Back to Journals » Infection and Drug Resistance » Volume 14
The First Saudi Study Investigating the Plasmid-borne Aminoglycoside and Sulfonamide Resistance among Acinetobacter baumannii Clinical Isolates Genotyped by RAPD-PCR: the Declaration of a Novel Allelic Variant Called aac(6ʹ)-SL and Three Novel Mutations in the sul1 Gene in the Acinetobacter Plasmid (s)
Authors El-Badawy MF, Abou-Elazm FI, Omar MS, El-Naggar ME , Maghrabi IA
Received 7 July 2021
Accepted for publication 9 September 2021
Published 12 November 2021 Volume 2021:14 Pages 4739—4756
DOI https://doi.org/10.2147/IDR.S324707
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Mohamed F El-Badawy,1 Fatma I Abou-Elazm,2 Mohamed S Omar,3 Mostafa E El-Naggar,4 Ibrahim A Maghrabi5
1Department of Microbiology and Immunology, Faculty of Pharmacy, University of Sadat City, Sadat City, Menoufia, 32897, Egypt; 2Department of Microbiology and Immunology, Faculty of Pharmacy, Misr University for Science and Technology, 6th of October City, Egypt; 3Department of Chemistry, Faculty of Science, Benha University, Benha, 13508, Egypt; 4Department of Pharmacology and Toxicology, Faculty of Pharmacy, University of Sadat City, Sadat City, Menoufia, 32897, Egypt; 5Department of Clinical Pharmacy, College of Pharmacy, Taif University, Taif, 21974, Saudi Arabia
Correspondence: Mohamed F El-Badawy
Department of Microbiology and Immunology, Faculty of Pharmacy, University of Sadat City, Sadat City, Menoufia, 32897 Egypt
Tel +20 103-205-9964
Email [email protected]
Background: Acinetobacter baumannii (A. baumannii) is one of the most important nosocomial pathogens responsible for a wide range of infections.
Aim: This study aimed to investigate the existence of the plasmidic genes encoding for aminoglycoside modifying enzymes (AMEs), 16S rRNA methyltransferases (RMT), and the altered dihydropetroate synthase (DHPS) encoded by the sul1 gene among A. baumannii clinical isolates collected from Taif, Kingdom of Saudi Arabia (KSA). The mutations in aac(6ʹ)-Ib and sul1 genes were also investigated.
Methods: Forty A. baumannii clinical isolates were investigated for their susceptibility to ten antibiotics. The plasmid DNA was extracted and screened for nine genes encoding for aminoglycoside resistance in addition to the sul1 gene. The clonal relatedness was determined by random amplified polymorphic DNA (RAPD)-PCR. Mutation in aac(6ʹ)-Ib and the sul1 genes were detected by capillary electrophoresis sequencing (CES).
Results: All isolates were A. baumannii in which 42.5% of them exhibited a high level of aminoglycoside resistance (HLAR). The most prevalent AMEs and RMT encoding genes were aph(3ʹ)-VI, the two aac(6ʹ) gene variants [aac(6ʹ)-Ib and aac(6ʹ)-SL], ant(3ʹʹ)-I, and armA in which 90%, 87.5%, 85%, and 45% of isolates tested positive, respectively. The other investigated aminoglycoside resistant encoding genes, namely aac(3)-II, aac(6ʹ)-II, and rmtB, were not detected. Only 15% of isolates harbored the sul1 gene. RAPD-PCR classified the 40 isolates into three clusters in which cluster II was the main cluster. DNA sequencing revealed that 34.29% (12/35) of isolates tested positive for aac(6ʹ)-Ib were found to harbor a common missense mutation in position 102 indicating a novel allelic variant named aac(6ʹ)-SL. Also, DNA sequencing revealed three missense mutations in the sul1 gene.
Conclusion: This is the first Saudi study to investigate the plasmid borne aminoglycoside and sulfonamide resistance genes among A. baumannii clinical isolates. A novel allelic variant for aac(6ʹ)-Ib was detected in addition to novel mutations in the sul1 gene.
Keywords: AMEs, armA, RAPD-PCR, A. baumannii, 16S rRNA
Introduction
Acinetobacter baumannii (A. baumannii) is one the most clinically important non-enteric Gram-negative pathogens that causes a wide range of nosocomial infections (NIs) especially among debilitated patients who are admitted to intensive care units (ICUs).1
In clinical settings, A. baumannii is the most pathogenic and commonly encountered species in the genus Acinetobacter followed by Acinetobacter calcoaceticus (A. calcoaceticus) and Acinetobacter lwoffii (A. lwoffii).2,3
A. baumannii was the only species in the genus Acinetobacter that intrinsically harbors blaOXA-51 which was then used for identification and differentiation of A. baumannii from the other Acinetobacter species.4,5
Multidrug-resistance (MDR), extensive drug resistance (XDR), and pan drug-resistance (PDR) patterns are common among A. baumannii due to the harboring of intrinsic resistance genes,2 genetic mutations, and/or horizontal gene transfer (HGT) by mobile genetic elements (MGEs) like insertion sequence elements (ISE), plasmids, and transposons.6
High morbidity and mortality by A. baumannii are commonly occured in hospital settings1 due to limited therapeutic options for treatment of infections caused by either MDR, XDR, and PDR strains7 so, in 2013, MDR A. baumannii has been termed by the center of disease control and prevention (CDC)8 as a “serious threat” and in 2016, carbapenem-resistant A. baumannii (CRAB) has been considered as a “level I: critical priority pathogen” in the World Health Organization (WHO) priority pathogen’s list.9
Aminoglycosides are broad-spectrum antibiotics that mainly achieve their action by selective inhibition of bacterial protein synthesis via selective binding to the 30S subunit of the bacterial ribosome.10 As aminoglycosides are positively charged molecules due to their amino groups, it has also been known that aminoglycosides disrupt bacterial cell membranes resulting in pore formation in the bacterial’s cell membrane resulting in bacterial cell death.11,12
Aminoglycosides have been widely used, in clinical settings after the emergence of β-lactam and quinolone-resistant strains, for the eradication of MDR Gram-negative isolates despite their nephrotoxic and ototoxic complications.13 According to the previously mentioned, the aminoglycoside resistant strains have emerged worldwide.14
Bacterial resistance to aminoglycosides can be mediated by different mechanisms like overexpression of efflux pumps, downregulation of outer membrane proteins (OMPs), ribosomal target modification, and enzymatic inactivation by aminoglycoside modifying enzymes (AMEs).5
Production of AMEs is the most important mechanism for aminoglycoside resistance in which the dissemination of AMEs occurs via MGEs that commonly harbor other resistance determinants5 to other antibiotic classes leading to the failure of the other antibiotics to cure infections, leading to the selection and the ease of dissemination of aminoglycoside resistant strains in hospital settings.15,16
The AMEs abolish the activity of aminoglycosides by their ability to acetylate, phosphorylate and adenylate the -OH or -NH2 of the 2-deoxystreptamine nucleus or the sugar moieties17 via aminoglycoside acetyltransferases (ACC), aminoglycoside nucleotidyltransferases (ANT), and aminoglycoside phosphotransferases (APH), respectively resulting in poor binding of the aminoglycosides to the bacterial ribosome with subsequent failure to achieve their action.18
