intronic miRNA.5 Mature miRNAs repress and/or degenerate the protein-coding messenger RNAs (mRNAs) posttrancriptionally through interaction with 3′-untranslated region of mRNAs,1–3,6 and this is outlined in Figure 2. Lin-4 is the first discovered miRNA during a genetic screen of the nematode Caenorhabditis elegans by Lee et al in 1993.7 Currently, more than 24,000 miRNAs have been discovered, spanning more than 200 species including flora, fauna, and some microorganisms,8 and more than 2,000 human mature miRNAs have been identified and reported in miRBase and miRBase Tracker.9–13 The regulation of mRNA by miRNA is a common biological phenomenon.14 Friedman et al15 reported that approximately 60% of human mRNA could be regulated by miRNAs.
  | Figure 2 The functional mechanism of miRNA. |
Dysregulation of miRNAs has been discovered in different solid tumors and leukemia.16 The study illustrating the aforementioned phenomenon demonstrated that miRNAs were frequently localized in common breakpoint regions related to tumors or in fragile sites, minimal regions of heterozygosity lost, and minimal amplification regions.17 The first report published in 2004 showed that during murine hematopoiesis, miRNAs were expressed specially and regulated dynamically.18 Several groups described the miRNAs expression profile and/or function during the normal and malignant hematopoiesis in murine and humans.18–24
Acute lymphoblastic leukemia (ALL) is the most common hematologic malignancy in children, and its incidence peaks from 2 to 5 years of age, while it is relatively rare in adults.25–28 The classification of ALL by French–American–British cooperative group based on morphology had been abandoned because it failed to meet clinical relevance. The current classification system based on morphology, immunology, cytogenetics, and molecular biology was introduced by World Health Organization, while immunophenotyping based on cell surface and cytoplasmic proteins is more widely applied. According to immunophenotyping, ALL could be classified into two types, T-ALL and B-ALL. The main markers of T-ALL include the terminal deoxynucleotidyl transferase (TdT), CD2+, CD3+, CD4+, CD5+, CD7+, and CD8+. B-ALL mainly includes three subtypes: early pre-B-cell, pre-B-cell, and mature B-cell. The main markers of early pre-B-cell include TdT+, HLA-DR+, CD19+, D34+, and CD10+/−. The main markers of pre-B-cell include TdT+, HLA-DR+, CD19+, CD10+, and CD20+, and the main markers of mature B-cell include HLA-DR+, CD19+, CD10+/−, CD20+, and surface Ig (sIg)+.29 Recently, besides the immunophenotyping of ALL, an increasing number of studies showed that the miRNA expression profiles in acute leukemia have cooperative interactions in the development of leukemia. Therefore, the miRNA expression profile can be used as biomarkers in diagnosis, differential diagnosis, prognosis, and therapy of hematologic cancers.30–32 In this review, the role of miRNA expression profiles as biomarkers in diagnosis, differential diagnosis, prognosis, and therapy of ALL is summarized.
The function of miRNAs in normal lymphopoiesis
Hematopoiesis is a process by which multipotent hematopoietic stem cells (HSCs) self-renew and differentiate into different lineages cells continuously. Lymphopoiesis is a part of the hematopoiesis process by which HSCs differentiate into lymphoid progenitors and finally into B- or T-lymphocytes. The development of T-cells occurs in the thymus and the development of B-cells has two stages inside and outside the bone marrow (BM) separately. Nonetheless, their development and activation at the periphery are controlled by complex protein signaling pathways, which are regulated by the miRNAs.33–35
miRNA-150 is expressed in both mature B- and T-cells. The lymphoid progenitors express the miRNA-150 to give rise to the mature B-cells and assist in the transition from progenitor B-cell (pro-B) to the precursor B-cell (pre-B) stage. And premature expression of miRNA-150 results in blocked transition from the pro-B-cell stage to the pre-B-cell stage.20,36–38 In the thymus, the expression of miRNA-150 may enhance T-cell development and its mechanisms, including enhancing key pathways of T-cell development (like the Notch Pathway) and suppressing alternative lineage differentiation (like B-cell differentiation) in progenitor cells.39 C-Myb is a confirmed target of miRNA-150, and is an essential transcription factor involved in early lymphoid development. Its targeted loss in B-cells leads to the maturation arrest from pro-B to pre-B-cell stage, and, simultaneously, miRNA-150 is found to be overexpressed.36,40
B-cell differentiation is regulated by the miR-155-PU.1 axis, and the mechanism is that miR-155 inhibits PU.1 expression, which leads to Pax5 downregulation and the initiation of the plasma cell differentiation pathway.41 Recent data show the role of miRNA-155 in the differentiation of T-cells into different effectors T-helper (Th) cell subsets. miRNA-155 regulates the differentiation of T-cells into Th type 1 cells, and its absence results in the direct differentiation from T-cells to Th type 2 cells.33,34,42–44
miRNA-181 is composed of three clusters located in different chromosomes.20,32,45 miRNA-181 expression is high in the early B-cell differentiation stage and subsequently decreases.18 In addition, miRNA-181 plays an important role in T-cell development and is expressed highly in double-positive T-cells. Its targets are BCL-2, CD69, EGR1, and T-cell receptor, all involved in positive T-cell selection.46–48
miRNA-17-92 cluster consists of six miRNAs: miRNA-17, miRNA-18a, miRNA-19a, miRNA-20a, miRNA-19b-1, and miRNA-92-149 and is highly expressed in the B- and T-lymphoid precursors and is decreased after maturation. As well as miRNA-150, absence of the cluster leads to the development disorders of B-cells from pro-B to pre-B-cell stage, due to the increased levels of the proapoptotic protein BIM that is the target of the cluster.20,50 Another study has also showed consistency with the aforementioned conclusion.51 It demonstrated that mice with targeted overexpression of the miRNA-17-92 cluster during lymphopoiesis develop severe lymphoproliferative disorders and autoimmunity.20,51 miRNAs relevant to normal lymphopoiesis are shown in Figure 3.33,52
The function of miRNAs in ALL
The function of miRNAs in diagnosis of ALL
ALL is a lymphoid malignancy, and around three quarters of childhood ALL cases contain one or more total alterations of chromosome, and it may involve B- or T- lineages and have lymphoid maturation arrest in distinct stages, leaving different immunophenotypes with different miRNA signatures.53–57 So, these signatures can help the diagnosis and classification diagnosis of ALL. Compared with normal pediatric BM samples, miRNA-100, miRNA-196b, and let-7e were expressed at a lower level in BM samples of pediatric ALL, while miRNA-128a and miRNA-181b were overexpressed. miRNA-100 was related to t(12; 21) positive ALL.58 A case-control study with 570 Chinese childhood ALL cases and 673 cancer-free controls suggested that miRNA-196a2 T>C polymorphism might increase the risk of ALL of children.59 B- and T-lineage ALL can be discriminated by the expression of miRNA-148, miRNA-151, and miRNA-424. Furthermore, B-lineage ALL subsets with special molecular lesions can be differentiated by a set of six miRNAs – miRNA-425-5p, miRNA-191, miRNA-146b, miRNA-128, miRNA-629, and miRNA-126 – which was highlighted by one-way analysis of variance.60 Zhang et al61 observed differential expression patterns of ALL which were composed of several known miRNAs and 20 newly identified miRNAs that had first been discovered at the genomic level in human ALL. These patterns constituted an ALL-specific miRNA signature for diagnosis. Gutierrez-Camino et al62 analyzed 118 single nucleotide polymorphisms (SNPs) presenting in pre-miRNAs and miRNA-processing genes. Eleven SNPs, including three SNPs presenting in three miRNA genes (miRNA-612, miRNA-499, and miRNA-449b) and eight SNPs presenting in six miRNA biogenesis pathway genes (TNRC6B, DROSHA, DGCR8, EIF2C1, CNOT1, and CNOT6) were significantly associated with ALL susceptibility. And, of those eleven SNPs, two SNPs presenting in miRNA-612 and miRNA-499 had a more significant association with ALL susceptibility.
