Back to Journals » Infection and Drug Resistance » Volume 15
The First Saudi Report of Novel and Common Mutations in the gyrA and parC Genes Among Pseudomonas Spp. Clinical Isolates Recovered from Taif Area
Authors El-Badawy MF, Eed EM, Sleem AS, El-Sheikh AAK, Maghrabi IA, Abdelwahab SF
Received 3 May 2022
Accepted for publication 1 July 2022
Published 16 July 2022 Volume 2022:15 Pages 3801—3814
DOI https://doi.org/10.2147/IDR.S372027
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Mohamed F El-Badawy,1,* Emad M Eed,2,* Asmaa S Sleem,3 Azza AK El-Sheikh,4 Ibrahim A Maghrabi,5 Sayed F Abdelwahab6
1Department of Microbiology and Immunology, Faculty of Pharmacy, University of Sadat City, Sadat City, Menoufia, 32897, Egypt; 2Department of Clinical Laboratory Sciences, College of Applied Medical Sciences, Taif University, Taif, 21944, Saudi Arabia; 3Department of Medical Microbiology and Immunology, Faculty of Medicine, Menoufia University, Menoufia, 32511, Egypt; 4Basic Health Sciences Department, College of Medicine, Princess Nourah bint Abdulrahman University, Riyadh, 11671, Saudi Arabia; 5Department of Clinical Pharmacy, College of Pharmacy, Taif University, Taif, 21944, Saudi Arabia; 6Department of Pharmaceutics and Industrial Pharmacy, College of Pharmacy, Taif University, Taif, 21944, Kingdom of Saudi Arabia
*These authors contributed equally to this work
Correspondence: Mohamed F El-Badawy, Department of Microbiology and Immunology, Faculty of Pharmacy, University of Sadat City, Sadat City, Menoufia, 32897, Egypt, Tel +20-103-205-9964, Email [email protected]
Background and Aims: Reports examine quinolone resistance mechanisms among Pseudomonas spp. are sporadic in the Kingdom of Saudi Arabia (KSA). We previously examined the genetic bases of plasmid-mediated quinolone resistance among Pseudomonas spp. clinical isolates. This study investigated chromosomally mediated quinolone resistance mechanisms via investigation of the mutations in the gyrA and parC genes.
Methods: The minimum inhibitory concentration (MIC) to different quinolones was determined. Twenty-nine quinolone resistant Pseudomonas spp. clinical isolates were included. The gyrA and parC genes were sequenced by Sanger capillary electrophoresis. Multiple sequence alignment for the translated gyrA and parC genes was performed to identify mutation sites.
Results: Of the 29 isolates, 27 isolates were P. aeruginosa and two were P. putida. The cluster analysis of the quinolone susceptibility pattern revealed seven susceptibility phenotypes (A-G) based on susceptibility patterns rather than the MIC values. Also, 22 different susceptibility phenotypes were detected based on MIC values. All isolates exhibited a missense mutation at position 83 (S83I/T/F) of the gyrA gene in addition to six missense mutations at positions outside the QRDR of this gene. In addition, 82.8% (24/29) of the isolates harbored a missense mutation in the parC gene at position 87 (S87L), along with six novel mutations outside the QRDR of the parC gene. Haplotyping of the gyrA, parC, and the overall QRDR revealed six, 10, and 13 different haplotypes, respectively.
Conclusion: This study documents the incidence of the commonly reported mutations in the gyrA and parC genes in addition to novel mutations in these genes among Pseudomonas spp. clinical isolates recovered from KSA. Together with our previous findings, these data provide an insight into the genetic background of quinolone resistance among Pseudomonas spp. clinical isolates in KSA.
Keywords: minimum inhibitory concentration, Pseudomonas, quinolone resistance determining region, quinolones, haplotyping
Introduction
Pseudomonas aeruginosa (P. aeruginosa) is a widely distributed opportunistic pathogen causing infections in certain high-risk patients, eg, those with immune suppression, cystic fibrosis, and severe burns.1,2 Also, P. aeruginosa is a frequent cause of healthcare-associated infections due to its intrinsic resistance to a wide range of antimicrobial agents and its possession of many regulatory and metabolic pathways that deal with antibiotic pressure.3,4
Quinolones are synthetic antibiotics that are frequently used to treat Pseudomonas infections. Unfortunately, due to the irrational use of quinolones, resistance to this group has been increasing over the last years with the emergence of multi-drug resistant (MDR) P. aeruginosa.5,6
Quinolones work by inhibiting the bacterial DNA supercoiling and relaxation process, which are crucial for the replication and transcription of DNA.7 Quinolones exert their effect via non-covalent binding and inhibition of two target enzymes namely, DNA gyrase and topoisomerase IV with some variations in the affinity of different quinolones to both enzymes.8
DNA gyrase and topoisomerase IV catalyze the bacterial DNA cleavage to allow negative supercoiling and re-ligation in an ATP-dependent manner. This process is crucial for DNA condensation, replication, and transcription initiation.9
DNA gyrase is a tetramer enzyme, composed of two GyrA and two GyrB subunits. The GyrA subunit binds to the bacterial DNA and cleaves its backbone while GyrB drives the ATP-dependent supercoiling process. Likewise, topoisomerase IV is composed of four subunits: two ParC and two ParE subunits, which have similar functions to GyrA and GyrB.10 Indeed, quinolones do not affect the DNA strand at the cleavage step but inhibit the re-ligation process, resulting in a sustained cleaved DNA backbone that inhibits DNA replication. Thus, quinolones are generally considered as bacteriostatic agents. However, their high dose can result in double-stranded DNA breaks with subsequent bactericidal effects.8