Resistance to aminoglycosides can also be achieved through ribosomal target modification via methylation of 16S rRNA by methyltransferases (MT) such as aminoglycosides resistance methyltransferase A (armA)14 and 16S rRNA methyltransferases B (rmtB).13,19
Sulfonamides are the oldest antimicrobial agents that were effectively used in the treatment of bacterial infection since 193220 in which they exert their antimicrobial activity by inhibition of bacterial's folic acid synthesis21 via their binding with dihydropteroate synthase (DHPS) due to their structural similarity with P-amino-benzoic acid (PABA) which is utilized by bacteria for the biosynthesis of folic acid.22
Resistance to sulfonamides can be either chromosomally or plasmid-mediated.21 Chromosomal -mediated sulfonamide resistance involves either a genetic mutation in the DHPS gene known as folP gene or the acquisition of an altered DHPS encoding gene known as sul gene23 via MGEs.24
As mentioned above, plasmid-mediated sulfonamide resistance is achieved by the sul gene which can be translocated between plasmids and chromosomes by MGEs.25 Plasmids harboring the sul gene can be disseminated between same/different bacterial species or different bacterial genera by either conjugation of transformation.24
There are three variants of the sul gene namely; sul1, sul2, and sul3.24 It was reported that the sul1 gene is carried on large conjugative plasmids and class 1 integrons, while the sul2 gene was found to be carried on small non-conjugative plasmids and large conjugative plasmids as well.25 The sul3 gene was reported in 2003 for the first time to be carried on a 54-kb conjugative plasmid in Escherichia coli (E. coli).26
Random amplified polymorphic DNA-polymerase chain reaction (RAPD-PCR) is a rapid molecular technique which is commonly used for molecular typing and epidemiological tracing to discriminate between bacterial isolates based on the use of short oligonucleotide primers ranging from 8 to 15 nucleotides27 that randomly bind to different regions on the whole bacterial genome with a subsequent amplification of many DNA fragments that migrate to different distances on agarose gel generating a complex DNA band patterns.27
Due to the lack of the published data about the genetic background of aminoglycoside and sulfonamide resistance among A. baumannii clinical isolates in the Kingdom of Saudi Arabia (KSA) so, the current study adopted such issue to investigate the the existence of AMEs, 16S rRNA methyltransferases, and altered DHPS plasmid encoding genes in A. baumanniiclinical isolates recovered from a large tertiary care hospital in the Western area, Taif, KSA. Also, the epidemiological typing using RAPD-PCR was performed to track the dissemination of the investigated isolates between the different hospital locations.
Materials and Methods
Ethical Statement
The current study was performed according to ethical approval No. 43-001 following the regulations of the ethics committee at Taif University that accredited by the national committee for bioethics with No. (HAO-02-T-105).
Bacterial Strains
The current study was conducted on 40 clinical isolates of A. baumannii that were recovered from the patients who were admitted to or attended various medical departments at a large tertiary care hospital in Taif, KSA during the period from October 2016 to May 2017. All isolates were recovered from the routinely investigated clinical specimens sent to the microbiology laboratory as a part of routine hospital laboratory procedures. The clinical isolates were recovered from sputum (n = 20), blood (n = 4), tracheal aspirate (n = 2), urine (n = 4), peritoneal fluid (n = 1), wound swab (n = 6), and catheter tip (n = 3). All strains were cryopreserved at −80 °C in tryptic soy broth (Scharlau, Spain) containing 15% glycerol to be used when needed.
Isolation and Identification
All strains were primarily isolated on blood agar then purified on MacConkey’s agar (Oxoid, UK). The isolates were provisionally identified to the genus and species level by the Vitek 2® system (BioMérieux, France) and API 20NE® (BioMérieux, France). Molecular confirmation to the species level was achieved via the amplification of the chromosomally- encoded gene that namely; blaOXA-51 using the specific primers (Table 1) that previously described.2
 |
Table 1 Primer Sequence and Cycling Conditions Used in PCR |
Antimicrobial Susceptibility Testing (AST)
All clinical isolates were tested for their susceptibility to ten different antibiotics namely; gentamicin, amikacin, streptomycin, sulfamethoxazole/trimethoprim, ciprofloxacin, levofloxacin, meropenem, tigecycline, polymyxin, and colistin representing 6 different antibiotic classes. Susceptibility testing was performed by the broth microdilution method as previously described28 where Klebsiella pneumoniae (K. pneumoniae) ATCC 700603 and Pseudomonas aeruginosa (P. aeruginosa) 27853 were employed as standard control strains. The susceptibility results were interpreted according to the breakpoints of the Clinical Laboratory Standards Institute (CLSI)29 for all tested antibiotics, while the breakpoint for streptomycin was interpreted according to Yang et al.30
Phenotypic Characterization of Antibiotic Resistance Pattern
The investigated clinical isolates were considered to have MDR phenotype when they resisted three different classes of antibiotics except for carbapenems, while XDR phenotype was considered when MDR isolate was resistant to meropenem. PDR phenotype was considered if XDR isolate was colistin and tigecycline resistant.2
Phenotypic Characterization of Aminoglycoside Resistance Level
Based on the definition proposed by Nie et al,31 Upadhyay et al32 and Doi et al33 all isolates that exhibited MIC value to gentamicin and amikacin ≥512 µg/mL were considered to have high level of aminoglycoside resistance (HLAR) phenotype.
Extraction of Total DNA
Total DNA (chromosomal and plasmid DNA) was extracted as previously described34 to be used in PCR reactions for the amplification of blaOXA-51 and the genotyping by RAPD-PCR technique. The extracted DNA was kept in a sterile DNase-free 0.5 mL tube at −20 °C until use.
Extraction of Plasmid DNA
Plasmid extraction was performed to investigate the existence of AMEs, 16S rRNA methyltransferases encoding genes and, the sul1 gene as well in which each isolate was cultured in 5 mL of lysogeny broth (LB) (Himedia®, India) to increase the plasmid yield. The inoculated LB tubes were then placed in the shaking incubator at 37 °C for 16 h. After that, the culture tubes were centrifuged at 13,200 rpm for 3 min. The bacterial cell pellet was then washed twice with phosphate buffer and any remaining buffer over the pellet was pipetted out. The plasmid DNA was extracted from the washed pellet by GeneJET® Plasmid Miniprep Kit (Thermo Fischer Scientific, USA) according to the manufacturer’s instructions.
