Back to Journals » OncoTargets and Therapy » Volume 14
The Emerging Role of Circ-SHPRH in Cancer
Authors Ju X, Tang Y, Qu R, Hao S
Received 25 April 2021
Accepted for publication 29 June 2021
Published 13 July 2021 Volume 2021:14 Pages 4177—4188
DOI https://doi.org/10.2147/OTT.S317403
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr William C. Cho
Xinyue Ju, Yan Tang, Rongfeng Qu, Shuhong Hao
Department of Hematology and Oncology, The Second Bethune Clinical Medical College of Jilin University, Changchun, Jilin, People’s Republic of China
Correspondence: Shuhong Hao Ziqiang Street No. 218, Changchun, Jilin, 130041, People’s Republic of China
Tel/Fax +86 431 81136427
Email [email protected]
Purpose: Circ-SHPRH is a circular RNA that can regulate the expression of target genes by sponging microRNAs (miRNAs) or translating tumor suppressor proteins. Recent studies have suggested that circ-SHPRH may play a role in the development of tumors and cancers. Hence, this paper aimed to review the biological characteristics, molecular mechanisms, and potential clinical significance of circ-SHPRH in a variety of tumors and to evaluate its potential as a new diagnostic and prognostic biomarker.
Methods: Numerous experiments were performed regarding the abnormal expression of circ-SHPRH in a variety of tumors, including hepatocellular carcinoma, gastric carcinoma, non-small cell lung cancer, osteosarcoma, colorectal cancer, cholangiocarcinoma, pancreatic ductal adenocarcinoma, retinoblastoma, and glioblastoma.
Results: Upregulation of circ-SHPRH reportedly inhibits tumor cell proliferation, migration, and invasion, leading to the inhibition of tumor development. The clinicopathological parameters and the functional characteristics of circ-SHPRH in multiple human tumors and cancers were summarized. Circ-SHPRH functions as a tumor suppressor gene and has great potential as a diagnostic and prognostic biomarker for different types of cancer.
Keywords: biomarker, cancer, circ-SHPRH, SHPRH-146AA
Introduction
Cancer is a major problem affecting human survival.1 According to the World Health Organization, 19.3 million people were diagnosed with cancer and 10 million died of cancer in 2020.2 At present, one in five people worldwide will suffer from cancer in their lifetime; moreover, the age of onset is decreasing, with atypical early symptoms and poor prognosis.2 Therefore, it is important to identify the effective diagnostic markers and potential therapeutic targets for cancer.
Currently, new therapeutic strategies,such as bacterial therapy, boron neutron capture therapy and targeted non-coding RNA (ncRNA) are under development and may be the treatment options for cancer in the future.3,4
In recent years, ncRNAs have become a popular topic in many researches.5 They result from the pervasive transcription of the mammalian genome,6 as the name suggests, and do not code for proteins.7 NcRNAs comprises the vast majority of cellular RNA and more than 90% of human RNA.8 They were originally thought to regulate gene expression only at the post-transcriptional level.9 However, recent studies have shown that ncRNAs are as important as proteins and play important roles in multiple cellular processes, including cell proliferation, migration, apoptosis, angiogenesis, inflammatory reaction and immune responses.10,11 In addition, ncRNAs can affect metabolic enzymes, transcription factors, and signaling pathways, especially the 50-AMP-activated protein kinase and the phosphatidylinositol 3-kinase /AKT/Mammalian target of rapamycin (mTOR) pathways, which promote glycolysis and regulates glucose metabolism, thereby promoting tumor progression.12
Circular RNAs (circRNAs), a new class of ncRNAs, were first discovered in the 1970s.13 In the past, they were thought to be a byproduct of splicing errors, with no biological function.14 However, with the wide application of high-throughput sequencing technology and bioinformatic methods, high levels of circRNA have been detected in tissues and cells of various eukaryotes, including Drosophila, melanogaster, nematodes, mice, monkeys, bovines, and humans.15–17,