The resistance to quinolones may either be chromosomally or plasmid-mediated. Chromosomally mediated quinolone resistance in P. aeruginosa may arise from (i) mutational alterations in the gyrA and/or parC genes with subsequent alteration in the binding affinity to GyrA and ParC proteins (target modification) that altogether forms the quinolone resistance determining region (QRDR), (ii) efflux pumps overexpression, and (iii) downregulation of certain outer membrane proteins (OMPs).11,12 These chromosomal mechanisms act separately or together leading to various degrees of quinolone resistance that range from reduced susceptibility to complete resistance.13 In this regard, and due to the fact that the pre-2019 Clinical Laboratory Standards Institute (CLSI) breakpoints for ciprofloxacin and levofloxacin against P. aeruginosa were too high to detect low level quinolone resistance,14,15 these breakpoints were recently amended. Now, the minimum inhibitory concentration (MIC) for ciprofloxacin, levofloxacin, norfloxacin, ofloxacin and gatifloxacin against P. aeruginosa are ≤0.5, ≤1, ≤4, ≤2 and ≤2 μg/mL, respectively.16
The QRDR is a hot spot for mutation that confer resistance to quinolones and fluoroquinolones. The QRDR of the GyrA protein is defined as the region from alanine at position 67 to glutamine at position 10617 while that of GyrB is from aspartate to lysine at positions 426 and 447, respectively (numbering is based on E. coli gyrase) as described.18 On the other hand, the QRDR of ParC protein is defined as the region from alanine to glutamine at positions 64 and 103, respectively, while that of ParE protein encompasses the area between aspartate and lysine at positions 420 and 441, respectively.19
Plasmid-mediated quinolone resistance (PMQR) has been previously described and many genes carried on plasmids were involved in that process, eg, qnrD, qnrS, and aac(6´)-Ib-cr.20 Chromosomally mediated quinolone resistance due to mutation(s) in the QRDR results in reduction or abolishing the affinity of quinolones to DNA gyrase and/or topoisomerase.12 These mutations are considered the major contributors to quinolone resistance in P. aeruginosa that were first reported in 1988.21
Previous studies showed that the most frequently reported mutation in the GyrA protein sequence variants was at positions 83 (threonine replacement by isoleucine) and 87 (aspartate replacement by asparagine, tyrosine or glycine). For ParC, the most reported sequence variations were at position 87, where serine is replaced by leucine.8
The target modification mediated resistance is usually accompanied by a high level of expression for efflux pumps that extrude quinolones outside the bacterial cell. To this end, clinically resistant Pseudomonas isolates often have multiple chromosomal mutations in gyrA and parC genes.22–24
In Saudi Arabia, few studies had investigated the quinolones resistance among Pseudomonas spp. in clinical settings, particularly in the western region20,25,26 with few mechanistic studies. We previously investigated PMQR among clinical isolates of Pseudomonas spp.20 The main objective of the current study was to investigate the sites of mutations in the QRDR among Saudi clinical isolates of quinolone-resistant Pseudomonas spp. that were previously20 confirmed to harbor at least one of the PMQR encoding genes.
Methods
Cases, Isolation, and Identification of the Clinical Isolates
A total of twenty-nine confirmed Pseudomonas spp. clinical isolates were retrieved from the laboratory culture collection of the Microbiology laboratory at the College of Pharmacy, Taif University to investigate chromosomal mutations in the QRDR. The 29 clinical isolates in this study were recovered from various clinical specimens including blood (n=1), catheter tip (n=1), axillary swab (n=1), sputum (n=15), bile (n=2), urine (n=6), and wound (n=3) infections. The clinical isolates were characterized and identified as previously described.20 In this regard, all isolates were primarily identified by Vitek2® system and API 20NE® (BioMérieux, France). The genus level was also molecularly confirmed via the amplification of the algD GDP gene using primer pairs and conditions as previously described.20 Also, the genus was confirmed and species level was identified via the amplification followed by sequencing of the two house keeping genes namely gyrA and parC genes as described below. The study protocol was approved by Taif University ethical committee (approval #38-35-0021), which is accredited by the National Committee for Bioethics (# HAO-02-T-105).
Antimicrobial Susceptibility Testing (AST)
Broth microdilution method was employed to investigate the susceptibility to the available generations of quinolones, where seven quinolones (Sigma-Aldrich, USA) were tested as described.20 The 1st generation quinolones was represented by nalidixic acid, while the 2nd generation quinolones was represented by ciprofloxacin, norfloxacin, and ofloxacin. The 3rd generation quinolones was represented by levofloxacin. Gemifloxacin and moxifloxacin were the representatives of the 4th generation quinolones. Escherichia coli (E.coli) ATCC 25922 and P. aeruginosa ATCC27853 were used as control strains. The susceptibility data were interpreted based on the guidelines of the CLSI27 except for gemifloxacin and moxifloxacin that were interpreted according to the two previous reports by Grillon et al and Stone et al, respectively,28,29 as there are no available data in the CLSI or other international antimicrobial committees about the MIC breakpoints for these two agents for Pseudomonas spp.
Cluster Analysis of Quinolone Susceptibility Profiles
The results of quinolone susceptibility were uploaded to the Bionumerics® 7.5 software (Applied Maths, Belgium) using the antibiotic susceptibility application utilizing the categorical value to calculate the similarity coefficient and unweighted pair group method with arithmetic mean (UPGMA) to perform cluster analysis. The results of cluster analysis were performed utilizing both the antibiotic MIC sensitive-intermediate-resistant (SIR) experiment type mode and the antibiotic MIC value experiment type mode.