Genotypic Detection of The Plasmid Mediated Aminoglycosides and Sulfonamide Resistance
The plasmid DNA from each isolate was used as a template to investigate seven different plasmid-borne AMEs encoding genes namely; ant(3″)-I, aph(3ʹ)-VI, aac(3)-II, aac(6ʹ)-II, aac(6ʹ)-Ib, aad(2)-Ia, aad(4)-Ia and two 16S rRNA methyltransferases encoding genes namely; armA and rmtB using the specific primers and cycling conditions listed in Table 1. Also, all isolates were investigated for the sul1 gene.
PCR
The PCR was performed in a 0.2 DNase-free PCR tube containing 30 μL of the total reaction mixture that was composed of 6 µL of 5x master mix (Solis BioDyne, Estonia), 4 µL of DNA template equivalent to about 12–15 ng, 0.9 µL from each of the forward (10 pmol/μL) and the reverse (10 pmol/μL) primer (Table 1).
The volume of the reaction mixture was completed to 30 µL by adding 18.2 µL of sterile DNase-free water. Five strains from the laboratory culture collection provided by the microbiology laboratory at Taif University were used as positive controls for the 11 investigated genes. E. coli ATCC 25922 was used as a negative control strain. For the RAPD-PCR technique, 1.8 μL of the HLWL74 primer35 was used.
Gene Amplification and Electrophoresis
Target genes were amplified using Mastercycler gradient® (Eppendorf, Germany), oligonucleotide primers (Korea, Seoul), and cycling conditions listed in Table 1. All the amplified PCR products were run on 1% agarose (Scharlau, Spain) gel containing 500 ng/mL ethidium bromide. The random amplified DNA fragments generated by the RAPD-PCR were run on 2.5% agarose gels to achieve well band separation.
Gel Extraction
All PCR products subjected to capillary electrophoresis sequencing (CES) were initially extracted from the agarose gel by a glass fiber membrane using gel extraction SV kit (MG®, Korea) according to the manufacturer's instructions with a maximum yield of 90%.
DNA Sequencing
The extracted amplified PCR products of the aac(6ʹ)-Ib and the sul1 genes were subjected to CES in one direction utilizing the forward amplification primers36,39 (Table 1) using 96 capillary type ABI PRISM® 3730XL DNA Analyzer (Applied Biosystems, USA).
DNA Sequence Correction
All the released DNA sequencing ab1 files were manually corrected using the FinchTv software40 version 1.5.0 (Geospiza Inc, USA).40 The corrected sequences were uploaded to the National Center for Biotechnology Information (NCBI)41 using the blastn service in which the query DNA sequence was identified and corrected.
Translation of The Sequenced aac(6ʹ)-Ib and sul1 Genes
The corrected DNA sequences of aac(6ʹ)-Ib and the sul1 genes were uploaded to open reading frame (ORF) finder website42 to obtain the corresponding amino acid sequences using the following search parameters; (i) minimal ORF length of 300 and 600 for aac(6ʹ)-Ib and the sul1 genes, respectively; (ii) genetic code was bacterial and archaeal; (iii) ORF start codon to use was ATG only.
NCBI Database Search Criteria and Reference Sequence (RefSeq) Selection
The corresponding amino acid sequences of the sequenced aac(6ʹ)-Ib and the sul1 genes were searched in the NCBI database against the corresponding amino acid sequences reported only in A. baumannii using the blastp service with selection criteria of sequence identity and query coverage of ≥98% in which AAG33663.1 and WP_063855115 were used as the amino acids RefSeq for aminoglycoside 6ʹ-N-acetyltransferase-Ib and sulfonamide-resistant DHPS Sul1, respectively.
Identification of the Mutation Sites in the aac(6ʹ)-Ib and the sul1 Genes
The translated query amino acid sequences were subject to multiple sequence alignment (MSA) against the selected reference sequence using the Jalview software43 utilizing the fast Fourier transform (MAFFT) web service.
GenBank Sequence Accession Numbers
The partial sequences of the mutated aac(6ʹ)-Ib and sul1 genes were submitted to the GenBank44 via the submission portal45 on the NCBI website in which each mutated gene has assigned a specific GenBank accession number and specific protein accession number.
Cluster Analysis of RAPD Fragments
The amplified DNA fragments generated by RAPD-PCR were analyzed as previously described36 using BioNumerics® 7.5 software46 in which the clonal relatedness between the 40 A. baumannii clinical isolates was determined based on the generated dendrogram using the unweighted pair-group method with arithmetic average (UPGMA) utilizing the Dice coefficient .
Results
Isolation and Identification
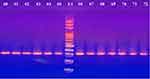
The present study revealed that all the recovered isolates belonged to the genus Acinetobacter based on the phenotypic profiles obtained from the Vitek2 and API 20NE systems, also all isolates were genetically confirmed to be A. baumannii in which 100% of isolates tested positive for the chromosomal -borne blaOXA-51 gene (Figure 1).
AST and Phenotypic Characterization of Resistance Pattern
As regards the tested aminoglycosides, the current study revealed that 55% (22/40), 57.5% (23/40), and 95% (38/40) of A. baumannii clinical were resistant to gentamicin, amikacin, and streptomycin, respectively.
Apart from tigecycline to which all isolates were sensitive, the susceptibility testing revealed that polymyxin B and colistin were the most effective agents following tigecycline in which 85% (34/40) and 67.5% (27/40) of isolates were sensitive, respectively.
Regarding the other tested antibiotics, it was found that meropenem and ciprofloxacin were the lowest effective agents in which 97.5% (39/40) of isolates were resistant for each. Also, the current study revealed that 70% (28/40) and 77.5% (31/40) of isolates were resistant to levofloxacin and sulfamethoxazole/trimethoprim, respectively.
Fortunately, no isolate exhibited a PDR pattern in which all isolates were sensitive to at least either tigecycline or colistin. On the other hand, it was found that 90% (36/40) of isolates exhibited an XDR pattern in which 67.5% (27/40) and 22.5% (9/40) of isolates were resistant to four and five different antibiotic classes, respectively. Only 5% of isolates exhibited MDR pattern in which only two isolates were resistant to only three antibiotic classes as shown in Table 2 and 3.
 |
Table 2 Susceptibility Pattern of A. baumannii Clinical Isolates |
The values of MIC50 for the tested aminoglycosides ranged from 16 to 1024 μg/mL in which the MIC50 for gentamicin, amikacin, and streptomycin were 16, 64, and 1024 μg/mL, respectively. On the other hand, the MIC90 for all of the tested aminoglycosides was 1024 μg/mL.
It was also found that the values of MIC50 and MIC90 for the tested polymyxins ranged from ≤0.5 to 2 μg/mL and 8 to 32 μg/mL, respectively.