CircRNA is generally generated by a precursor messenger RNA (pre-mRNA) through typical splicing and post-head-tail splicing18 It is a covalently closed continuous loop, without 3′ or 5′ free ends.19 CircRNAs perform four main functions. First, circRNA regulate transcription.20 In 2015, Li et al discovered exon-intron circRNAs, a class of circRNAs related to RNA polymerase II in human cells, which can interact with U1 snRNP to promote the transcription of their parent genes. Second, circRNAs compete as endogenous RNA or microRNA (miRNA) sponges.21 In 2013, Hansen et al found that a circRNA that was highly expressed in human and mouse brains acted as a sponge for miR-7, and the testis-specific circRNA sex-determining region Y, acted as a sponge for miR-138, suggesting that the miRNA sponge effect realized by circRNA formation was a common phenomenon. Third, protein translation of circRNAs;22 in 2017, Pamudurti et al provided strong evidence that a subset of circRNAs were translated in vivo . Fourth, circRNAs interact with RNA-binding proteins (RBPs);23 in 2018, Zang et al discussed in detail the relationship between circRNAs and RBPs, as well as their interactions, which deepened the understanding of the function of circRNAs. Because of their special closed-loop structure, circRNAs are resistant to exonucleases, making them highly stable.24 In addition, circRNAs are critical for evaluating various tissues, organs, and developmental stages in mammals.21 Furthermore, they are specifically expressed in body fluids,25–27 especially in the blood;27 therefore, it has the potential to serve as a powerful biomarker for cancers.28,29
In recent years,increasing evidence has confirmed the involvement of circRNAs in the occurrence and development of many diseases, such as viral infection, diabetes, tumor (ovarian, thyroid, breast and stomach cancer). 30,31 Moreover circRNAs are closely related to the clinicopathological characteristics and prognosis of tumors.32–37 For example, higher Circ-HIPK3 expression is associated with epithelial ovarian cancer lymph node infiltration, high Federation International of Gynecology and Obstetrics staging, and poor disease free survival and overall survival.33 In gastric cancer, low levels of Circ-PVT1 are associated with advanced T (T4) tumors and nerve invasion. When the expression of Circ-PVT1 is downregulated, the proliferation of gastric cancer cells is inhibited.34 Overexpression of Circ-ITCH inhibits the proliferation, migration, and invasion of breast cancer tissues and cells. Low Circ-ITCH expression is associated with higher incidence of lymph node metastasis and the formation of larger tumors, along with more advanced tumor node metastasis (TNM) stages in breast cancer.35
Circ-SHPRH is a circRNA that has been extensively studied recently. In this review, we focused on the biological functions and mechanisms of action of circ-SHPRH (hsa_circ_0001649) in various tumors, and evaluated its potential as a new potential diagnostic and prognostic biomarker.
Formation and Function of Circ-SHPRH
Circ-SHPRH (circBase ID: hsa_circ_0001649, circ2TraitsID: hsa_circ_001599) is a transcriptional product of the SNF2 histone linker PHD. The gene encoding RING helicase (SHPRH) is located at chr6: 146209155–146216113. Mature circ-SHPRH is derived from exons 26–29 of SHPRH gene and contains only 440 nucleotides by intron shearing.38,39 A schematic diagram of the structure is shown in Figure 1.
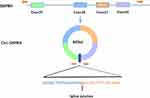 |
Figure 1 Schematic diagram depicting circ-SHPRH formation. |
Recent studies have shown that circ-SHPRH plays a critical role in many biological processes, including cell growth, migration, and invasion.38 In addition, it is a potential marker for many diseases and cancers,40 such as colorectal cancer (CRC),41 hepatocellular carcinoma (HCC),42 and non-small-cell-lung cancer (NSCLC).43 According to the current research, circ-SHPRH mainly acts through the following two aspects: (1) as an miRNA sponge; and (2) through its protein-coding function.
Circ-SHPRH can regulate the expression of its target by acting as miRNA “ sponge”.44 For example, circ-SHPRH can inhibit the expression of Wnt3A and β-catenin by downregulating miR-20a, thereby inhibiting the proliferation, migration, and invasion of gastric cancer cells.45 In lung cancer, circ-SHPRH plays an antitumor role by downregulating the expression of tumor-promoting miRNAs, miR-331-3p and miR-8-5p.45 The miRNAs that can be absorbed by circ-SHPRH are shown in Figure 2.
Recently, some circRNAs have been found to play a role in protein translation.46,47 SHPRH-146AA is a tumor suppressor protein encoded by circ-SHPRH.39,48,49 Zhang et al combined deep RNA-seq with computational methods to predict potential coding circRNAs, and found that a circRNA was downregulated in glioma compared to normal brain tissue. This circRNA, which was designated as circ-SHPRH, has an open reading frame and an internal ribosomal entry site. The whole circ-SHPRH was translated into a suspected 146-amino acid (AA) protein (called “SHPRH-146AA”), and the protein level of SHPRH-146AA was increased after transfection with the circ-SPHRH vector.50 These results suggest that circ-SHPRH has a protein-coding function.