Chromosomal DNA Extraction, PCR, Amplified Genes’ Extraction and Purification
Chromosomal DNA was extracted by InstaGene Matrix kit (Bio-Rad, USA) according to the manufacturer’s instructions. The amount of DNA used in each PCR reaction was 20–100 ng/total reaction volume. The target gene-specific primer pairs and cycling conditions are listed in Table 1 using Dr.MAX DNA Polymerase (Doctor protein INC, Korea) and DNA Engine Tetrad 2 Peltier Thermal Cycler (Bio-Rad, USA). Two μL of each amplified PCR product were loaded on 1.5% agarose gel and allowed to run for 20 min at 300V and 200A. The separated DNA bands of interest were excised from the agarose gel to be purified using multiscreen® PCR96 (Millipore SAS, Molsheim, France) as PCR cleanup plates.
 |
Table 1 Primer Sequences, Amplicon Size and PCR Conditions of the Target Genes |
DNA Sequencing
The purified PCR products of the gyrA and parC genes were Sanger sequenced in one direction by capillary electrophoresis sequencing (CES) utilizing the forward primer used in PCR amplification, the BigDye terminator v3.1 sequencing kit, and the ABI PRISM® 3730XL automated sequencer (Applied Biosystems, Foster City, CA). Nucleotide sequences were determined at the Macrogen sequencing facility (Macrogen Inc., Seoul, Korea).
Sequence Correction and Translation of the gyrA and parC Genes
All the gyrA and parC gene sequences were corrected using the molecular evolutionary genetic analysis (MEGA®) 6 software30 (Bio design institute, Tempe) as previously described.31 The corrected DNA sequences were then uploaded to the National Center for Biotechnology Information (NCBI)32,33 using the blastn service to identify the query sequence.
The corrected DNA sequences of the gyrA and parC genes were uploaded to the website of open reading frame (ORF) finder in which the correct reading frame was the amino acid sequence that was obtained using the following parameters; (i) minimal ORF length nucleotide (nt) of 150 and 75 for the gyrA and parC genes, respectively; (ii) genetic code was 11. Bacterial, Archaeal, and Plant plastid; (iii) ORF start codon to use was ATG only.
NCBI Database Search Criteria and Reference Sequence (RefSeq) Selection
The corresponding amino acids of the sequenced gyrA and parC genes were subjected to blastp in the NCBI database against P0AES4.1 and BAA37152.1 RefSeq for DNA gyrase subunit A (GyrA protein) and topoisomerase IV subunit (ParC protein), respectively.
Identification of the Mutation Sites in the gyrA and parC Genes
Multiple sequence alignment (MSA) of the translated corrected sequences of the gyrA and parC genes was performed as previously described using the Jalview software,34 in which the fast Fourier transform (MAFFT) web service was applied.
GenBank Sequence Accession Numbers
All the DNA and the amino acid sequences of the investigated gyrA and parC genes were submitted to the GenBank35 via the online submission portal36 in which each gene has been assigned a specific GenBank accession number.
Multiple Sequence Alignment (MSA) and Phylogenetic Analysis of the Sequenced Genes
The amino acids obtained from the sequenced genes were subjected to MSA to get the clonal relationship between the sequenced genes and the selected RefSeq for each gene using the Jalview software utilizing MAFFT web service to generate the dendrogram.34 All the aligned amino acid sequences for each of gyrA and parC genes were subjected to phylogenetic analysis using the neighbor-joining method using % identity from MAFFT WS alignment of Cut&Paste Input-FASTA function in Jalview software to generate the phylogenetic relatedness dendrogram.34
Results
Isolation and Identification of Pseudomonas Species
The present study revealed that all the recovered isolates belonged to the genus Pseudomonas based on the data obtained by API 20NE and Vitek2®systems. 93.1% (n=27) of the 29 isolates were P. aeruginosa, while 6.9% (two isolates) were P. putida. Molecular confirmation of the phenotypically identified isolates via the amplification of the algD GDP gene revealed that all isolates belonged to the genus Pseudomonas, where all isolates tested positive (Figure 1). Also, the amplification and sequencing of gyrA and parC genes confirmed these data (Figure 2).
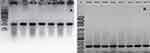 |
Figure 2 (A) PCR amplicons of the gyrA gene; (B) PCR amplicons of the parC gene. lanes MF66 to MF72 represent the isolate numbers. Abbreviation: L, 100 bp DNA ladder. |
Antimicrobial Susceptibility Testing
This study revealed that all the investigated isolates were resistant to at least one of the tested quinolones, where 100% of the isolates were resistant to nalidixic acid (Table 2, Figures 3 and 4). Among the tested quinolones, moxifloxacin was the lowest effective to which 96.6% (28/29) of the isolates were resistant. On the other hand, gemifloxacin exhibited the best activity among the tested quinolones, where only 69.0% of the isolates (20/29) were resistant. Concerning the other tested quinolones, it was found that 86.2% of the isolates (25/29) were resistant to each of ciprofloxacin, norfloxacin, ofloxacin, and levofloxacin. High-level quinolone resistance was observed among all the tested isolates, where the MIC50 and MIC90 of the tested fluoroquinolones ranged from 16 to 64 and 32 to >1024, respectively (Table 2).
 |
Table 2 Quinolone Susceptibility Pattern Among Pseudomonas Spp. Clinical Isolates |
 |
Figure 3 Cluster analysis of quinolone susceptibility profile based on Sensitive Intermediate-Resistant (SIR) interpretation. |
Cluster Analysis of Quinolone Susceptibility Profile Based on SIR Interpretation and MIC Values
The cluster analysis of the quinolone susceptibility pattern based on SIR classification (Figure 3) revealed seven quinolone susceptibility phenotypes (A-G) distributed among two main clusters (cluster I and II) in which four isolates belonged to cluster I, while the remaining 25 isolates belonged to cluster II. Cluster I was divided into two clades (A and B). Cluster II was divided into five clades (C to G) in which clade G included 20 isolates that exhibited the same resistance profile that was characterized by the resistance to all tested quinolones.