Regarding sulfamethoxazole/trimethoprim, it was found that the values of MIC50 and MIC90 were 13.4/256 μg/mL and 53.8/1025 μg/mL, respectively. In addition, obvious elevated values of MIC50 and MIC90 for ciprofloxacin and levofloxacin were observed (Table 2).
Phenotypic Characterization of Aminoglycoside Resistance Level
The current study exhibited that 42.5% of isolates showed HLAR in which the MIC values of 17 isolates were ≥512 µg/mL for gentamicin and amikacin, while 52.5% of isolates exhibited LLAR in which 21 isolates were streptomycin-resistant and exhibited MIC value <512 µg/mL to gentamicin and amikacin. On the other hand, 5% (2/40) of isolates were sensitive to all of the tested aminoglycosides.
Genotypic Detection of Plasmid Mediated Aminoglycosides and Sulfonamide Resistance
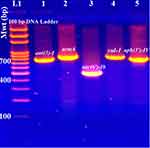
The current study revealed that aph(3ʹ)-VI and aac(6ʹ)-Ib gene variants [aac(6ʹ)-Ib and aac(6ʹ)-SL] (Figure 2 and Supplementary Figures 1–3) were the most prevalent plasmid encoding AMEs in which 90% (36/40) and 87.5% (35/40) of isolates tested positive, respectively (Table 3). On the other hand, ant(3″)-I and armA (Figure 2 and Supplementary Figures 4–6) ranked the 3rd and the 4th most prevalent plasmid-borne aminoglycoside resistance encoding genes in which 85% (34/40) and 45% (18/40) of isolates tested positive, respectively.
 |
Table 3 Resistance and Genetic Profile of A. baumannii Clinical Isolates |
The lowest detected gene responsible for aminoglycoside modification by adenylation was aad(4ʹ)-Ia in which 5% (2/40) of isolates tested positive. The other investigated aminoglycoside resistance encoding genes were not detected in which all isolates tested negative for aad (2ʹʹ)-Ia, aac(6ʹ)-II, aac(3)-II, and rmtB.
As regards the investigation of plasmid-mediated sulfonamide resistance, the current study revealed that 15% (6/40) of the isolates were found to harbor the altered DHPS encoding gene namely; the sul1 gene (Figure 2 and Supplementary Figures 7 and 8).
Correlated Phenotypic and Genotypic Resistance Profiles
The current study revealed 19 different resistance genotypes (Table 4) among the 38 aminoglycoside resistant isolates in which the resistance genotype A (blaOXA-51, ant(3″)-I, aph(3ʹ)-VI, aac(6ʹ)-Ib) was the most prevalent to which 4–6 antibiotics were resistant. On the other hand, the resistance genotype B (blaOXA-51, ant(3″)-I, aph(3ʹ)-VI, aac(6ʹ)-SL) and the resistance genotype C (blaOXA-51, ant(3″)-I, aph(3ʹ)-VI, armA, aac(6ʹ)-Ib) were the second prevalent resistance genotypes to which 4–6 and 6–9 antibiotics were resistant, respectively.
 |
Table 4 Combined Phenotypic and Genotypic Resistance Profile Among Aminoglycosides Resistant A. baumannii Clinical Isolates |
Detection of the Novel aac(6ʹ)-SL Allelic Variant in The Acinetobacter Plasmid
Only 34.29% (12/35) of the isolates which tested positive for acc(6ʹ)-Ib were found to harbor a common missense mutation in position 102 in which leucine (hydrophobic) was substituted for serine (neutral-polar)amino acid in position 102 indicating a novel allelic variant that was named as aac(6ʹ)-SL due to the presence of serine (S) amino acid instead of leucine in position 102 (L102S) and the normal presence of leucine (L) in position 117 (Figure 3).
Detection of the Mutation Sites in sul1 Gene in The Acinetobacter Plasmid
The sequencing of the sulfonamide-resistant DHPS encoding gene; namely sul1revealed a common novel misense mutation in 66.6% (4/6) of isolates tested positive in which cysteine (neutral-polar) was substituted for glycine (hydrophobic) in position 98 (C98G). Other two novel missense mutations were detected in only one isolate (Acb_96) in which aspartate (acidic-charged) was substituted for asparagine (neutral-polar) in position 40 (D40N), secondly; alanine (hydrophobic) was substituted for threonine (neutral-polar) in position 233 (A232T) as shown in Figure 4.
GenBank Accession Numbers of the the Novel aac(6ʹ)-SL Allelic Variant
The mutated aac(6ʹ)-Ib genes that detected in the current study and named acc(6')SL allelic variants were assigned the following gene accession numbers by the GenBank: MZ820065 MZ820066, MZ820067, MZ820071, MZ820072, MZ820064, MZ820068, MZ820063, MZ820069, MZ820070, MZ820073 and, MZ820074 for the isolates No. Acb73, Acb75, Acb81, Acb75, Acb80, Acb82, Acb83, Acb90, Acb91, Acb97, Acb108, and Acb109, repectively.
GenBank Accession Numbers of sul1 Genes
The mutated sul1 genes that detected in the present study were assigned the following gene accession numbers by the Genbank: MZ751055, MZ751056, MZ751057, and MZ751058 for the isolates No. Acb62, Acb81, Acb82, and Acb96, repectively.
Cluster Analysis of RAPD Fragments
The amplified fragments generated by RAPD-PCR (Figure 5 and Supplementary Figure 9) were able to classify the 40 clinical isolates of A. baumannii into three main clusters (Figure 6) in which 92.5% (37/40) of the isolates belonged to cluster II, while 5% (2/40) of the isolates were related to cluster I exhibiting identical band profile. Only one isolate was related to cluster III.
As cluster II was the main cluster to which 37 isolates were related, this cluster was found to include 24 different banding profiles in which 14 isolates exhibited 14 single different unique profiles with different RAPD-PCR banding patterns, whereas 23 isolates showed 10 different combined profiles (include more than one isolate within the same profile) exhibiting specific identical RAPD-PCR banding pattern, for instance, the isolates Acb_66, Acb_98, Acb_68, Acb_108 were included in one combined profile (Figure 6) exhibiting an identical RAPD-PCR banding pattern, also the isolates Acb_110 and Acb_67, Acb_107 and Acb_99, Acb_71 and Acb_73, Acb_90 and Acb_69, Acb_94 and Acb_65, Acb_62 and Acb_63, Acb_70, Acb_74 and Acb_76, Acb_75 and Acb_82, Acb_80 and Acb_81 exhibited nine different combined profiles with a specific identical RAPD-PCR banding pattern for each.