Furthermore, different subtypes of ALL can be distinguished. Schotte et al63,64 compared the miRNA expression levels of seven major subtypes of pediatric ALL, which were T-cell, MLL-rearranged, TEL-AML1-positive, E2A-PBX1-positive, hyperdiploid ALL, BCR-ABL-positive, and “B-other” ALLs. They obtained the differential expression of the special miRNAs, such as miRNA-708, which were expressed 250- to 6,500-fold higher in the 57 TEL-AML1, BCR-ABL, E2A-PBX1, hyperdiploid, and B-other cases than in the 20 MLL-rearranged and 15 T-ALL cases (0.0001<P<0.01). Then, they analyzed expression levels of 397 miRNAs in 81 cases of pediatric ALL and 17 normal hematopoietic control cases, demonstrating the unique miRNA signatures of each subtype. Additionally, the miRNA signature of TEL-AML1-positive and hyperdiploid cases overlapped partly, which may suggest a common underlying biology. Mavrakis et al65 reported that five miRNAs – miRNA-19b, miRNA-20a, miRNA-26a, miRNA-92, and miRNA-223, were identified as being capable of promoting T-ALL development in a mouse model and accounting for the majority of miRNA expression in human T-ALL, which could be used to reveal the pattern of gene interactions of T-ALL.
In addition, miRNA expression profiles may reveal new subset of ALL. A new subset of ALL with T-cell origin, which has similar a gene expression profile as acute myeloid leukemia (AML), was identified by comparing the mRNA and miRNA expression profiles with other cases. It has significantly higher levels of miRNA-223 expression than the other subsets, which suggests an unfavorable clinical course.66 Other groups also have performed analogous studies to discover miRNAs expression signatures of ALL.67–70
Differential diagnosis from AML
As well as gene expression profile,71 differential miRNAs expression can be utilized to define myeloid or lymphoid lineage leukemia and distinguish ALL from AML. De Leeuw et al72 reported five of the most lineage-discriminative miRNAs – miRNA-23a, miRNA-27a, miRNA-199b, miRNA-221, and miRNA-223 – which could distinguish ambiguous lineage acute leukemia either as AML or ALL. With a bead-based miRNA-expression profiling assay, Mi et al73 suggested that there were 27 differently expressed miRNAs between ALL and AML in a large-scale genome-wide miRNA expression profiling assay. Compared with AML, let-7b and miRNA-223 were downexpressed and miRNA-128a and -128b were overexpressed in ALL. Also, no less than two miRNAs of these four miRNAs could discriminate ALL from AML with an accuracy rate more than 95%. Using quantitative PCR (qPCR), Wang et al74 separated patients with ALL from those with AML based on differential expression of 16 miRNAs, including previously reported eight miRNAs and newly identified eight miRNAs. The documented information of miRNAs in diagnosis and differential diagnosis of ALL is listed in Table 1.
Prognostic impact of miRNAs in ALL
miRNA signatures can be used not only in the diagnosis the ALL, but also in the prognosis of patients. Several miRNAs, involved in cell proliferation and apoptosis regulation, may interfere with either oncogenic or tumor-suppressor pathways and are implicated in leukemogenesis, influencing the prognosis of patients. For instant, Ohyashiki et al75 reported that cellular miRNA-92a expression was significantly increased in a subset of ALL cells, and ALL patients with overexpression of miRNA-92a had poor prognoses. Compared with peripheral blood mononuclear cells from healthy volunteers, the cell-to-plasma ratio of miRNA-92a expression was particularly higher in both ALL and AML cells. Nemes et al76 suggested that expression level of miRNAs could be used as indicators of prognosis in children with ALL, such as higher expression of miR-128b at diagnosis predicted a better prognosis and prednisolon response. A study of 147 patients with acute leukamia (AL) and 100 healthy individuals showed that AL (including both ALL and AML) patients with high miRNA-24 expression tended to have shorter overall survival (P<0.05).77
High miRNA-16 expression was involved in hyperleukocytosis and poor cytogenetic groups. In B-cell ALLs, patients with miRNA-16 above quartile 75 had a significantly shorter disease-free survival (DFS), and in T-cell ALLs, a significant trend that was a survival shortening from the lowest to the highest miRNA-16 levels was revealed for both DFS and overall survival.78 Another study with 38 cases of T-LBL/ALL patients and 15 cases of reactive hyperplasia of lymph nodes as controls conducted by Tong et al79 claimed that although the overall survival rate in miRNA-16 high-expression group decreased compared with control group, the miRNA-16 expression correlated with BCL-2 protein (r=0.51, P<0.05), and the prognosis in BCL-2-positive expression group was better than that in the negative expression group, indicating that BCL-2 may be also a factor influencing prognosis. The study with 70 cases of T-LBL/ALL and 30 cases of reactive lymph node as controls conducted by Li et al80 reported that the high-expression group of miRNA-16 had longer overall survival than the low-expression group and that the prognosis of BCL-2 negative was better than BCL-2 positive. So, further studies are required to elucidate the definite role of miRNA-16 on the prognosis of ALL and the relationship between miRNA-16 and BCL-2.