On the other hand, the cluster analysis of the quinolone susceptibility according to the MIC values revealed a complex but more accurate, descriptive, and discriminatory profile where the 29 isolates exhibited 22 different susceptibility phenotypes (Figure 4). Only 18 isolates exhibited single unique profile, while 11 isolates were distributed among four different profiles (A-D), in which profile A and B included three and four isolates, respectively, while profiles C and D included two isolates each. Profile A included isolates #MF70, MF73, and MF77, while the profile B included isolates #MF69, MF75, MF80, and MF81. On the other hand, profile C included isolates #MF72 and MF91, and profile D contained isolates #MF67 and MF84.
Detection of the Frequently Reported Missense Mutations Within the QRDR Frame (Positions 83 and 87) of gyrA Gene
This study revealed that all isolates exhibited a missense mutation at position 83 of GyrA protein in which serine (S) was substituted by either isoleucine (I), threonine (T), or phenylalanine (F). The most common missense mutation in the current study was at position 83 (S83I) in which 82.8% (24/29) of the isolates tested positive (Figures 4 and 5). The second frequent form of missense mutation in the current study was at position 83 (S83T) where 13.8% (4/29) of the isolates exhibited such mutation. The last and the least frequent form of mutation was at position 83 (S83F) that was detected in only one isolate. On the other hand, this study revealed that 13.8% (4/29) of the isolates exhibited the previously reported missense mutation at position 87 that involved the substitution of aspartate (D) with asparagine (N): (D87N).
Detection of Seven Different Novel Missense Mutations in the gyrA Gene Outside the QRDR Frame
This study revealed that all isolates were found to harbor six missense mutations at positions 112, 116, 125, 127, 130, and 135 (Figures 4 and 5) in which isoleucine (I) was substituted by valine (V) at position 112 (I112V), serine (S) was substituted by histidine (H) at position 116 (S116H), or Aspragine (S116N), isoleucine (I) was substituted by valine (V) at position 125 (I125V), leucine (L) was substituted by methionine (M) at position 127 (L127M), isoleucine (I) was substituted by leucine (L) at position 130 (I130L), and methionine (M) was substituted by leucine (L) at position 135 (M135L). Only one isolate (MF66) exhibited a missense mutation at position 77 in which tyrosine (Y) was substituted by valine (V) (Figure 5).
Identification of the Mutation Sites in the parC Gene Within the QRDR Frame (Positions 65, 66, and 87)
This study demonstrated that 82.8% (24/29) of the Pseudomonas spp. clinical isolates harbored the previously reported missense mutation at position 87 (Figures 4 and 6) in which serine (S) was substituted for leucine (L); (S87L). Importantly, this study showed two novel missense mutations at two different sites (65 and 66) in which the 1st novel mutation was detected in two forms at position 65, where 82.8% (24/29) and 6.9% (2/29) of the isolates exhibited a substitution of serine (S) with isoleucine (I); (S65I) (the 1st form) and serine with phenylalanine (F); (S65F) (the 2nd form). The 2nd novel mutation was observed at position 66 in which lysine (K) was substituted by glutamine (Q); (K66Q) in 86.2% (25/29) of the isolates (Figures 4 and 6).
Detection of Four Novel Missense Mutations at Three Different Sites in the ParC Protein Outside the QRDR Frame
This study demonstrated four novel mutations in 20.7% (5/29) of isolates at three different positions outside the QRDR frame of the ParC protein at positions 138, 139, and 140 in which two forms of mutations at position 138 were detected, the 1st form was substitution of valine (V) by glycine (V138G), the 2nd form was substitution of valine by glutamic acid (E (V138E). The mutations at the other two sites were substitution of leucine (L) by alanine (A) at positions 139 and 140 (L139A and L140A).
Haplotyping of the Sequenced gyrA and parC Genes
The current study demonstrated six different haplotypes (A-F) for the gyrA gene in which the haplotype D (S83I/I112V/S116N/ I125V/ L125M/ I130L/M135L) was the most prevalent one that was detected in 62.1% (18/29) of the isolates (Figures 4 and 5). Regarding the parC gene, ten different haplotypes (A-J) were detected with haplotype A (S65I/(K66Q/S87L) being the most prevalent; represented by 58.6% (17/29) of the isolates (Figures 4 and 6).
Combined QRDR Haplotypes for the gyrA and parC Genes
According to the identified mutation sites in both gyrA and parC genes, 13 QRDR haplotypes (A-M) were detected (Figure 4). QRDR genotype E (gyrA; S83I, I112V, S116N, I125V, L127M, I130L, M135L/parC; S65I, K66Q, S87L) was the most prevalent, where 41.4% (12/29) of the isolates expressed such a haplotype. On the other hand, genotypes C and D ranked the 2nd prevalent haplotypes (two isolates each). The other haplotypes (A-B and F-M) were equally distributed (3.5%, one isolate each, Figure 4).