Discussion
The problem of antibiotic resistance represents one of the biggest obstacles facing the health system in most countries of the world, if not all, as this problem increases the financial burden of the health system due to the prolonged stay of the infected patients in the hospitals,34 especially in ICUs, as the patients infected with MDR bacterial strains usually suffer from life-threatening conditions47 that may require admission to to ICUs and placing them on ventilators leading to an increase in the cost of treatment . A. baumannii is one of the most important members of those MDR bacterial strains and therefore the research on A. baumannii occupies a great importance worldwide, especially in KSA.2
Based on reviewing the available cited literature and according to the Saudi review conducted by Yezli et al48 in 2014 and Ibrahim et al49 in 2019 there is no data about the resistance pattern and the genetic background of the plasmid-mediated aminoglycoside and sulfonamide resistance among A. baumannii clinical isolates in the Taif area so, we were prompted to investigate such a topic in an attempt to contribute in building the data concerning such an issue.
By reviewing the cited literature we found that gentamicin, tobramycin, and amikacin were officially introduced to KSA in 1975, 1983, and 1984, respectively.50 Based on the data retrieved from previous literature, it was found that the emergence of gentamicin resistant strains began to appear in KSA slowly from low-level to high-level of resistance in which Moaz et al50 and Memish et al51 reported that the resistance level to gentamicin was 26.6% (254/959) during 1983 and 1984 and 31.2% (1930/6189) in 2009, respectively among Saudi P. aeruginosa clinical isolates without any data about the resistance among A. baumannii.56
It is not surprising to report a high incidence of aminoglycoside resistance among A. baumannii clinical isolates in which the recent studies confirmed the presence of certain intrinsic aminoglycoside resistance genes on the Acinetobacter chromosome. For instance, in 201752 it was reported that the gene encoding for ANT (3″)-II is located on the Acinetobacter chromosome and was found to be transferred horizontally among the Acinetobacter spp. by homologous recombination. Based on the previous data, intrinsic and acquired resistance to aminoglycosides via AMEs is easy to occur via either intrinsic and/or acquired resistance.
The current study demonstrated that 55% of A. baumannii clinical isolates were gentamicin resistant, this finding is relatively consistent to a high extent with the findings of Haseeb et al53 and Abdalhamid et al54 who reported that 46% and 54.6% of A. baumannii clinical isolates recovered from Makkah and Dammam were gentamicin resistant, respectively.
About amikacin resistance, our study showed a low rate of amikacin resistance as compared with the recent Saudi study conducted by Almaghrabi et al55 in which 74.5% of A. baumannii clinical isolates collected from Aseer region in the southwest of the Kingdom were amikacin resistant, while only 27.5% of isolates included in the present study were amikacin resistant indicating that rate of amikacin resistance in the west region of the Kingdom is much lower than that reported in the southwest region at the time of the study.
Based on the definition proposed by Nie et al,31 Upadhyay et al32 and Doi et al33 a HLAR in A. baumannii is considered when the MIC value to gentamicin and amikacin ≥512 µg/mL so, the present study exhibited that 42.5% of isolates showed HLAR in which the MIC values for 17 isolates were ≥512 µg/mL to either gentamicin and/or amikacin, this finding is lower than the report of Upadhyay et al32 who showed that 79.2% of investigated A. baumannii clinical isolates exhibited HLAR.
Due to the scarcity of the studies dealing with the genetic background of AMEs among A. baumannii clinical isolates in KSA so, in this section, the findings of the current study were discussed in comparison with the findings of the other studies that were performed in the countries near to KSA like the Gulf countries and the countries from which most laborers in the KSA are hired from, like Egypt, India, and Pakistan.
The current study revealed a high prevalence of genes encoding for AMEs in which all isolates that exhibited HLAR resistance were found to harbor at least one of plasmid encoding genes for AMEs and/or 16S rRNA methylase, this finding is closely related to the finding of the recent Indian32 study which reported that 83.8% of HLAR A. baumannii clinical isolates harbored AMEs and/or 16s methyltransferase encoding genes.
The molecular investigation of AMEs in the present study showed that aph (3ʹ)-VI and aac(6ʹ)-Ib were the most prevalent AMEs encoding genes in which 90% and 87.50% of isolates tested positive, respectively, a closely related finding was reported by Polotto et al56 in which aph (3ʹ)-VI and aac(6ʹ)-Ib were the most prevalent AMEs encoding genes that were detected in 55% and 47% of A. baumannii clinical isolates, respectively.
As regards the prevalence of nucleotidyl transferases encoding genes, the current study revealed that 85%, 5%, and no isolates were found to harbor ant(3′′)-I, aad(4ʹ)-Ia, and aad(2ʹ)-Ia, respectively, these findings are highly consistent with the findings of Nie et al31 who reported a high prevalence rate for ant(3′′)-I in which 95.1% of their isolates tested positive, while all isolates tested negative for aad(2ʹ)-Ia and aad(4ʹ)-Ia, these findings are similar to our findings that demonstrated a low prevalence rate for aad(4ʹ)-Ia and failure to demonstrate aad(2ʹ)-Ia. Based on the previouly mentioned data, the authors of the current study concluded that ant(3′′)-I is the most commonly detected variant of ANTs encoding genes among A. baumannii.
In contrast to the findings of the current study and the findings of Nie et al31 a relatively low prevalence rate of 33% for ant(3ʹ)-Ia was reported by the recent Iranian study conducted by Jouybari et al57 among A. baumannii clinical isolates.
Concerning the genes encoding for 16S rRNA methylases, only armA gene was detected in the current study in which 45% (18/40) of the isolates tested positive, while rmtB was not detected. Closely related findings were conducted by the previous Chinese study31 which reported that 59.54% (103/173) of A. baumannii clinical isolates tested positive for armA, while no isolates tested positive for rmtB .
The current study demonstrated that the existence of AMEs encoding genes is not an evidence for the incidence of HLAR pattern at least in this study in which aph(3ʹ)-VI, aac(6ʹ)-Ib, ant(3')-I, and aad(4ʹ)-Ia were detected in 76.19% (16/21), 71.43% (15/21), 90.48% (19/21), and 9.52% (2/21) of isolates that did not exhibit a HLAR pattern. In addition to, 80.95% (17/21) of isolates that did not exhibit a HLAR were found to harbor 2–4 AMEs encoding genes assuring that that the co-existence of AMEs is not a condition for the incidence of HLAR pattern.
On the contrary, the current study demonstrated that 94.11% (16/17) of isolates that showed HLAR pattern were found to harbor an armA gene indicating that there is a close relationship between the existence of armA gene and the incidence of HLAR, this conclusive finding was also reported by Doi et al.33
In the current study, 19 different genetic aminoglycoside resistance profiles were detected. In the same context, 22 aminoglycoside resistance genetic profiles were reported by the Iranian study that was conducted by Jouybari et al57 indicating that the Iranian A. baumannii clinical isolates are more genetically diverse than Saudi isolates.
The diverse aminoglycoside genetic profiles of the investigated isolates in the current study suggest that foreign labor recruited from the neighboring countries may contribute to the introduction of genetically diverse isolates.