The function of miRNAs in therapy of ALL
Glucocorticoids
Glucocorticoids (GCs) induce apoptosis in lymphoid lineage cells and, therefore, are used in the therapy of ALL and related malignancies.81 However, a proportion of patients with ALL are insensitive to prednisone. Here, eight miRNAs can help to distinguish the patients sensitive from those insensitive to prednisone, which are miRNA-18a, miRNA-532, miRNA 218, miRNA-625, miRNA-193a, miRNA-638, miRNA-550, and miRNA-633.82 And, suppose the patients with MLL-rearranged ALL were insensitive to GCs, miRNA-128b and miRNA-221 may serve as GCs sensitizers potentially. Both miRNAs are downregulated in MLL-rearranged ALL. The restoration of miRNA-128b downregulates target genes including MLL, AF4, and both MLL-AF4, and AF4-MLL fusion oncogenes, and the restoration of miRNA-221 downregulates CDKN1B cooperatively. Thus, the sensitivity of two cultured lines of MLL-AF4 ALL cells to GCs is strengthened.83 In a subsequent study, Kotani et al84 illustrated that one novel mutation of miRNA-128b significantly reduced its processing, and the resultant downregulation of mature miRNA-128b gave rise to GCs resistance due to the failure to downregulate the fusion oncogenes. Harada et al85 transiently overexpressed pre-miRNA-17, an miRNA precursor, in the SUP-B15 cell line by electroporation and monitored the dexamethasone-induced levels of apoptosis using annexin/propidium iodide (PI) staining. They found that overexpression of miRNA-17 reduced dexamethasone-induced cell death. By inhibition of miRNA-17 through locked nucleic acid (LNA) inhibitor, sensitivity to dexamethasone was increased. Therefore, by regulating miRNAs, therapeutic effect of GCs may be improved.
Tyrosine-kinase inhibitors (TKI)
For the ALL with BCR-ABL fusion gene, the application of TKI may be a promising strategy, but the prognosis remains suboptimal.86 BCR-ABL1 and ABL1 are the direct targets of miRNA-203, which is silenced by genetic and epigenetic mechanisms in hematopoietic malignancies expressing either ABL1 or BCR-ABL1, and the restoration of miRNA-203 expression reduces ABL1 and BCR-ABL1 levels and inhibits cell proliferation.86,87 The inhibition of DNMT3A by forced expression of miRNA-217 may benefit in preventing drug resistance to TKI treatment in Philadelphia-chromosome-positive ALL patients. Hence, it may indicate another therapeutic strategy for BCR-ABL-positive ALL.88
Demethylation
Demethylation may be a potential therapeutic strategy for ALL.89 In the MLL-AF4 ALL, miRNA-143 is epigenetically repressed by promoter hypermethylation in MLL-AF4-positive primary blasts and cell lines, but not in normal BM cells and MLL-AF4-negative primary blasts. Meanwhile, miRNA-143 was identified as a regulator of MLL-AF4 expression, and its restoration could induce apoptosis, negatively contributing to leukemia cell growth. Therefore, upregulation of miRNA-143 expression has therapeutic promise for MLL-AF4 B-cell ALL.90 Other documented information of miRNAs for prognosis and/or treatment of ALL is listed in Table 2, and miRNA-196b is presented as an example to illustrate the interaction between miRNA and its target in Figure 4.
  | Table 2 Other documented information of miRNAs for prognosis and/or treatment of ALL |
  | Figure 4 The mechanism between miRNA-196b and their targets. |
Future directions
miRNAs promote lymphoblastic leukemogenesis through different mechanisms such as enhancing the expression of oncogenes or suppressing apoptosis. However, many problems need to be solved in the future.
Some miRNAs involved in the control of lymphopoiesis are deregulated in ALL, for instance, miRNA-128a, miRNA-126, and miRNA-146 are deregulated miRNAs in ALL and they also play roles in lymphopoiesis. As miRNAs are frequently localized in common breakpoint regions related to tumors or in fragile sites, one may speculate that miRNAs that play a role in lymphopoiesis are prone to be deregulated in lymphocyte original cancers like ALL. For example, a study reported that many of the miRNAs deregulated in multiple lymphoma are also intimately involved in lymphocyte biology under physiological conditions.91 While one should keep in mind that it is a very young field, the documented miRNAs involved in lymphomagenesis or deregulated miRNAs involved in cancer like ALL are incomplete as is our knowledge about their function. Therefore, further studies are needed to verify this speculation.
In the aforementioned studies, different methods were used in detect the miRNAs in cells or plasma, including bead-based array, planar array, and qPCR.60,73,74 Consequently, the lack of uniform methods for detecting miRNAs leads to the inability to compare results between different researches. Second, different studies may share few similar miRNA profiles when comparing the differences in origin of normal and aberrant cells. For instance, normal CD34+ cells can be obtained under different conditions, such as after growth factor mobilization versus collected directly from the BM with no mobilization.3 Therefore, it is possible that the growth factor changed the expression profile of miRNAs in CD34+ cells as well as the change of mRNAs expression reported previously.3,92 Additionally, some studies use unselected peripheral blood mononuclear cells or BM mononuclear cells from normal donors as controls instead of CD34+ cells.73,75 From the aforementioned points, better and uniform methods for the detection of miRNAs will help to understand normal and aberrant lymphopoiesis more thoroughly. Also, the uniformity of collecting cells in experimental and control groups separately will help make the results more accurate, enabling comparability among different studies to be obtained. Therefore, standardization of the related studies is the most imperative problem that must be settled in the future.