Circ-SHPRH in Various Human Tumors
In recent years, as further studies on circ-SHPRH have been conducted, abnormal expression of circ-SHPRH has been found to be associated with a variety of tumors, including HCC, gastric carcinoma (GC), NSCLC, osteosarcoma (OS), cholangiocarcinoma (CCA), pancreatic ductal adenocarcinoma (PDCA), retinoblastoma (RB), and glioblastoma (GBM). In addition, decreased circ-SHPRH expression was associated with the formation of larger tumors, tumor thrombus, and tumor satellites, along with shorter survival time in patients with cancer. The correlation between clinicopathological parameters of different types of cancer and circ-SHPRH expression is shown in Table 1. Moreover, circ-SHPRH upregulation can result in the inhibition of tumor development by regulating the biological behavior of tumor cells; thereby, acting as a tumor suppressor gene. The specific mechanisms and functional characteristics of circ-SHPRH in different types of cancers are summarized in Table 2.
 |
Table 1 Clinicopathological Parameters of Circ-SHPRH in Human Tumors |
 |
Table 2 Functional Characteristics of Circ-SHPRH in Multiple Human Cancers |
Hepatocellular Carcinoma
HCC ranks sixth in incidence and fourth in mortality among all cancers worldwide, with a poor prognosis and an estimated median survival time of less than 2 years.51,52 In recent years, research on circRNAs in liver cancer has been carried out in full force to provide ideas for the early diagnosis of liver cancer, and to identify potential therapeutic target.53 Recently, circ-SHPRH has been found to play an antitumor role in liver cancer.42,54,55
Qin et al evaluated the clinical value of circ-SHPRH in liver cancer, and the results revealed that the expression of circ-SHPRH in HCC tissues is lower than that in adjacent non-cancerous HCC tissues. Moreover, circ-SHPRH expression is related to tumor size and the formation of tumor thrombus. The receiver operating characteristic (ROC) curve was plotted to evaluate whether circ-SHPRH expression can be used as a biomarker to distinguish cancerous tissues from adjacent non-cancerous liver tissues, the area under the ROC curve was 0.63, and the sensitivity and specificity were 0.81, and 0.69, respectively, suggesting that circ-SHPRH may be a new potential diagnostic biomarker for liver cancer.42 In HCC cells, siRNA-mediated circ-SHPRH knockdown significantly increased mRNA levels of several matrix metalloproteinases (MMPs), such as MMP9, MMP10, and MMP13. MMP9, MMP10, and MMP13 degrade various components of the basement membrane, and promote liver metastases.56–58 This further supported the earlier findings that low levels of circ-SHPRH were positively correlated with HCC metastasis.
Similary, another study also found lower expression of circ-SHPRH in liver cancer tissues than in normal liver tissues.Moreover, low expression of circ-SHPRH is associated with poor overall survival patients with HCC, suggesting that circ-SHPRH can serve as a potential prognostic biomarker in patients with HCC.54 Zhang et al studied the biological function and mechanism of circ-SHPRH in HCC cells and found that circ- SHPRH was downregulated in HCC cells and found that circ-SHPRH overexpression facilitates apoptosis and inhibits the proliferation and migration of HCC cells.
Furthermore, Su et al demonstrated that circ- SHPRH expression is significantly lower in liver cancer cell lines and tumor tissues than in normal liver tissues, and the low expression of circ-SHPRH was related to the degree of tumor differentiation and tumor satellites. Both in vivo and in vitro experiments have shown that circ-SHPRH overexpression inhibits the proliferation and migration of HCC cells. The regulation of circ-SHPRH in HCC cells was achieved by targeting SHPRH, and the results indicate that there is a positive correlation between circ-SHPRH and SHPRH in HCC samples.55 Further studies showed that circ-SHPRH acted as a competitive endogenous RNA sponge for miR-127-5p, miR-612, and miR-4688, thereby activating SHPRH.
Gastric Carcinoma
Gastric cancer is the fifth most common cancer and the third most common cause of cancer-related deaths.59 Most patients with atypical early symptoms were diagnosed with advanced gastric cancer at admission, with high malignancy and poor prognosis.60,61 A growing number of studies have shown that circRNAs play an important role in the progression of gastric cancer and exhibit great potential as useful marker and therapeutic targets.62–64 Circ-SHPRH has recently been associated with the occurrence and development of gastric cancer, and its specific roles and mechanisms are as follows.