In addition to what was mentioned in the previous paragraph, the ease of transmission of AMEs and 16S rRNA methylase encoding genes via plasmids or other MGEs like integrons and transposons is another possibility that explains the wide diversity of aminoglycoside genetic profiles that detected in the current study. All the previous possibilities limit the effectiveness of aminoglycosides in the treatment of infection caused by aminoglycoside resistant isolates so, effective measures should be implemented to overcome the spread of antibiotic resistant strains, especially in the hospital environment by (i) following infection control measures, (ii) following the antibiotic stewardship policy, (iii) avoiding the irrational use of broad-spectrum antibiotics except for cases which require the use of these antibiotics.58
As the current study revealed that ciprofloxacin was the least effective agent to which 97.5% of isolates were resistant so, the authors decided to sequence the amplified aac(6ʹ)-Ib gene to investigate if the previously reported aac(6ʹ)-Ib-cr variant (responsible for ciprofloxacin acetylation)59 is the cause for that finding, but instead it was found that only 34.29% (12/35) of isolates tested positive for aac(6ʹ)-Ib have showed a new allelic variant that was named aac(6ʹ)-SL due to the substitution of leucine (L) by serine (S) in position 102 (L102S).
The detected aac(6ʹ)-SL variant was considered to be a novel variant for aac(6ʹ)-Ib in which all the previously reported aac(6ʹ)-Ib allelic variants59 associated with ciprofloxacin resistance was found to have leucine (L) in position 117 and either arginine (R) or tryptophan (W) in position 102 which differ from our findings that revealed the presence of serine in position 102 and leucine in position 117.
The authors of the current study thought that the newly detected aac(6ʹ)-SL allelic variant may greatly contribute to 100% ciprofloxacin resistance among the isolates that horboured such novel allelic variant based on the fact that, the missense mutation in the position 102 in the wild type of aac(6')-Ib gene that resulted in the previously reported aac(6')-Ib-cr variant has been proved to acetylate ciprofloxacin with subsequent faliure of ciprofloxacin to achieve its action and hence ciprofloxacin resistance.
As regards to sulfonamide resistance, the present study revealed that 77.5% (31/40) of the investigated isolates were resistant to sulfamethoxazole/trimethoprim, in spite of the existence of the plasmid borne sul1 gene in 19.53%(6/31) of the sulfamethoxazole/trimethoprim resistant isolates indicating that the resistance to sulfonamid in the isolates tested negative for the sul1gene is greatly contributed to the existence of other resistance determinants -like efflux pump system60 and/or other sul gene variants like sul2 and/or sul3.21
In contrast to the current study, which revealed that 15% of isolates tested positive for the sul1 gene, a relatively high prevalence for the sul1 gene was reported in A. baumannii according to the recent Iranian study conducted by Tavakol et al61 who declared that 63.63% A. baumannii isolates were found to harbor the sul1 gene indicating that the sul1 gene is less prevalent in Saudi Arabia than Iran among A.baumanniiduring the period of the study.
The molecular epidemiological investigation by RAPD-PCR revealed that 92.5% of the investigated isolates were confirmed to be nosocomially transmitted as they were included under one cluster (cluster II) with high clonal similarity, despite that these isolates were recovered from different hospital locations at different time intervals of approximately 7 months starting from 19 October 2016 to 14 May 2017, which confirms the circulation of these isolates in the hospital environment during the period of the study .
Limitation of the Study
Gene cloning and expression experiments for the detected novel aac(6ʹ)-SL allelic variant is required to confirm the role of this novel gene variant in ciprofloxacin resistance.
Conclusion
This is the first Saudi study to shed light on the plasmid-borne aminoglycoside resistance via screening of plasmid-encoding genes for AMEs and 16S rRNA methyltransferases among A. baumannii clinical isolates. The current study demonstrated a close association between the existence of the armA gene and the occurrence of HLAR also, the coexistence of one or more AMEs encoding genes is not a prerequisite for the incidence of a HLAR pattern. aph (3ʹ)-VI and aac(6ʹ)-Ib were the most prevalent AMEs encoding genes that circulate in the hospital environment, while the armA gene was the only detected 16S rRNA methylases encoding genes among clonally related nosocomially transmitted A. baumannii clinical isolates fingerprinted by RAPD-PCR. This is the first study that addresses the detection of a novel allelic variant that we named aac(6ʹ)-SL and three novel mutations in the sul1 gene.
Acknowledgment
The current work was supported by Taif University Researchers Supporting Project number (TURSP-2020/18), Taif University, Taif, Saudi Arabia.
Disclosure
The authors declare no conflicts of interest in this work.
References
1. Williams CL, Neu HM, Alamneh YA, et al. Characterization of Acinetobacter baumannii copper resistance reveals a role in virulence. Front Microbiol. 2020;11:16. doi:10.3389/fmicb.2020.00016
2. El-Badawy MF, Abdelwahab SF, Alghamdi SA, Shohayeb MM. Characterization of phenotypic and genotypic traits of carbapenem-resistant Acinetobacter baumannii clinical isolates recovered from a tertiary care hospital in Taif, Saudi Arabia. Infect Drug Resist. 2019;12:3113. doi:10.2147/IDR.S206691
3. Wong D, Nielsen TB, Bonomo RA, Pantapalangkoor P, Luna B, Spellberg B. Clinical and pathophysiological overview of Acinetobacter infections: a century of challenges. Clin Microbiol Rev. 2017;30(1):409–447. doi:10.1128/CMR.00058-16
4. Turton JF, Woodford N, Glover J, Yarde S, Kaufmann ME, Pitt TL. Identification of Acinetobacter baumannii by detection of the blaOXA-51-like carbapenemase gene intrinsic to this species. J Clin Microbiol. 2006;44:2974–2976. doi:10.1128/JCM.01021-06
5. Khurshid M, Rasool MH, Ashfaq UA, et al. Acinetobacter baumannii sequence types harboring genes encoding aminoglycoside modifying enzymes and 16SrRNA methylase; a Multicenter Study from Pakistan. Infect Drug Resis. 2020;13:2855. doi:10.2147/IDR.S260643
6. López-Durán PA, Fonseca-Coronado S, Lozano-Trenado LM, et al. Nosocomial transmission of extensively drug resistant Acinetobacter baumannii strains in a tertiary level hospital. PLoS One. 2020;15:e0231829. doi:10.1371/journal.pone.0231829
7. Du X, Xu X, Yao J, et al. Predictors of mortality in patients infected with carbapenem-resistant Acinetobacter baumannii: a systematic review and meta-analysis. Am J Infect Control. 2019;47:1140–1145. doi:10.1016/j.ajic.2019.03.003