Considering that miRNAs are the underlying mechanism in the development of human disease, regulation of miRNA function may have therapeutic utility. Although some miRNAs are indentified to have therapeutic effect on some disease, the strategy to interrupt the function of miRNA is limited. In vitro, transfection with miRNA mimics or miRNA inhibitors into cells is a common way to increase miRNA expression or to decrease miRNA expression, respectively, while safety concern and degradation limit their utility in vivo. Many strategies like chemical modifications, LNA, and phosphorothioate linkages have been developed to increase stability and safety.93 Antagomirs, which are improved miRNA inhibitors, are antisense single-stranded oligonucleotides that are chemically modified, cholesterol conjugated, etc. Antagomirs could silence miRNAs after combining with them and are stable enough to be administrated by intravenous injection.94 Besides the stability of the miRNA mimics or miRNA inhibitors themselves, delivery methods also play a significant role.95 The advantages of miRNA mimics expressed from plasmid vectors or viral vectors have longer expression compared with transfection of lipid reagents or electroporation.96 Some drugs were also reported to potentially modulate miRNAs expression in diseases, but further studies of their pharmacodynamics, pharmacokinetics, safety, etc are required.97
Off-target effects, which are brought about by interactions between the RNA interference (RNAi) molecules and nontarget genes, or other cellular components, RNAi molecules, mainly include small interfering RNA (siRNA), short hairpin RNA, and miRNA. Off-target effects could be generally classified as specific off-target effects, also known as miRNA-like off-target effects, and nonspecific off-target effects.98 Many strategies are studied to diminish or eliminate the undesired effects, such as designing new vectors and chemical modification of RNAi molecules.99,100 Compared with siRNAs, few studies are related to off-target effects of miRNAs, and therefore, further studies are needed in the future.
Conclusion
There are still many problems that need to be solved before the clinical application of miRNAs in ALL, such as comprehensive understanding of the role of related miRNAs both in physiology and pathology of ALL, standard detecting methods, effective and specific-targeting delivery methods, and acceptable off-target effect. miRNA-based therapeutics is an attractive research area and is a promising field to improve the treatment of cancers like ALL and other diseases.
Acknowledgment
This work was supported by the National Natural Science Foundation of the People’s Republic of China (grant numbers 81170492 and 81370673), National High Technology Research and Development Program 863 of the People’s Republic of China (grant number 2012AA022703), National Key Basic Research Program 973 of the People’s Republic of China (grant number 2010CB732404), Key Medical Projects of Jiangsu Province (grant number BL2014078), and Key Discipline of Jiangsu Province (2011–2015).
Disclosure
The authors report no conflicts of interest in this work.
References
Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–355. | ||
Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. | ||
Marcucci G, Mrozek K, Radmacher MD, Garzon R, Bloomfield CD. The prognostic and functional role of microRNAs in acute myeloid leukemia. Blood. 2011;117(4):1121–1129. | ||
Jung HJ, Suh Y. Circulating miRNAs in ageing and ageing-related diseases. J Genet Genomics. 2014;41(9):465–472. | ||
Monteys AM, Spengler RM, Wan J, et al. Structure and activity of putative intronic miRNA promoters. RNA. 2010;16(3):495–505. | ||
Lai EC. Micro RNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat Genet. 2002;30(4):363–364. | ||
Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–854. | ||
Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42(Database issue):D68–D73. | ||
Weilner S, Grillari-Voglauer R, Redl H, Grillari J, Nau T. The role of microRNAs in cellular senescence and age-related conditions of cartilage and bone. Acta Orthop. 2015;86(1):92–99. | ||
Sethi S, Ali S, Sethi S, Sarkar FH. MicroRNAs in personalized cancer therapy. Clin Genet. 2014;86(1):68–73. | ||
Zhong S, Ma T, Zhang X, et al. MicroRNA expression profiling and bioinformatics analysis of dysregulated microRNAs in vinorelbine-resistant breast cancer cells. Gene. 2015;556(2):113–118. | ||
Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol Med. 2014;20(8):460–469. | ||
Van Peer G, Lefever S, Anckaert J, et al. miRBase Tracker: keeping track of microRNA annotation changes. Database (Oxford). 2014;2014:pii bau080. | ||
Sanghvi VR, Mavrakis KJ, Van der Meulen J, et al. Characterization of a set of tumor suppressor microRNAs in T cell acute lymphoblastic leukemia. Sci Signal. 2014;7(352):ra111. | ||
Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19(1):92–105. | ||
Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6(11):857–866. | ||
Calin GA, Sevignani C, Dumitru CD, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101(9):2999–3004. | ||
Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303(5654):83–86. | ||
Wang Q, Huang Z, Xue H, et al. MicroRNA miR-24 inhibits erythropoiesis by targeting activin type I receptor ALK4. Blood. 2008;111(2):588–595. | ||
Havelange V, Garzon R. MicroRNAs: emerging key regulators of hematopoiesis. Am J Hematol. 2010;85(12):935–942. | ||