Sun et al enrolled 25 patients to study the role and mechanism of circ-SHPRH in gastric cancer tissues and cells and found that the level of circ-SHPRH is significantly lower than that in adjacent normal tissues. Cell counting Kit-8 (CCK-8) assay and flow cytometry showed that circ-SHPRH upregulation significantly inhibits cell proliferation and promotes apoptosis . Transwell migration assay revealed that the circ-SHPRH upregulation inhibits cell migration and invasion.65 MiR-20a is highly expressed in GC,66 it inhibits autophagy and promote tumor cell proliferation67,68 Therefore, it could be a diagnostic and prognostic marker of GC. The authors further aimed to clarify the correlation between the function of circ-SHPRH in GC and miR-20a. Circ-SHPRH upregulation was shown to significantly reduce the expression of miR-20a, indicating that miR-20a is negatively regulated by circ-SHPRH. Further studies showed that upregulation of circ-SHPRH restrained the ERK and Wnt/β-catenin pathways by downregulating miR-20a.
Similarly, Li et al also found that the expression of circ-SHPRH is lower in gastric cancer tissues than that in paracancerous tissues. In addition, preoperative and postoperative whole blood samples of 20 patients were collected to detect the expression of circ-SHPRH, and the results revealed that circ-SHPRH was upregulated in postoperative serum samples of patients. Thus, circ-SHPRH may be considered as a panel for the early detection of GC.69 Analysis of circ-SHPRH levels and clinicopathological data revealed that the expression of circ-SHPRH is associated with pathological differentiation, but not with other clinicopathological factors, including age, sex, TNM stage, lymphatic metastasis, CEA, CA19-9, and CA-724 levels. Additionally, satisfactory results were obtained by ROC analysis. Sensitivity and specificity were 0.711 and 0.816, respectively, and the area under the curve was 0.834, suggesting that circ-SHPRH may be used as a new diagnostic biomarker for GC with high accuracy, specificity, and sensitivity.
Lung Cancer
Worldwide, lung cancer cases and deaths are increasing.70 According to a global survey, lung cancer is estimated to have accounted for 2.2 million new cases and 1.79 million deaths in 2020,71 which was a significant increase from 2018,51 thereby making it the most common cause of death.51,71 NSCLC accounts for approximately 85% of all lung cancer cases.72 Therefore, there is an urgent need to identify new biomarkers, and develop molecularly targeted treatments for NSCLC.
Liu et al measured the relative expression of circ-SHPRH in 53 paired NSCLC tissues and their matched normal tissues by quantitative real-time polymerase chain reaction (qRT-PCR). The results showed that circ-SHPRH expression is lower in tumor tissue specimens than in normal tissue specimens and is correlated with advanced TNM stage and positive lymph nodes. In addition, multivariate Cox regression analysis confirmed that circ-SHPRH can serve as an independent prognostic biomarker in patients with NSCLC. In both in vivo and in vitro experiments, the upregulation of circ-SHPRH was found to inhibit the proliferation, migration, and invasion of tumor cells.43 Bioinformatics analysis was further used to predict circ-SHPRH targets, and miR-140-3p, miR-203, miR-223, miR-331-3p, miR-338-5p, and miR-488 were identified as potential targets. However, circ-SHPRH overexpression was found to only inhibit the expressions of miR-331-3p and miR-338-5p . The correlation between circ-SHPRH and these two miRNAs was further verified by luciferase reporter assay. The results confirmed that miR-331-3p and miR-338-5p are miRNA targets of circ-SHPRH. Further research showed that miR-331-3p and miR-338-5p upregulation could partially reverse the tumor inhibition effect of circ-SHPRH, suggesting that circ-SHPRH plays a role in NSCLC by sponging miR-331-3p and miR-338-5p. The role and mechanism of circ-SHPRH in small -cell lung cancer have not been reported, and further exploration is needed.
Osteosarcoma
OS is the most common primary malignant bone tumor in children and young adults, with a 5-year event-free survival rate of approximately 70% and an overall survival rate of less than 20% in patients with metastatic or recurrent OS.73 For decades, the prognosis of patients with overall survival has not improved.73,74 Therefore, in the future considerable efforts should be directed at the identification of new biomarkers and new OS recognition targets.