Phylogenetic Relatedness Between the Sequenced gyrA Genes
The phylogenetic analysis of the sequenced gyrA genes revealed six different GyrA protein sequences that belonged to two main clusters (cluster I and cluster II) involving six different clades (Figure 7). The GeneBank accession numbers for the 29 sequenced gyrA gene samples are OL795503-OL795531 (https://www.ncbi.nlm.nih.gov/nuccore/OL795503). It was found that 93.1% (27/29) of the gyrA gene sequences belonged to cluster I that was classified into two main clades (A and B) with clade B having three subclades (B.1, B.2. and B.3). Subclade B.3 was the major subclade within-cluster I with eighteen isolates that exhibited identical amino acid sequences (Figure 7). Also, four sequences were clustered under clade B.1. exhibiting identical amino acid sequences. In addition, four sequences were clustered under clade A. Only 6.9% (2/29) of the sequences belonged to cluster II that involved two clades (C and D; Figure 7).
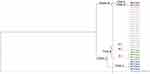 |
Figure 7 Phylogenetic relatedness between the sequenced gyrA genes and the P0AES4.1 reference sequence using neighbor-joining method utilizing % identity from MAFFTWS alignment. |
Phylogenetic Relatedness Between the Sequenced parC Genes
The cluster analysis of the sequenced parC genes revealed nine different ParC protein sequences distributed among two main clusters (cluster I and cluster II) involving four main clades (A-D); two clades within each cluster (Figure 8). The GeneBank accession numbers for the 29 sequenced parC gene samples are OL795886-OL795914 (https://www.ncbi.nlm.nih.gov/nuccore/OL795886). The current study demonstrated that 58.6% (17/29) of the ParC protein sequences (Figure 8) were contained within subclade B.1. On the other hand, only three sequences (MF69, MF75, and MF79) exhibited identical sequence profiles within subclade C.1 in cluster I (Figure 8).
 |
Figure 8 Phylogenetic relatedness between the sequenced parC genes and the BAA37152.1 reference sequence using the neighbor-joining method utilizing % identity from MAFFTWS alignment. |
Discussion
This study examined the genetic mutations in the QRDR of 29 quniolone-resistant Pseudomonas spp. isolates (27 P. aeruginosa and two P. putida) that were recovered from clinical cases admitted or attended to a tertiary care hospital located in Taif area, Saudi Arabia. These isolates were previously confirmed to have at least one of the PMQR genes.20
The cluster analysis of the quinolone susceptibility pattern revealed seven quinolone susceptibility phenotypes (A-G) classified into two main clusters (I and II). Also, 22 different susceptibility phenotypes were determined based on MIC values. All isolates exhibited a missense mutation at position 83 of gyrA gene (S83I/T/F) and harbored six additional missense mutations outside the QRDR frame at positions 112, 116, 125, 127, 130, and 135. In addition, 82.8% (24/29) of the isolates harbored the common mutation in the parC gene (S87L).
Six additional novel mutations outside the QRDR frame were found in the parC gene. Six, 10 and 13 different gyrA, parC and combined QRDR haplotypes were found with haplotype D, A and E being the most prevalent, respectively. This study concludes that the previously reported and novel QRDR mutaions are highly diverse in gyrA and parC genes among Pseudomonas spp. in Saudi Arabia. These findings together with our previous findings20 provide an insight into the genetic makeup of quinolone resistance among Saudi Pseudomonas spp. clinical isolates at least in the western region of the Kingdom.
To the best of our knowledge, there are no reports from Saudi Arabia that investigated the genetic mechanisms of quinolone resistance among Pseudomonas isolates by examining the chromosoal mutations in both the gyrA and parC genes. Several aspects of these findings deserve further discussion.
Pseudomonas spp. resistance to fluoroquinolones is usually mediated by mutations in the amino acids located in the QRDRs of gyrA, gyrB, parC and parE genes.37 The most commonly reported mutations in GyrA are S83I and D87N while that in ParC is S87L.
According to the results of the current study, the sequencing of the gyrA and parC genes confirmed the presence of several mutations in the QRDRs in all the investigated isolates. These were represented by missense mutations at position 83 (S83I/T/F) of the gyrA gene in addition to the mutation in the parC gene (S87L), where 82.8% (24/29) of the isolates harbored this mutation. Similar mutations in those positions of gyrA and parC, and QRDRs were reported among Pseudomonas isolates in Tunisia,7 Palestine,38 Estonia,2 Iran,33,39,40 South China,41 USA,42 Germany,24 Japan,43,44 and New Zealand.45
It is to be noted that GyrA wild-type amino acid at position 83 of P. aeruginosa was reported as either serine or threonine in several studies as most comparisons were done with E. coli GyrA.2,13,43–47
Regardless of whether threonine or serine is the wild type, we have isoleucine mutation in 24 (83%) of the isolates in this study.
To the best of our knowledge, the following mutations in the GyrA protein are considered novel: Y77V, S83F, I112V, S116H, S116N, I125V, L127M, I130L and M135L. On the other hand, and also to the best of our knowledge, the following mutations in the ParC protein are considered novel: S65I, S65F, K66Q, V138G, V138E, L139A and L140A.
In this study, the correlation of quinolone resistance as determined by MIC and SIR values with the presence of mutations in the QRDRs of the gyrA and parC genes of the 29 clinical isolates, and the combination of both were examined herein and their phylogenetic analysis is shown in Figures 4–8.