Cheng J, Guo S, Chen S, et al. An extensive network of TET2-targeting MicroRNAs regulates malignant hematopoiesis. Cell Rep. 2013;5(2):471–481. | ||
Raghavachari N, Liu P, Barb JJ, et al. Integrated analysis of miRNA and mRNA during differentiation of human CD34+ cells delineates the regulatory roles of microRNA in hematopoiesis. Exp Hematol. 2014;42(1):14–27.e11–e12. | ||
Wojtowicz EE, Walasek MA, Broekhuis MJ, et al. MicroRNA-125 family members exert a similar role in the regulation of murine hematopoiesis. Exp Hematol. 2014;42(10):909–918.e901. | ||
Hu Y, Xiong Q, Yang Y, et al. Integrated analysis of gene expression and microRNA regulation in three leukemia-related lymphoblastic cell lines. Gene. 2015;564(1):39–52. | ||
Inaba H, Greaves M, Mullighan CG. Acute lymphoblastic leukaemia. Lancet. 2013;381(9881):1943–1955. | ||
Faderl S, Albitar M. Insights into the biologic and molecular abnormalities in adult acute lymphocytic leukemia. Hematol Oncol Clin North Am. 2000;14(6):1267–1288. | ||
Pui CH, Evans WE. Treatment of acute lymphoblastic leukemia. N Engl J Med. 2006;354(2):166–178. | ||
Vitale A, Guarini A, Chiaretti S, Foa R. The changing scene of adult acute lymphoblastic leukemia. Curr Opin Oncol. 2006;18(6):652–659. | ||
Randolph TR. Advances in acute lymphoblastic leukemia. Clin Lab Sci. 2004;17(4):235–245. | ||
Li Q, Liu L, Li W. Identification of circulating microRNAs as biomarkers in diagnosis of hematologic cancers: a meta-analysis. Tumour Biol. 2014;35(10):10467–10478. | ||
Cui J. MiR-16 family as potential diagnostic biomarkers for cancer: a systematic review and meta-analysis. Int J Clin Exp Med. 2015;8(2):1703–1714. | ||
Tanaka M, Oikawa K, Takanashi M, et al. Down-regulation of miR-92 in human plasma is a novel marker for acute leukemia patients. PloS One. 2009;4(5):e5532. | ||
Slavov SN, Gimenes Teixeira HL, Rego EM. The role of micro-ribonucleic acids in normal hematopoiesis and leukemic T-lymphogenesis. Braz J Med Biol Res. 2010;43(7):619–626. | ||
O’Connell RM, Rao DS, Chaudhuri AA, Baltimore D. Physiological and pathological roles for microRNAs in the immune system. Nat Rev Immunol. 2010;10(2):111–122. | ||
Johanson TM, Skinner JP, Kumar A, Zhan Y, Lew AM, Chong MM. The role of microRNAs in lymphopoiesis. Int J Hematol. 2014;100(3):246–253. | ||
Xiao C, Calado DP, Galler G, et al. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell. 2007;131(1):146–159. | ||
Zhou B, Wang S, Mayr C, Bartel DP, Lodish HF. miR-150, a microRNA expressed in mature B and T cells, blocks early B cell development when expressed prematurely. Proc Natl Acad Sci U S A. 2007;104(17):7080–7085. | ||
He Y, Jiang X, Chen J. The role of miR-150 in normal and malignant hematopoiesis. Oncogene. 2014;33(30):3887–3893. | ||
Ghisi M, Corradin A, Basso K, et al. Modulation of microRNA expression in human T-cell development: targeting of NOTCH3 by miR-150. Blood. 2011;117(26):7053–7062. | ||
Thomas MD, Kremer CS, Ravichandran KS, Rajewsky K, Bender TP. c-Myb is critical for B cell development and maintenance of follicular B cells. Immunity. 2005;23(3):275–286. | ||
Lu D, Nakagawa R, Lazzaro S, et al. The miR-155-PU.1 axis acts on Pax5 to enable efficient terminal B cell differentiation. J Exp Med. 2014;211(11):2183–2198. | ||
Thai TH, Calado DP, Casola S, et al. Regulation of the germinal center response by microRNA-155. Science. 2007;316(5824):604–608. | ||
Banerjee A, Schambach F, DeJong CS, Hammond SM, Reiner SL. Micro-RNA-155 inhibits IFN-gamma signaling in CD4+ T cells. Eur J Immunol. 2010;40(1):225–231. | ||
Seddiki N, Brezar V, Ruffin N, Levy Y, Swaminathan S. Role of miR-155 in the regulation of lymphocyte immune function and disease. Immunology. 2014;142(1):32–38. | ||
Yang Z, Wan X, Gu Z, et al. Evolution of the mir-181 microRNA family. Comput Biol Med. 2014;52:82–87. | ||
Li QJ, Chau J, Ebert PJ, et al. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell. 2007;129(1):147–161. | ||
Neilson JR, Zheng GX, Burge CB, Sharp PA. Dynamic regulation of miRNA expression in ordered stages of cellular development. Genes Dev. 2007;21(5):578–589. | ||
Verduci L, Azzalin G, Gioiosa S, et al. microRNA-181a enhances cell proliferation in acute lymphoblastic leukemia by targeting EGR1. Leuk Res. 2015;39(4):479–485. | ||
Rao R, Nagarkatti PS, Nagarkatti M. Delta(9) Tetrahydrocannabinol attenuates Staphylococcal enterotoxin B-induced inflammatory lung injury and prevents mortality in mice by modulation of miR-17-92 cluster and induction of T-regulatory cells. Br J Pharmacol. 2015;172(7):1792–1806. | ||
Ventura A, Young AG, Winslow MM, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132(5):875–886. | ||
Xiao C, Srinivasan L, Calado DP, et al. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat Immunol. 2008;9(4):405–414. | ||
Bhagavathi S, Czader M. MicroRNAs in benign and malignant hematopoiesis. Arch Pathol Lab Med. 2010;134(9):1276–1281. | ||
Mullighan CG. Genomic characterization of childhood acute lymphoblastic leukemia. Semin Hematol. 2013;50(4):314–324. | ||
Rego EM, Garcia AB, Carneiro JJ, Falcao RP. Immunophenotype of normal and leukemic bone marrow B-precursors in a Brazilian population. A comparative analysis by quantitative fluorescence cytometry. Braz J Med Biol Res. 2001;34(2):183–194. | ||
Rego EM, Tone LG, Garcia AB, Falcao RP. CD10 and CD19 fluorescence intensity of B-cell precursors in normal and leukemic bone marrow. Clinical characterization of CD10 (+strong) and CD10 (+weak) common acute lymphoblastic leukemia. Leuk Res. 1999;23(5):441–450. | ||