Sun et al evaluated the expression of circ-SHPRH in 26 pairs of OS tissues and their corresponding normal adjacent non-tumor tissues. The expression of circ-SHPRH in OS was lower than that in normal tissues. In addition, circ-SHPRH overexpression inhibites the growth of OS cells and promoted apoptosis. Furthermore, co-transfection with miR-338-5p,miR-647, and miR-942 mimics significantly restores the viability and significantly reduces the apoptosis of circ-SHPRH-overexpression cells, suggesting that the biological function of circ-SHPRH depends on its ability to sponge these miRNAs.75 These miRNAs may regulate cell proliferation and survival by regulating autophagy pathways.76
The signal transducer and activator of transcription (STAT) signaling pathway is involved in tumor cell recognition and tumor-driven immune escape and plays an important role in tumorigenesis and cancer development.77 STAT3 and STAT5 are the major intermediaries of the STAT signaling pathway, which is involved in tumor initiation and progression.77,78 Sun et al further investigated the effect of circ-SHPRH on the STAT signaling pathway, and western blotting revealed that circ-SHPRH upregulation significantly decreases the levels of STAT3 and STAT5, whereas, their levels were significantly increased after co-transfection with miR-338-5p, miR-647, and miR-942 mimics. These results suggest that circ-SHPRH inhibited the STAT pathway by sponging miR-338-5p, miR-647, and miR-942.75
Colorectal Cancer
CRC is the fourth most deadly cancer worldwide, with nearly 900,000 deaths per year;The global burden of CRC is expected to increase by 60%, accounting for an estimated 2.2 million new cases and 1.1 million deaths by 2030.79,80 Therefore, a full understanding of the occurrence and development of CRC is beneficial to the development of clinical work in the future.
Ji et al evaluated 64 pairs of tumor tissues and their matched paracancerous tissues. The results showed that circ-SHPRH expression in CRC was significantly lower than that in the corresponding non-tumor tissues and circ-SHPRH expression is closely correlated with pathological differentiation. To evaluate the diagnostic value of circ-SHPRH in CRC, a ROC curve was generated. The area under the ROC curve was 0.857, the sensitivity was 0.828, and the specificity was 0.781, indicating high sensitivity and specificity. In addition, circ-SHPRH expression is upregulated after surgery compared with that before surgery. These results suggest that circ-SHPRH plays a tumor suppressor role in CRC and can be used as a new diagnostic biomarker for CRC.41
Cholangiocarcinoma
Although CCA is a relatively rare tumor, its early clinical presentation is atypical, it is highly invasive and it has a poor prognosis.81,82 Intra-and inter-tumor heterogeneity and chemotherapy resistance make it more difficult to treat.83 Therefore, it is important to elucidate the pathogenesis of this tumor in order to develop more effective treatments.
Recent studies have shown that circ-SHPRH expression is lower in bile duct carcinoma tissues and cells than in normal bile duct tissue and epithelial cells.84 Fisher’s exact tests revealed that circ-SHPRH downregulation is closely associated with larger tumor size and lower differentiation grade in CCA. Through in vivo and in vitro experiments, Xu et al evaluated the biological function of circ-SHPRH and confirmed that the overexpression of circ-SHPRH inhibits cell proliferation and tumor occurrence. Moreover, the migration and invasion abilities of tumor cells are disrupted and apoptosis is induced.84
Pancreatic Ductal Adenocarcinoma
Although the time to progression of PDAC is estimated to be more than 20 years, it is generally diagnosed at advanced stages and has a poor prognosis.85,86 Therefore, there is an urgent need to identify effective diagnostic markers and understand its molecular mechanisms.
Jiang et al used qRT-PCR to detect the expression of circ-SHPRH in 58 PDAC tissues and matched normal tissues, and the results showed that the expression of circ-SHPRH in PDAC tissues is significantly lower than that in normal tissues. Moreover, Fischer’s precise test revealed that patients with low circ-SHPRH expression had more advanced tumor stages and lower grades of differentiation. To further evaluate the prognostic value of circ-SHPRH, the Kaplan-Meier method was used for analyzing the survival. Patients with high circ-SHPRH expression had good 5-year overall survival rates.87 Multivariate analysis further confirmed that circ-SHPRH can serve as an independent prognostic marker of overall survival in PDAC patients. In terms of biological function studies, CCK8 assay showed that circ-SHPRH overexpression significantly inhibits proliferation, and colony formation test validated that the circ-SHPRH upregulation weakens the cloning ability of PDAC cells. Caspase-3 and caspase-9 have been identified as apoptotic markers in previous studies.88,89 In the circ-SHPRH overexpression group, the number of apoptotic cells is significantly increased and caspase-3 and caspase-9 are activated, suggesting that circ-SHPRH may play an anti-cancer role by promoting PDAC apoptosis . Collectively, these findings provide a potential therapeutic target for the treatment of this deadly disease.
Retinoblastoma
RB is the most common intraocular malignancy in children, accounts for 3% of all childhood cancers, and seriously affect the quality of children’s lives.90 Although recent studies on the genetics of RB have helped improve the overall clinical management of this malignancy,90 there is still a need to identify more effective biomarkers and therapeutic targets in RB.