8. Control CfD. Prevention Antibiotic Resistance Threats in the United States. Atlanta, GA: CDC; 2013.
9. Shrivastava SR, Shrivastava PS, Ramasamy J. World health organization releases global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. J Med Soc. 2018;32:76. doi:10.4103/jms.jms_25_17
10. Kong J, Wu Z-X, Wei L, Chen Z-S, Yoganathan S. Exploration of antibiotic activity of aminoglycosides, in particular ribostamycin alone and in combination with ethylenediaminetetraacetic acid against pathogenic bacteria. Front Microbiol. 2020;11:1718. doi:10.3389/fmicb.2020.01718
11. Kotra LP, Haddad J, Mobashery S. Aminoglycosides: perspectives on mechanisms of action and resistance and strategies to counter resistance. Antimicrob Agents Chemother. 2000;44(12):3249–3256. doi:10.1128/AAC.44.12.3249-3256.2000
12. Magnet S, Blanchard JS. Molecular insights into aminoglycoside action and resistance. Chem Rev. 2005;105(2):477–498. doi:10.1021/cr0301088
13. Wachino J-I, Doi Y, Arakawa Y. Aminoglycoside resistance: updates with a focus on acquired 16S ribosomal RNA methyltransferases. Infect Dis Clin. 2020;34(4):887–902. doi:10.1016/j.idc.2020.06.002
14. Galimand M, Sabtcheva S, Courvalin P, Lambert T. Worldwide disseminated armA aminoglycoside resistance methylase gene is borne by composite transposon Tn1548. Antimicrob Agents Chemother. 2005;49:2949–2953. doi:10.1128/AAC.49.7.2949-2953.2005
15. Krause KM, Serio AW, Kane TR, Connolly LE. Aminoglycosides: an overview. Cold Spring Harb Perspect Med. 2016;6(6):a027029. doi:10.1101/cshperspect.a027029
16. Vakulenko SB, Mobashery S. Versatility of aminoglycosides and prospects for their future. Clin Microbiol Rev. 2003;16(3):430–450. doi:10.1128/CMR.16.3.430-450.2003
17. El-Badawy MF, Tawakol WM, El-Far SW, et al. Molecular identification of aminoglycoside-modifying enzymes and plasmid-mediated quinolone resistance genes among Klebsiella pneumoniae clinical isolates recovered from Egyptian patients. InterJ Microbiol. 2017;2017. doi:10.1155/2017/8050432
18. Aishwarya KVL, Geetha PV, Eswaran S, Mariappan S, Sekar U. Spectrum of aminoglycoside modifying enzymes in gram-negative bacteria causing human infections. J Lab Phys. 2020;12:27.
19. McGann P, Chahine S, Okafor D, et al. Detecting 16S rRNA methyltransferases in Enterobacteriaceae by use of Arbekacin. J Clin Microbiol. 2016;54:208–211. doi:10.1128/JCM.02642-15
20. Bickel MH. The development of sulfonamides (1932–1938) as a focal point in the history of chemotherapy. Gesnerus. 1988;45:67–86. doi:10.1163/22977953-04501006
21. Byrne-Bailey K, Gaze W, Kay P, Boxall A, Hawkey P, Wellington E. Prevalence of sulfonamide resistance genes in bacterial isolates from manured agricultural soils and pig slurry in the United Kingdom. Antimicrob Agents Chemother. 2009;53(2):696–702. doi:10.1128/AAC.00652-07
22. Sköld O. Sulfonamide resistance: mechanisms and trends. Drug Resist Updat. 2000;3(3):155–160. doi:10.1054/drup.2000.0146
23. Rolbiecki D, Harnisz M, Korzeniewska E, Jałowiecki Ł, Płaza G. Occurrence of fluoroquinolones and sulfonamides resistance genes in wastewater and sludge at different stages of wastewater treatment: a preliminary case study. Appl Sci. 2020;10(17):5816. doi:10.3390/app10175816
24. Jiang H, Cheng H, Liang Y, et al. Diverse mobile genetic elements and conjugal transferability of sulfonamide resistance genes (sul1, sul2, and sul3) in Escherichia coli isolates from Penaeus vannamei and pork from large markets in Zhejiang, China. Front Microbiol. 2019;10:1787. doi:10.3389/fmicb.2019.01787
25. Wu S, Dalsgaard A, Hammerum AM, Porsbo LJ, Jensen LB. Prevalence and characterization of plasmids carrying sulfonamide resistance genes among Escherichia coli from pigs, pig carcasses and human. Acta Vet Scand. 2010;52:47. doi:10.1186/751-0147-52-47
26. Perreten V, Boerlin P. A new sulfonamide resistance gene (sul3) in Escherichia coli is widespread in the pig population of Switzerland. Antimicrob Agents Chemother. 2003;47(3):1169–1172. doi:10.1128/AAC.47.3.1169-1172.2003
27. Butler JM. Non-human DNA. Advanced Topics in Forensic DNA Typing: Methodology. Elsevier; 2012:473–495. doi:10.1016/B978-0-12-374513-2.00016-6
28. Qaiyumi S. Macro- and microdilution methods of antimicrobial susceptibility testing. In: Schwalbe R, Steele-Moore L, Goodwin AC, editors. Antimicrobial Susceptibility Testing Protocols. 1st edition. Boca Raton: CRC Press; 2007:75–79.
29. Cockerill FR. Performance Standards for Antimicrobial Susceptibility Testing: Twenty-First Informational Supplement. Wayne, PA: CLSI; 2011:29.
30. Yang C-H, Su P-W, Moi S-H, Chuang L-Y. Biofilm formation in Acinetobacter baumannii: genotype-phenotype correlation. Molecules. 2019;24(10):1849. doi:10.3390/molecules24101849
31. Nie L, Lv Y, Yuan M, et al. Genetic basis of high level aminoglycoside resistance in Acinetobacter baumannii from Beijing, China. Acta Pharm Sin B. 2014;4(4):295–300. doi:10.1016/j.apsb.2014.06.004
32. Upadhyay S, Khyriem AB, Bhattacharya P, Bhattacharjee A, Joshi SR. High-level aminoglycoside resistance in Acinetobacter baumannii recovered from intensive care unit patients in Northeastern India. Indian J Med Microbiol. 2018;36(1):43–48. doi:10.4103/ijmm.IJMM_17_225
33. Doi Y, Adams JM, Yamane K, Paterson DL. Identification of 16S rRNA methylase-producing Acinetobacter baumannii clinical strains in North America. Antimicrob Agents Chemother. 2007;51(11):4209–4210. doi:10.1128/AAC.00560-07
34. El-Badawy MF, El-Far SW, Althobaiti SS, Abou-Elazm FI, Shohayeb MM. The first Egyptian report showing the co-existence of blaNDM-25, blaOXA-23, blaOXA-181, and blaGES-1 among carbapenem-resistant K. pneumoniae clinical isolates genotyped by BOX-PCR. Infect Drug Resist. 2020;13:1237. doi:10.2147/IDR.S244064
35. Aguado V, Vitas A, Garcia-Jalon I. Random amplified polymorphic DNA typing applied to the study of cross-contamination by Listeria monocytogenes in processed food products. J Food Prot. 2001;64(5):716–720. doi:10.4315/0362-028X-64.5.716
36. El-Badawy MF, Tawakol WM, Maghrabi IA, Mansy MS, Shohayeb MM, Ashour MS. Iodometric and molecular detection of ESBL production among clinical isolates of E. coli fingerprinted by ERIC-PCR: the first Egyptian report declares the emergence of E. coli O25b-ST131clone harboring bla GES. Microb Drug Resist. 2017;23(6):703–717. doi:10.1089/mdr.2016.0181
37. Georgios M, Egki T, Effrosyni S. Phenotypic and molecular methods for the detection of antibiotic resistance mechanisms in gram negative nosocomial pathogens. Tren Infect Dis. 2014:139–162. doi:10.5772/57582.