Schabath R, Ratei R, Ludwig WD. The prognostic significance of antigen expression in leukaemia. Best Pract Res Clin Haematol. 2003;16(4):613–628. | ||
Li WY, Chen XM, Xiong W, Guo DM, Lu L, Li HY. Detection of microvesicle miRNA expression in ALL subtypes and analysis of their functional roles. J Huazhong Univ Sci Technol Med Sci. 2014;34(5):640–645. | ||
de Oliveira JC, Scrideli CA, Brassesco MS, et al. Differential miRNA expression in childhood acute lymphoblastic leukemia and association with clinical and biological features. Leuk Res. 2012;36(3):293–298. | ||
Tong N, Xu B, Shi D, et al. Hsa-miR-196a2 polymorphism increases the risk of acute lymphoblastic leukemia in Chinese children. Mutat Res. 2014;759:16–21. | ||
Fulci V, Colombo T, Chiaretti S, et al. Characterization of B- and T-lineage acute lymphoblastic leukemia by integrated analysis of MicroRNA and mRNA expression profiles. Genes Chromosomes Cancer. 2009;48(12):1069–1082. | ||
Zhang H, Yang JH, Zheng YS, et al. Genome-wide analysis of small RNA and novel MicroRNA discovery in human acute lymphoblastic leukemia based on extensive sequencing approach. PLoS One. 2009;4(9):e6849. | ||
Gutierrez-Camino A, Lopez-Lopez E, Martin-Guerrero I, et al. Noncoding RNA-related polymorphisms in pediatric acute lymphoblastic leukemia susceptibility. Pediatr Res. 2014;75(6):767–773. | ||
Schotte D, Chau JC, Sylvester G, et al. Identification of new microRNA genes and aberrant microRNA profiles in childhood acute lymphoblastic leukemia. Leukemia. 2009;23(2):313–322. | ||
Schotte D, De Menezes RX, Moqadam FA, et al. MicroRNA characterize genetic diversity and drug resistance in pediatric acute lymphoblastic leukemia. Haematologica. 2011;96(5):703–711. | ||
Mavrakis KJ, Van Der Meulen J, Wolfe AL, et al. A cooperative microRNA-tumor suppressor gene network in acute T-cell lymphoblastic leukemia (T-ALL). Nat Genet. 2011;43(7):673–678. | ||
Chiaretti S, Messina M, Tavolaro S, et al. Gene expression profiling identifies a subset of adult T-cell acute lymphoblastic leukemia with myeloid-like gene features and over-expression of miR-223. Haematologica. 2010;95(7):1114–1121. | ||
Ju X, Li D, Shi Q, Hou H, Sun N, Shen B. Differential microRNA expression in childhood B-cell precursor acute lymphoblastic leukemia. Pediatr Hematol Oncol. 2009;26(1):1–10. | ||
Agueli C, Cammarata G, Salemi D, et al. 14q32/miRNA clusters loss of heterozygosity in acute lymphoblastic leukemia is associated with up-regulation of BCL11a. Am J Hematol. 2010;85(8):575–578. | ||
Hasani SS, Hashemi M, Eskandari-Nasab E, Naderi M, Omrani M, Sheybani-Nasab M. A functional polymorphism in the miR-146a gene is associated with the risk of childhood acute lymphoblastic leukemia: a preliminary report. Tumour Biol. 2014;35(1):219–225. | ||
Duyu M, Durmaz B, Gunduz C, et al. Prospective evaluation of whole genome microRNA expression profiling in childhood acute lymphoblastic leukemia. Biomed Res Int. 2014;2014:967585. | ||
Golub TR, Slonim DK, Tamayo P, et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286(5439):531–537. | ||
de Leeuw DC, van den Ancker W, Denkers F, et al. MicroRNA profiling can classify acute leukemias of ambiguous lineage as either acute myeloid leukemia or acute lymphoid leukemia. Clin Cancer Res. 2013;19(8):2187–2196. | ||
Mi S, Lu J, Sun M, et al. MicroRNA expression signatures accurately discriminate acute lymphoblastic leukemia from acute myeloid leukemia. Proc Natl Acad Sci U S A. 2007;104(50):19971–19976. | ||
Wang Y, Li Z, He C, et al. MicroRNAs expression signatures are associated with lineage and survival in acute leukemias. Blood Cells Mol Dis. 2010;44(3):191–197. | ||
Ohyashiki JH, Umezu T, Kobayashi C, et al. Impact on cell to plasma ratio of miR-92a in patients with acute leukemia: in vivo assessment of cell to plasma ratio of miR-92a. BMC Res Notes. 2010;3:347. | ||
Nemes K, Csoka M, Nagy N, et al. Expression of certain leukemia/lymphoma related microRNAs and its correlation with prognosis in childhood acute lymphoblastic leukemia. Pathol Oncol Res. 2015;21(3):597–604. | ||
Organista-Nava J, Gomez-Gomez Y, Illades-Aguiar B, et al. High miR-24 expression is associated with risk of relapse and poor survival in acute leukemia. Oncol Rep. 2015;33(4):1639–1649. | ||
Kaddar T, Chien WW, Bertrand Y, et al. Prognostic value of miR-16 expression in childhood acute lymphoblastic leukemia relationships to normal and malignant lymphocyte proliferation. Leuk Res. 2009;33(9):1217–1223. | ||
Tong LG, Wu WZ, Zhang YP, et al. Expression of miR-16 in patients with T lymphoblastic lymphoma/acute lymphoblastic leukemia. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2014;22(1):99–103. | ||
Li J, Li P, Wang JF, Xi YF. Significance of microRNA-16 and bcl-2 expression in T lymphoblastic lymphoma/leukemia and its relation with prognosis. Zhonghua Bing Li Xue Za Zhi. 2013;42(11):748–752. | ||
Rainer J, Ploner C, Jesacher S, et al. Glucocorticoid-regulated microRNAs and mirtrons in acute lymphoblastic leukemia. Leukemia. 2009;23(4):746–752. | ||
Xu L, Liang YN, Luo XQ, Liu XD, Guo HX. Association of miRNAs expression profiles with prognosis and relapse in childhood acute lymphoblastic leukemia. Zhonghua Xue Ye Xue Za Zhi. 2011;32(3):178–181. | ||
Kotani A, Ha D, Hsieh J, et al. miR-128b is a potent glucocorticoid sensitizer in MLL-AF4 acute lymphocytic leukemia cells and exerts cooperative effects with miR-221. Blood. 2009;114(19):4169–4178. | ||