As shown, all the clinical isolates harbored several missense mutations in the gyrA, and parC genes. However, these mutations did not necessarily lead to an increase in the MIC values especially for the fluoroquinolones but various degrees of resistance. Similar findings were reported.13
In contrast, combinations of mutations in the QRDRs of gyrA and parC always resulted in a fluoroquinolone-resistant phenotype. Similar findings were reported elsewhere.24,45
As mentioned above, quinolones’ resistance is either chromosomally or plasmid-mediated through different mechanisms.11,12 These mechanisms act separately or in combination leading to various degrees of quinolone resistance that range from reduced susceptibility to complete resistance.13
In this study, the S83F mutation in the gyrA gene was greatly associated with fluoroquinolone resistance, where isolate #MF89 exhibited full resistance to all the tested fluoroquinolones despite the absence of S83I mutation suggesting that the presence of S83F in this isolate along with the presence of S87L mutation in the parC gene may be the leading cause of resistance (Figure 4).
We, also, found that S83T mutation in the gyrA gene was associated with the fluoroquinolone resistance. In this regard, isolates #MF68 and MF91 exhibited complete resistance to all investigated quinolones despite the absence of S83I and S87L mutations in the gyrA and parC genes, respectively. In addition, S83T mutation in the gyrA gene was detected in isolates #MF85 and MF93 that exhibited resistance to two to five tested quinolones despite the absence of S83I and S87L mutations in the gyrA and parC genes, respectively.
On the other hand, this study showed that all the following mutations including I112V, S116N, I125V, L125M, I130L, and M135L in the gyrA gene were not associated with the fluoroquinolone resistance, where isolate #MF89 exhibited a complete sensitivity pattern to all the tested fluoroquinolones despite the presence of these mutations. However, this isolate was resistant to nalidixic acid (Figure 4).
In addition, this study demonstrated that the S65I and K66Q mutations in the parC gene did not result in fluoroquinolone resistance as isolate #MF89 was sensitive to all tested fluoroquinolones despite the presence of these mutations along with the S83I mutation in the gyrA gene.
Moreover, in this study, the S83I and S87L mutations in the gyrA and parC genes, respectively, were not a condition for fluoroquinolone resistance but instead, they were associated with nalidixic acid resistance, where isolate #MF89 was sensitive to all tested quinolones except nalidixic and was found to harbor these mutations. Taken together with our previous findings,20 these data show the diversity of quinolone resistance mechanisms among Pseudomonas isolates in Saudi Arabia.
In the context of the significance of this study in disease biology, the findings of the current study strongly suppose that the detected common and novel mutations in the current study in both gyrA and parC genes among the Pseudomonas spp. clinical isolates may indirectly affect the disease biology by decreasing or abolishing susceptibility of Pseudomonas spp. to either old or newer generations of fluoroquinolones which subsequently redirects the prescribing clinician to the use of the other antibacterial agents on a large scale that may lead to the emergence of new resistant strains to the newly prescribed antibiotics that may be preserved as a last resort for treatment of either multiple drug-resistant (MDR) pan-drug resistant (PDR) isolates.
The previous hypothesis increases the antibiotic selective pressure via stimulation of other resistance mechanism leading to the emergence of multiple MDR or even PDR Pseudomonas isolates that may circulate in the hospital environment leading to either increased hospital stays with subsequent increased morbidity and mortality rates or transmission of the newly evolved resistance mechanisms via horizontal gene transfer (HGT).
Conclusions
This study revealed the previously and frequently reported mutations in the QRDR and novel QRDR mutations in gyrA and parC genes that are highly diverse among Pseudomonas spp. These data and our previous findings provide an understanding of the genetic bases of quinolone resistance among Pseudomonas isolates for the first time from Saudi Arabia.
Acknowledgments
Taif University Researchers Supporting Project Number (TURSP-2020/51), Taif, University, Taif, Saudi Arabia and Princess Nourah bint Abdulrahman University Researchers Supporting Project number (PNURSP2022R91), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia.
Disclosure
The authors declare no conflicts of interest in this work.
References
1. Kocsis B, Gulyas D, Szabo D. Diversity and distribution of resistance markers in Pseudomonas aeruginosa international high-risk clones. Microorganisms. 2021;9(2):359. doi:10.3390/microorganisms9020359
2. Telling K, Laht M, Brauer A, et al. Multidrug resistant Pseudomonas aeruginosa in Estonian hospitals. BMC Infect Dis. 2018;18(1):513. doi:10.1186/s12879-018-3421-1
3. Dulyayangkul P, Satapoomin N, Avison MB, Charoenlap N, Vattanaviboon P, Mongkolsuk S. Over-expression of hypochlorite inducible Major Facilitator Superfamily (MFS) pumps reduces antimicrobial drug susceptibility by increasing the production of MexXY Mediated by ArmZ in Pseudomonas aeruginosa. Front Microbiol. 2020;11:592153. doi:10.3389/fmicb.2020