38. Mir AR, Bashir Y, Dar FA, Sekhar M. Identification of genes coding aminoglycoside modifying enzymes in E. coli of UTI patients in. India Sci World J. 2016;2016. doi:10.1155/2016/1875865
39. Ranjbar R, Masoudimanesh M, Dehkordi FS, Jonaidi-Jafari N, Rahimi E. Shiga (Vero)-toxin-producing Escherichia coli isolated from the hospital foods; virulence factors, o-serogroups and antimicrobial resistance properties. Antimicrob Resist Infect Control. 2017;6(1):1–11. doi:10.1186/s13756-13016-10163-y
40. Patterson J, Chamberlian B, Thayer D. FinchTv Software40 Version 1.5.0. Geospiza Inc; 2019. Available from: https://digitalworldbiology.com/FinchTV.
41. National Center for Biotechnology Information, U.S. National Library of Medicine.Homepage. Available from: https://www.ncbi.nlm.nih.gov/.
42. National Center for Biotechnology Information, U.S. National Library of Medicine. Open Reading Frame Finder. Available from: https://www.ncbi.nlm.nih.gov/orffinder.
43. Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. Jalview version 2-a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi:10.1093/bioinformatics/btp033
44. National Center for Biotechnology Information, U.S. National Library of Medicine. GenBank Overview. Available from: www.ncbi.nlm.nih.gov/genbank/.
45. National Center for Biotechnology Information, U.S. National Library of Medicine. Submission Portal. Available from: https://submit.ncbi.nlm.nih.gov/.
46. BioNumerics: bioNumerics version (the version you are using) created by applied maths NV. Available from: https://www.applied-maths.com.
47. El-Badawy MF, Alrobaian MM, Shohayeb MM, Abdelwahab SF. Investigation of six plasmid-mediated quinolone resistance genes among clinical isolates of pseudomonas: a genotypic study in Saudi Arabia. Infect Drug Resist. 2019;12:915–923. doi:10.2147/IDR.S203288
48. Yezli S, Shibl AM, Livermore DM, Memish ZA. Prevalence and antimicrobial resistance among gram-negative pathogens in Saudi Arabia. J Chemother. 2014;26:257–272. doi:10.1179/1973947814Y.0000000185
49. Ibrahim ME. Prevalence of Acinetobacter baumannii in Saudi Arabia: risk factors, antimicrobial resistance patterns and mechanisms of carbapenem resistance. Ann Clin Microbiol Antimicrob. 2019;18:1–12. doi:10.1186/s12941-018-0301-x
50. Moaz A, Shannon K, Phillips I. Mechanisms of gentamicin resistance in gram-negative bacilli in Riyadh, Kingdom of Saudi Arabia. J Antimicrob Chemother. 1989;24:689–698. doi:10.1093/jac/24.5.689
51. Memish ZA, Shibl AM, Kambal AM, Ohaly YA, Ishaq A, Livermore DM. Antimicrobial resistance among non-fermenting gram-negative bacteria in Saudi Arabia. J Antimicrob Chemother. 2012;67:1701–1705. doi:10.1093/jac/dks091
52. Zhang G, Leclercq SO, Tian J, et al. A new subclass of intrinsic aminoglycoside nucleotidyltransferases, ANT (3”)-II, is horizontally transferred among Acinetobacter spp. by homologous recombination. PLoS Genet. 2017;13:e1006602. doi:10.1371/journal.pgen
53. Haseeb A, Faidah HS, Bakhsh AR, et al. Antimicrobial resistance among pilgrims: a retrospective study from two hospitals in Makkah, Saudi Arabia. Int J Infect Dis. 2016;47:92–94. doi:10.1016/j.ijid.2016.06.006
54. Abdalhamid B, Hassan H, Itbaileh A, Shorman M. Characterization of carbapenem-resistant Acinetobacter baumannii clinical isolates in a tertiary care hospital in Saudi Arabia. New Microbiol. 2014;37:65–73.
55. Almaghrabi MK, Joseph MR, Assiry MM, Hamid ME. Multidrug-resistant Acinetobacter baumannii: an emerging health threat in aseer region, Kingdom of Saudi Arabia. Can J Infect Dis Med Microbiol. 2018;2018. doi:10.1155/2018/9182747
56. Polotto M, Casella T, Tolentino FM, et al. Investigation of carbapenemases and aminoglycoside modifying enzymes of Acinetobacter baumannii isolates recovered from patients admitted to intensive care units in a tertiary-care hospital in Brazil. Rev Soc Bras Med Trop. 2020;53. doi:10.1590/0037-8682-0094-2019
57. Jouybari MA, Ahanjan M, Mirzaei B, Goli HR. Role of aminoglycoside-modifying enzymes and 16S rRNA methylase (ArmA) in resistance of Acinetobacter baumannii clinical isolates against aminoglycosides. Rev Soc Bras Med Trop. 2021;54. doi:10.1590/0037-8682-0599-2020
58. Lee C-R, Cho IH, Jeong BC, Lee SH. Strategies to minimize antibiotic resistance. Inter J Environ Res Pub Health. 2013;10:4274–4305. doi:10.3390/ijerph10094274
59. Kim D-W, Thawng CN, Choi J-H, Lee K, Cha C-J. Polymorphism of antibiotic-inactivating enzyme driven by ecology expands the environmental resistome. ISME J. 2018;12:267–276. doi:10.1038/ismej.2017.168
60. Kyriakidis I, Vasileiou E, Pana ZD, Tragiannidis A. Acinetobacter baumannii antibiotic resistance mechanisms. Pathogens. 2021;10(3):373. doi:10.3390/pathogens10030373
61. Tavakol M, Momtaz H, Mohajeri P, Shokoohizadeh L, Tajbakhsh E. Genotyping and distribution of putative virulence factors and antibiotic resistance genes of Acinetobacter baumannii strains isolated from raw meat. Antimicrob Resist Infect Control. 2018;7(1):1–11. doi:10.1186/s13756-018-0405-2
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.