Kotani A, Ha D, Schotte D, den Boer ML, Armstrong SA, Lodish HF. A novel mutation in the miR-128b gene reduces miRNA processing and leads to glucocorticoid resistance of MLL-AF4 acute lymphocytic leukemia cells. Cell Cycle. 2010;9(6):1037–1042. | ||
Harada M, Pokrovskaja-Tamm K, Soderhall S, Heyman M, Grander D, Corcoran M. Involvement of miR17 pathway in glucocorticoid-induced cell death in pediatric acute lymphoblastic leukemia. Leuk Lymphoma. 2012;53(10):2041–2050. | ||
Faber J, Gregory RI, Armstrong SA. Linking miRNA regulation to BCR-ABL expression: the next dimension. Cancer Cell. 2008;13(6):467–469. | ||
Bueno MJ, Perez de Castro I, Gomez de Cedron M, et al. Genetic and epigenetic silencing of microRNA-203 enhances ABL1 and BCR-ABL1 oncogene expression. Cancer Cell. 2008;13(6):496–506. | ||
Nishioka C, Ikezoe T, Yang J, Nobumoto A, Tsuda M, Yokoyama A. Downregulation of miR-217 correlates with resistance of Ph(+) leukemia cells to ABL tyrosine kinase inhibitors. Cancer Sci. 2014;105(3):297–307. | ||
Stumpel DJ, Schotte D, Lange-Turenhout EA, et al. Hypermethylation of specific microRNA genes in MLL-rearranged infant acute lymphoblastic leukemia: major matters at a micro scale. Leukemia. 2011;25(3):429–439. | ||
Dou L, Zheng D, Li J, et al. Methylation-mediated repression of microRNA-143 enhances MLL-AF4 oncogene expression. Oncogene. 2011;31(40):507–517. | ||
Di Lisio L, Sanchez-Beato M, Gomez-Lopez G, et al. MicroRNA signatures in B-cell lymphomas. Blood Cancer J. 2012;2(2):e57. | ||
Steidl U, Kronenwett R, Rohr UP, et al. Gene expression profiling identifies significant differences between the molecular phenotypes of bone marrow-derived and circulating human CD34+ hematopoietic stem cells. Blood. 2002;99(6):2037–2044. | ||
Lennox KA, Behlke MA. A direct comparison of anti-microRNA oligonucleotide potency. Pharm Res. 2010;27(9):1788–1799. | ||
Krutzfeldt J, Rajewsky N, Braich R, et al. Silencing of microRNAs in vivo with “antagomirs.” Nature. 2005;438(7068):685–689. | ||
Castano IM, Curtin CM, Shaw G, Murphy JM, Duffy GP, O’Brien FJ. A novel collagen-nanohydroxyapatite microRNA-activated scaffold for tissue engineering applications capable of efficient delivery of both miR-mimics and antagomiRs to human mesenchymal stem cells. J Control Release. 2015;200:42–51. | ||
Henry JC, Azevedo-Pouly AC, Schmittgen TD. MicroRNA replacement therapy for cancer. Pharm Res. 2011;28(12):3030–3042. | ||
Agirre X, Vilas-Zornoza A, Jimenez-Velasco A, et al. Epigenetic silencing of the tumor suppressor microRNA Hsa-miR-124a regulates CDK6 expression and confers a poor prognosis in acute lymphoblastic leukemia. Cancer Res. 2009;69(10):4443–4453. | ||
Singh S, Narang AS, Mahato RI. Subcellular fate and off-target effects of siRNA, shRNA, and miRNA. Pharm Res. 2011;28(12):2996–3015. | ||
Yang N, Mahato RI. GFAP promoter-driven RNA interference on TGF-beta1 to treat liver fibrosis. Pharm Res. 2011;28(4):752–761. | ||
Ui-Tei K, Naito Y, Zenno S, et al. Functional dissection of siRNA sequence by systematic DNA substitution: modified siRNA with a DNA seed arm is a powerful tool for mammalian gene silencing with significantly reduced off-target effect. Nucleic Acids Res. 2008;36(7):2136–2151. | ||
Bhatia S, Kaul D, Varma N. Functional genomics of tumor suppressor miR-196b in T-cell acute lymphoblastic leukemia. Mol Cell Biochem. 2011;346(1–2):103–116. | ||
Loughran SJ, Kruse EA, Hacking DF, et al. The transcription factor Erg is essential for definitive hematopoiesis and the function of adult hematopoietic stem cells. Nat Immunol. 2008;9(7):810–819. | ||
Wu L, Li H, Jia CY, et al. MicroRNA-223 regulates FOXO1 expression and cell proliferation. FEBS Lett. 2012;586(7):1038–1043. | ||
Sun W, Shen W, Yang S, Hu F, Li H, Zhu TH. miR-223 and miR-142 attenuate hematopoietic cell proliferation, and miR-223 positively regulates miR-142 through LMO2 isoforms and CEBP-beta. Cell Res. 2010;20(10):1158–1169. | ||
Ferrando AA, Herblot S, Palomero T, et al. Biallelic transcriptional activation of oncogenic transcription factors in T-cell acute lymphoblastic leukemia. Blood. 2004;103(5):1909–1911. | ||
Ferrando AA, Neuberg DS, Staunton J, et al. Gene expression signatures define novel oncogenic pathways in T cell acute lymphoblastic leukemia. Cancer Cell. 2002;1(1):75–87. | ||
Lee YS, Dutta A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 2007;21(9):1025–1030. | ||
Yamada N, Noguchi S, Kumazaki M, et al. Epigenetic regulation of microRNA-128a expression contributes to the apoptosis-resistance of human T-cell leukaemia jurkat cells by modulating expression of fas-associated protein with death domain (FADD). Biochim Biophys Acta. 2014;1843(3):590–602. | ||
Lessard J, Sauvageau G. Bmi-1 determines the proliferative capacity of normal and leukaemic stem cells. Nature. 2003;423(6937):255–260. | ||
Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304(5670):594–596. | ||
Coskun E, von der Heide EK, Schlee C, et al. The role of microRNA-196a and microRNA-196b as ERG regulators in acute myeloid leukemia and acute T-lymphoblastic leukemia. Leuk Res. 2011;35(2):208–213. | ||
Popovic R, Riesbeck LE, Velu CS, et al. Regulation of mir-196b by MLL and its overexpression by MLL fusions contributes to immortalization. Blood. 2009;113(14):3314–3322. | ||
Bhatia S, Kaul D, Varma N. Potential tumor suppressive function of miR-196b in B-cell lineage acute lymphoblastic leukemia. Mol Cell Biochem. 2010;340(1–2):97–106. | ||