4. Raman G, Avendano EE, Chan J, Merchant S, Puzniak L. Risk factors for hospitalized patients with resistant or multidrug-resistant Pseudomonas aeruginosa infections: a systematic review and meta-analysis. Antimicrob Resist Infect Control. 2018;7(1):79. doi:10.1186/s13756-018-0370-9
5. Viasus D, Puerta-Alcalde P, Cardozo C, et al. Predictors of multidrug-resistant Pseudomonas aeruginosa in neutropenic patients with bloodstream infection. Clin Microbiol Infect. 2020;26(3):345–350. doi:10.1016/j.cmi.2019.07.002
6. Hernandez-Garcia M, Garcia-Castillo M, Garcia-Fernandez S, et al. Presence of chromosomal crpP-like genes is not always associated with ciprofloxacin resistance in Pseudomonas aeruginosa clinical isolates recovered in ICU patients from Portugal and Spain. Microorganisms. 2021;9(2):388. doi:10.3390/microorganisms9020388
7. Ben Nejma M, Sioud O, Mastouri M. Quinolone-resistant clinical strains of Pseudomonas aeruginosa isolated from University Hospital in Tunisia. 3 Biotech. 2018;8(1):1. doi:10.1007/s13205-017-1019-8
8. Rehman A, Patrick WM, Lamont IL. Mechanisms of ciprofloxacin resistance in Pseudomonas aeruginosa: new approaches to an old problem. J Med Microbiol. 2019;68(1):1–10. doi:10.1099/jmm.0.000873
9. Ahmed MN, Porse A, Sommer MOA, Hoiby N, Ciofu O. Evolution of antibiotic resistance in biofilm and planktonic Pseudomonas aeruginosa populations exposed to subinhibitory levels of ciprofloxacin. Antimicrob Agents Chemother. 2018;62(8). doi:10.1128/AAC.00320-18
10. Fan Z, Chen H, Li M, et al. Pseudomonas aeruginosa polynucleotide phosphorylase contributes to ciprofloxacin resistance by regulating PrtR. Front Microbiol. 2019;10:1762. doi:10.3389/fmicb.2019.01762
11. Hernández-García M, García-Castillo M, García-Fernández S, et al. Presence of chromosomal crpP-like genes is not always associated with ciprofloxacin resistance in Pseudomonas aeruginosa clinical isolates recovered in ICU patients from Portugal and Spain. Microorganisms. 2021;9(2):388. doi:10.3390/microorganisms9020388
12. Chávez-Jacobo VM, Hernández-Ramírez KC, Romo-Rodríguez P, et al. CrpP is a novel ciprofloxacin-modifying enzyme encoded by the Pseudomonas aeruginosa pUM505 plasmid. Antimicrob Agents Chemother. 2018;62(6):e02629–17. doi:10.1128/AAC.02629-17
13. Guan X, Xue X, Liu Y, et al. Plasmid-mediated quinolone resistance–current knowledge and future perspectives. J Int Med Res. 2013;41(1):20–30. doi:10.1177/0300060513475965
14. Van TT, Minejima E, Chiu CA, Butler-Wu SM. Don’t get wound up: revised fluoroquinolone breakpoints for Enterobacteriaceae and Pseudomonas aeruginosa. J Clin Microbiol. 2019;57(7):e02072–18. doi:10.1128/JCM.02072-18
15. Llanes C, Köhler T, Patry I, Dehecq B, Delden CV, Plésiat P. Role of the MexEF-OprN Efflux system in low-level resistance of Pseudomonas aeruginosa to ciprofloxacin. Antimicrob Agents Chemother. 2011;55(12):5676–5684. doi:10.1128/AAC.00101-11
16. CLSI. Performance Standards for Antimicrobial Susceptibility Testing.
17. Yoshida H, Bogaki M, Nakamura M, Nakamura S. Quinolone resistance-determining region in the DNA gyrase gyrA gene of Escherichia coli. Antimicrob Agents Chemother. 1990;34(6):1271–1272. doi:10.1128/AAC.34.6.1271
18. Yoshida H, Bogaki M, Nakamura M, Yamanaka LM, Nakamura S. Quinolone resistance-determining region in the DNA gyrase gyrB gene of Escherichia coli. Antimicrob Agents Chemother. 1991;35(8):1647–1650. doi:10.1128/AAC.35.8.1647
19. Champoux JJ. DNA topoisomerases: structure, function, and mechanism. Annu Rev Biochem. 2001;70(1):369–413. doi:10.1146/annurev.biochem.70.1.369
20. El-Badawy MF, Alrobaian MM, Shohayeb MM, Abdelwahab SF. Investigation of six plasmid-mediated quinolone resistance genes among clinical isolates of Pseudomonas: a genotypic study in Saudi Arabia. Infect Drug Resist. 2019;12:915. doi:10.2147/IDR.S203288
21. Robillard NJ, Scarpa AL. Genetic and physiological characterization of ciprofloxacin resistance in Pseudomonas aeruginosa PAO. Antimicrob Agents Chemother. 1988;32(4):535–539. doi:10.1128/AAC.32.4.535
22. Lee JK, Lee YS, Park YK, Kim BS. Alterations in the GyrA and GyrB subunits of topoisomerase II and the ParC and ParE subunits of topoisomerase IV in ciprofloxacin-resistant clinical isolates of Pseudomonas aeruginosa. Int J Antimicrob Agents. 2005;25(4):290–295. doi:10.1016/j.ijantimicag.2004.11.012
23. Lucchetti-Miganeh C, Redelberger D, Chambonnier G, et al. Pseudomonas aeruginosa genome evolution in patients and under the hospital environment. Pathogens. 2014;3(2):309–340. doi:10.3390/pathogens3020309
24. Bruchmann S, Dotsch A, Nouri B, Chaberny IF, Haussler S. Quantitative contributions of target alteration and decreased drug accumulation to Pseudomonas aeruginosa fluoroquinolone resistance. Antimicrob Agents Chemother. 2013;57(3):1361–1368. doi:10.1128/AAC.01581-12
25. Memish ZA, Shibl AM, Kambal AM, Ohaly YA, Ishaq A, Livermore DM. Antimicrobial resistance among non-fermenting Gram-negative bacteria in Saudi Arabia. J Antimicrob Chemother. 2012;67(7):1701–1705. doi:10.1093/jac/dks091
26. Alamri A, Hamid ME, Abid M, et al. Trend analysis of bacterial uropathogens and their susceptibility pattern: a 4-year (2013–2016) study from Aseer region, Saudi Arabia. Urol Ann. 2018;10(1):41. doi:10.4103/UA.UA_68_17
27. Wayne P. Clinical and Laboratory Standards Institute: performance standards for antimicrobial susceptibility testing: 20th informational supplement. CLSI Document M100-S20; 2010.