Using qRT-PCR, a recent study found that circ-SHPRH expression is downregulated in RB tissues and cells compared with that in normal retinal tissues and cells. Further analysis of the clinical value of circ-SHPRH in RB showed that circ-SHPRH downregulation is significantly correlated with larger tumor size and more advanced international classification stage for intraocular RB.91 Patients with decreased circ-SHPRH expression have poor 5-year overall survival after surgery. Multivariate Cox regression analysis revealed that decreased circ-SHPRH expression is an independent prognostic indicator of overall survival with RB. The biological functions of circ-SHPRH in RB cells were further explored and results showed that circ-SHPRH overexpression inhibites the proliferation and cloning ability of RB cells, which was further confirmed by xenotransplantation experiments. In addition, the overexpression of circ-SHPRH promotes RB apoptosis . This was further confirmed by flow cytometry, which showed that the overexpression of circ-SHPRH increased the activities of caspase-3 and caspase-9, while simultaneously decreasing the expression of Bcl-2 and increasing the expression of Bax.
Furthermore,studies have reported that the Akt/mTOR signaling pathway participates in the initiation and promotion of diseases, including tumors, through the autophagy pathway, and plays an important role in the regulation of cell proliferation, angiogenesis, transcription, translation and metabolism.92,93 Therefore, Xing et al further explored whether circ-SHPRH plays a role in RB through the Akt/mTOR signaling pathway, and found that p-Akt and p-mTOR are negatively correlated with the expression of circ-SHPRH, but had little effect on the expression of total Akt and total mTOR, suggesting that circ-SHPRH may affect RB apoptosis and proliferation by regulating the Akt/mTOR pathway.91
Glioblastoma
GBM is the most aggressive—although less common—form of intrinsic brain tumor.94
Therefore, it is important to better understand the pathogenesis and progression of gliomaand identify the epigenetic factors (P53,EGFR, microRNAs) that contribute to this tumor.95 As circ-SHPRH is a novel cancer-related circRNA that is downregulated in several cancer types and might be an important diagnostic and prognostic biomarker for various cancers,38 its pathological role in GBM has also been evaluated.
Chen et al evaluated 14 candidate circRNAs and detected a higher expression of six circRNAs (circNT5E, circFOXO3, circ_0001946, circ_0029426, circSHPRH, and circMMP9) in patients with GBM than in healthy volunteers. Further research demonstrated that circFOXO3, circ_0029426, and circ-SHPRH may serve as potential biomarkers to predict the occurrence of GBM. Risk score analysis revealed that the combination of three circRNAs could distinguish patients with GBM from healthy controls, with an area under the curve value of 0.906.96
Contrary to the strategy employed by Chen et al, Wang et al evaluated 64 paired glioma tumor tissues and matched adjacent non-tumor tissue samples and found significantly lower circ-SHPRH expression in tumor tissues and cells than in paired non-tumor tissues.97 The relationship between circ-SHPRH expression and clinical characteristics of glioma was further explored, and the results showed that circ-SHPRH downregulation is strongly associated with larger tumor size and advanced WHO grade. Futhermore, Kaplan-Meier analysis revealed that patients with decreased expression of circ-SHPRH exhibit a low survival rate. Combined results of univariate and multivariate Cox regression models determined circ-SHPRH to be an independent prognostic index for patients with glioma. In vivo and in vitro experiments revealed that circ-SHPRH overexpression inhibits proliferation and increased apoptosis of tumor cells.Additonally, circ-SHPRH overexpression activates Caspase-3 and Caspase-9, decreases the expression of the anti-apoptotic related protein Bcl-2, and increases the expression of the pro-apoptotic related protein Bax, as indicated by western blotting.98 These results suggest that the Bcl-2/caspase-3 pathway plays a key role in circ-SHPRH-mediated apoptosis. In vivo experiments revealed that the tumor weight in the circ-SHPRH overexpression group was significantly lower than that in the negative control group. Additionally, the xenograft formed by circ-SHPRH-overexpressing tumor cells exhibited lower Ki67 expression than tumor cells transfected with empty vector, further confirming that circ-SHPRH plays a tumor-suppressive role in glioma.