Schotte D, Lange-Turenhout EA, Stumpel DJ, et al. Expression of miR-196b is not exclusively MLL-driven but is especially linked to activation of HOXA genes in pediatric acute lymphoblastic leukemia. Haematologica. 2010;95(10):1675–1682. | ||
Tassano E, Acquila M, Tavella E, Micalizzi C, Panarello C, Morerio C. MicroRNA-125b-1 and BLID upregulation resulting from a novel IGH translocation in childhood B-Cell precursor acute lymphoblastic leukemia. Genes Chromosomes Cancer. 2010;49(8):682–687. | ||
Okamura S, Arakawa H, Tanaka T, et al. p53DINP1, a p53-inducible gene, regulates p53-dependent apoptosis. Mol Cell. 2001;8(1):85–94. | ||
Mets E, Van Peer G, Van der Meulen J, et al. MicroRNA-128-3p is a novel oncomiR targeting PHF6 in T-cell acute lymphoblastic leukemia. Haematologica. 2014;99(8):1326–1333. | ||
Yoo NJ, Kim YR, Lee SH. Somatic mutation of PHF6 gene in T-cell acute lymphoblatic leukemia, acute myelogenous leukemia and hepatocellular carcinoma. Acta Oncol. 2012;51(1):107–111. | ||
Porstner M, Winkelmann R, Daum P, et al. miR-148a promotes plasma cell differentiation and targets the germinal center transcription factors Mitf and Bach2. Eur J Immunol. 2015;45(4):1206–1215. | ||
Guo W, Liu R, Ono Y, et al. Molecular characteristics of CTA056, a novel interleukin-2-inducible T-cell kinase inhibitor that selectively targets malignant T cells and modulates oncomirs. Mol Pharmacol. 2012;82(5):938–947. | ||
Chiaretti S, Guarini A, De Propris MS, et al. ZAP-70 expression in acute lymphoblastic leukemia: association with the E2A/PBX1 rearrangement and the pre-B stage of differentiation and prognostic implications. Blood. 2006;107(1):197–204. | ||
Faraoni I, Laterza S, Ardiri D, Ciardi C, Fazi F, Lo-Coco F. MiR-424 and miR-155 deregulated expression in cytogenetically normal acute myeloid leukaemia: correlation with NPM1 and FLT3 mutation status. J Hematol Oncol. 2012;5:26. | ||
Kunze K, Gamerdinger U, Lessig-Owlanj J, et al. Detection of an activated JAK3 variant and a Xq26.3 microdeletion causing loss of PHF6 and miR-424 expression in myelodysplastic syndromes by combined targeted next generation sequencing and SNP array analysis. Pathol Res Pract. 2014;210(6):369–376. | ||
Starnes LM, Sorrentino A, Pelosi E, et al. NFI-A directs the fate of hematopoietic progenitors to the erythroid or granulocytic lineage and controls beta-globin and G-CSF receptor expression. Blood. 2009;114(9):1753–1763. | ||
Lu KV, Chang JP, Parachoniak CA, et al. VEGF inhibits tumor cell invasion and mesenchymal transition through a MET/VEGFR2 complex. Cancer Cell. 2012;22(1):21–35. | ||
Koller K, Das S, Leuschner I, Korbelius M, Hoefler G, Guertl B. Identification of the transcription factor HOXB4 as a novel target of miR-23a. Genes Chromosomes Cancer. 2013;52(8):709–715. | ||
Xishan Z, Xianjun L, Ziying L, Guangxin C, Gang L. The malignancy suppression role of miR-23a by targeting the BCR/ABL oncogene in chromic myeloid leukemia. Cancer Gene Ther. 2014;21(9):397–404. | ||
Arabanian LS, Fierro FA, Stolzel F, et al. MicroRNA-23a mediates post-transcriptional regulation of CXCL12 in bone marrow stromal cells. Haematologica. 2014;99(6):997–1005. | ||
Kong KY, Owens KS, Rogers JH, et al. MIR-23A microRNA cluster inhibits B-cell development. Exp Hematol. 2010;38(8):629–640.e621. | ||
Scheibner KA, Teaboldt B, Hauer MC, et al. MiR-27a functions as a tumor suppressor in acute leukemia by regulating 14-3-3theta. PLoS One. 2012;7(12):e50895. | ||
Frenquelli M, Muzio M, Scielzo C, et al. MicroRNA and proliferation control in chronic lymphocytic leukemia: functional relationship between miR-221/222 cluster and p27. Blood. 2010;115(19):3949–3959. | ||
Rommer A, Steinleitner K, Hackl H, et al. Overexpression of primary microRNA 221/222 in acute myeloid leukemia. BMC Cancer. 2013;13:364. | ||
Mei Y, Gao C, Wang K, et al. Effect of microRNA-210 on prognosis and response to chemotherapeutic drugs in pediatric acute lymphoblastic leukemia. Cancer Sci. 2014;105(4):463–472. | ||
Xiang Y, Ma N, Wang D, et al. MiR-152 and miR-185 co-contribute to ovarian cancer cells cisplatin sensitivity by targeting DNMT1 directly: a novel epigenetic therapy independent of decitabine. Oncogene. 2014;33(3):378–386. | ||
Zhu H, Miao MH, Ji XQ, Xue J, Shao XJ. miR-664 negatively regulates PLP2 and promotes cell proliferation and invasion in T-cell acute lymphoblastic leukaemia. Biochem Biophys Res Commun. 2015;459(2):340–345. | ||
Li XJ, Luo XQ, Han BW, Duan FT, Wei PP, Chen YQ. MicroRNA-100/99a, deregulated in acute lymphoblastic leukaemia, suppress proliferation and promote apoptosis by regulating the FKBP51 and IGF1R/mTOR signalling pathways. Br J Cancer. 2013;109(8):2189–2198. | ||
Li X, Li D, Zhuang Y, Shi Q, Wei W, Ju X. Overexpression of miR-708 and its targets in the childhood common precursor B-cell ALL. Pediatric Blood Cancer. 2013;60(12):2060–2067. | ||
Han BW, Feng DD, Li ZG, et al. A set of miRNAs that involve in the pathways of drug resistance and leukemic stem-cell differentiation is associated with the risk of relapse and glucocorticoid response in childhood ALL. Hum Mol Genet. 2011;20(24):4903–4915. | ||
Mets E, Van der Meulen J, Van Peer G, et al. MicroRNA-193b-3p acts as a tumor suppressor by targeting the MYB oncogene in T-cell acute lymphoblastic leukemia. Leukemia. 2015;29(4):798–806. |
 © 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.