28. Grillon A, Schramm F, Kleinberg M, Jehl FJPO. Comparative activity of ciprofloxacin, levofloxacin and moxifloxacin against Klebsiella pneumoniae, Pseudomonas aeruginosa and Stenotrophomonas maltophilia assessed by minimum inhibitory concentrations and time-kill studies. PLoS One. 2016;11(6):e0156690. doi:10.1371/journal.pone.0156690
29. Stone TJ, Summers K, Williamson J, Palavecino E, Palavecino E. 1600. Closing the gap on moxifloxacin breakpoints for Stenotrophomonas maltophilia. Open Forum Infect Dis. 2020;7(Supplement_1):S796–S796. doi:10.1093/ofid/ofaa439.1780
30. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–2729. doi:10.1093/molbev/mst197
31. El-Badawy MF, Tawakol WM, Maghrabi IA, Mansy MS, Shohayeb MM, Ashour MS. Iodometric and molecular detection of ESBL production among clinical isolates of E. coli fingerprinted by ERIC-PCR: the first Egyptian report declares the emergence of E. coli O25b-ST131clone harboring blaGES. Microb Drug Resist. 2017;23(6):703–717. doi:10.1089/mdr.2016.0181
32. NIH; NLM; NCBI; 2022.Welcome to the NCBI [homepage]. Available from:: https://www.ncbi.nlm.nih.gov/.
33. Farahi RM, Ali AA, Gharavi S. Characterization of gyrA and parC mutations in ciprofloxacin-resistant Pseudomonas aeruginosa isolates from Tehran hospitals in Iran. Iran J Microbiol. 2018;10(4):242–249.
34. Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25(9):1189–1191. doi:10.1093/bioinformatics/btp033
35. NIH; NLM; NCBI. GenBank Overview; 2022. Available from: http://www.ncbi.nlm.nih.gov/genbank/.
36. NIH; NLM; NCBI. Submission Portal; 2022. Available from: https://submit.ncbi.nlm.nih.gov/.
37. Mouneimne H, Robert J, Jarlier V, Cambau E. Type II topoisomerase mutations in ciprofloxacin-resistant strains of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1999;43(1):62–66. doi:10.1128/AAC.43.1.62
38. Adwan G, Omar G. Phenotypic and molecular characterization of fluoroquinolone resistant Pseudomonas aeruginosa isolates in Palestine. Braz J Bio. 2021;82. doi:10.1590/1519-6984.239868.
39. Arabameri N, Heshmatipour Z, Eftekhar Ardebili S, Jafari Bidhendi Z. The role of gene mutations (gyrA, parC) in resistance to ciprofloxacin in clinical isolates of Pseudomonas aeruginosa. Iran J Pathol. 2021;16(4):426–432. doi:10.30699/IJP.2021.520570.2542.
40. Nouri R, Ahangarzadeh Rezaee M, Hasani A, Aghazadeh M, Asgharzadeh M. The role of gyrA and parC mutations in fluoroquinolones-resistant Pseudomonas aeruginosa isolates from Iran. Braz J Microbiol. 2016;47(4):925–930. doi:10.1016/j.bjm.2016.07.016
41. Yang X, Xing B, Liang C, Ye Z, Zhang Y. Prevalence and fluoroquinolone resistance of Pseudomonas aeruginosa in a hospital of South China. Int J Clin Exp Med. 2015;8(1):1386–1390.
42. Domitrovic TN, Hujer AM, Perez F, et al. Multidrug resistant Pseudomonas aeruginosa causing prosthetic valve endocarditis: a genetic-based chronicle of evolving antibiotic resistance. Open Forum Infect Dis. 2016;3(4):ofw188. doi:10.1093/ofid/ofw188
43. Yonezawa M, Takahata M, Matsubara N, Watanabe Y, Narita H. DNA gyrase gyrA mutations in quinolone-resistant clinical isolates of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1995;39(9):1970–1972. doi:10.1128/AAC.39.9.1970
44. Nakano M, Deguchi T, Kawamura T, et al. Mutations in the gyrA and parC genes in fluoroquinolone-resistant clinical isolates of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1997;41(10):2289–2291. doi:10.1128/AAC.41.10.2289
45. Rehman A, Jeukens J, Levesque RC, Lamont IL. Gene-gene interactions dictate ciprofloxacin resistance in Pseudomonas aeruginosa and facilitate prediction of resistance phenotype from genome sequence data. Antimicrob Agents Chemother. 2021;65(7):e0269620. doi:10.1128/AAC.02696-20
46. Koide K, San LL, Pachanon R, et al. Amino acid substitution Ser83Ile in GyrA of DNA gyrases confers high-level quinolone resistance to nontyphoidal salmonella without loss of supercoiling activity. Microb Drug Resist. 2021;27(10):1397–1404. doi:10.1089/mdr.2020.0437
47. Shaheen A, Tariq A, Iqbal M, et al. Mutational diversity in the quinolone resistance-determining regions of type-II topoisomerases of Salmonella Serovars. Antibiotics. 2021;10(12):1455. doi:10.3390/antibiotics10121455
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.