We mentioned earlier that circ-SHPRH can be translated into a 146-AA protein.39,48,49 Further analysis revealed that both SHPRH and SHPRH-146AA are abundantly expressed in normal human brains and downregulated in GBM. Using clinical data, it was also found that patients with higher expression of SHPRH-146AA or SHPRH survived longer than those with lower expression of SHPRH-146AA or SHPRH. In terms of biological function, the overexpression of SHPRH-146AA reduces the malignant behavior and tumorigenicity in vitro and in vivo. Mechanistically, SHPRH-146AA can prolong the half-life of full-length SHPRH and protect full-length SHPRH from degradation by ubiquitin proteasomes. Moreover, stabilized SHPRH functioning as an E3 ligase, sequentially ubiquitinates proliferating cell nuclear antigen, leading to inhibited cell proliferation and tumorigenicity. These findings suggest that SHPRH-146AA serves as a decoy molecule that can protect full-length SHPRH from degradation, thereby promoting the tumor inhibitory function of SHPRH.
Conclusion and Prospects
In this review, we outlined the biological functions and mechanisms of circ-SPHRH in various tumors and gained a deeper understanding of its role in tumorigenesis and cancer development. In addition, the association of circ-sPHRH with several clinicopathological features of tumors has been revealed, as well as the possibility of using circ-sPHRH as a new target and potential prognostic marker.
However, studies on circ-SPHRH are limited. For example,the listed experimental data were obtained from small samples. Moreover, there are few studies on the role of circ-SPHRH in certain types of tumors, such as breast cancer and ovarian cancer.Furthermore, its role in other disease types, such as endometriosis and blood diseases, requires further exploration. Furthermore, functional studies have only focused on the role of circ-SPHRH as miRNA sponges. Although there is evidence that circ-SPHRH encodes proteins, limited relevant references are available. Thus, further studies are warranted. Other possible functions of circ-SPHRH should also be explored in the future.
Abbreviations
HCC, hepatocellular carcinoma; GC, gastric carcinoma; NSCLC, non-small cell lung cancer; OS, osteosarcoma; CRC, colorectal cancer; CCA, cholangiocarcinoma; PDCA, pancreatic ductal adenocarcinoma; RB, retinoblastoma; GBM, glioblastoma; ncRNA, non-coding RNA; mTOR, Mammalian target of rapamycin; circRNAs, Circular RNAs; pre-mRNA, precursor messenger RNA; RBP, RNA-binding proteins; TNM, tumor node metastasis; ROC, the receiver operating characteristic; MMP, matrix metalloproteinase; CCK-8, Cell counting Kit-8; qRT-PCR, quantitative real-time polymerase chain reaction; STAT, the signal transducer and activator of transcription.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This work was funded by the Science and Technology Department of Jilin Province, P. R. C. [grant number 20190201050JC] and the Science and Technology Department of Jilin Province, P. R. C. [grant number 20200201543JC].
Disclosure
The authors report no conflicts of interest in this work.
References
1. Siegel RL, Miller KD, Fuchs HE, et al. Cancer Statistics, 2021. CA Cancer J Clin. 2021;71(1):7–33.
2. International Agency for Research on Cancer Latest global cancer data,2020. GLOBOCAN database. Available from: https://www.iarc.fr/faq/latest-global-cancer-data-2020-qa/.
3. Marzhoseyni Z, Shojaie L, Tabatabaei SA, et al. Streptococcal bacterial components in cancer therapy. Cancer Gene Ther. 2021.
4. Mirzaei HR, Sahebkar A, Salehi R, et al. Boron neutron capture therapy: moving toward targeted cancer therapy. J Cancer Res Ther. 2016;12(2):520–525.
5. Slack FJ, Chinnaiyan AM. The role of non-coding RNAs in oncology. Cell. 2019;179(5):1033–1055.
6. Sauvageau M. Diverging RNPs: toward understanding lncRNA-protein interactions and functions. Adv Exp Med Biol. 2019;1203:285–312.
7. Ding B, Lou W, Xu L, et al. Non-coding RNA in drug resistance of hepatocellular carcinoma. Biosci Rep. 2018;38(5):BSR20180915.
8. Niu ZS, Wang WH, Dong XN, et al. Role of long noncoding RNA-mediated competing endogenous RNA regulatory network in hepatocellular carcinoma. World J Gastroenterol. 2020;26(29):4240–4260.
9. Wei JW, Huang K, Yang C, et al. Non-coding RNAs as regulators in epigenetics. Oncol Rep. 2017;37(1):3–9.
10. Xie N, Liu G. ncRNA-regulated immune response and its role in inflammatory lung diseases. Am J Physiol. 2015;309(10):L1076–1087.
11. Hashemian SM, Pourhanifeh MH, Fadaei S, et al. Non-coding RNAs and exosomes: their role in the pathogenesis of sepsis. Mol Ther Nucleic Acids. 2020;21:51–74.
12. Mirzaei H, Hamblin MR. Regulation of glycolysis by non-coding RNAs in cancer: switching on the warburg effect. Mol Ther Oncolytics. 2020;19:218–239.
13. Sanger HL, Klotz G, Riesner D, et al. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci U S A. 1976;73(11):3852–3856.
14. Xie R, Zhang Y, Zhang J, et al. The role of circular RNAs in immune-related diseases. Front Immunol. 2020;11:545.
15. Gruner H, Cortés-López M, Cooper DA, et al. CircRNA accumulation in the aging mouse brain. Sci Rep. 2016;6:38907.
16. Abdelmohsen K, Panda AC, De S, et al. Circular RNAs in monkey muscle: age-dependentchanges. Aging. 2015;7(11):903–910.
17. Hall H, Medina P, Cooper DA, et al. Transcriptome profiling of aging Drosophila photoreceptors reveals gene expression trends that correlate with visual senescence. BMC Genomics. 2017;18(1):894.
18. Cai Y, Lei X, Chen Z, et al. The roles of cirRNA in the development of germ cells. Acta Histochem. 2020;122(3):151506.
19. Liang G, Yang Y, Niu G, et al. Genome-wide profiling of Sus scrofa circular RNAs across nine organs and three developmental stages. DNA Res. 2017;24(5):523–535.
20. Li Z, Huang C, Bao C, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22(3):256–264.
21. Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388.
22. Pamudurti NR, Bartok O, Jens M, et al. Translation of CircRNAs. Mol Cell. 2017;66(1):9–21.e7.
23. Zang J, Lu D, Xu A. The interaction of circRNAs and RNA binding proteins: an important part of circRNA maintenance and function. J Neurosci Res. 2020;98(1):87–97.
24. Qu S, Yang X, Li X, et al. Circular RNA: a new star of noncoding RNAs. Cancer Lett. 2015;365(2):141–148.
25. Dong WW, Li HM, Qing XR, et al. Identification and characterization of human testis derived circular RNAs and their existence in seminal plasma. Sci Rep. 2016;6:39080.
26. Jafari Ghods F. Circular RNA in Saliva. Adv Exp Med Biol. 2018;1087:131–139.
27. Lv C, Sun L, Guo Z, et al. Circular RNA regulatory network reveals cell-cell crosstalk in acute myeloid leukemia extramedullary infiltration. J Transl Med. 2018;16(1):361.
28. Kulcheski FR, Christoff AP, Circular MR. RNAs are miRNA sponges and can be used as a new class of biomarker. J Biotechnol. 2016;238:42–51.
29. Li P, Chen S, Chen H, et al. Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clinica chimica acta. 2015;444:132–136.
30. Abbaszadeh-Goudarzi K, Radbakhsh S, Pourhanifeh MH, et al. Circular RNA and diabetes: epigenetic regulator with diagnostic role. Curr Mol Med. 2020;20(7):516–526.
31. Nahand JS, Jamshidi S, Hamblin MR, et al. Circular RNAs: new epigenetic signatures in viral infections. Front Microbiol. 2020;11:1853.
32. Li Z, Ruan Y, Zhang H, et al. Tumor-suppressive circular RNAs: mechanisms underlying their suppression of tumor occurrence and use as therapeutic targets. Cancer Sci. 2019;110(12):3630–3638.
33. Liu N, Zhang J, Zhang LY, et al. CircHIPK3 is upregulated and predicts a poor prognosis in epithelial ovarian cancer. Eur Rev Med Pharmacol Sci. 2018;22(12):3713–3718.
34. Chen J, Li Y, Zheng Q, et al. Circular RNA profile identifies circPVT1 as a proliferative factor and prognostic marker in gastric cancer. Cancer Lett. 2017;388:208–219.
35. Wang ST, Liu LB, Li XM, et al. Circ-ITCH regulates triple-negative breast cancer progression through the Wnt/β-catenin pathway. Neoplasma. 2019;66(2):232–239.
36. Borran S, Ahmadi G, Rezaei S, et al. Circular RNAs: new players in thyroid cancer. Pathol Res Pract. 2020;216(10):153217.
37. Shabaninejad Z, Vafadar A, Movahedpour A, et al. Circular RNAs in cancer: new insights into functions and implications in ovarian cancer. J Ovarian Res. 2019;12(1):84.
38. Li F, Huang Q, Gong Z, et al. Diagnostic and prognostic roles of circ-SHPRH for solid cancers: a meta-analysis. Onco Targets Ther. 2019;12:4351–4357.
39. Zhang M, Huang N, Yang X, et al. A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene. 2018;37(13):1805–1814.
40. Haque S, Harries LW. Circular RNAs (circRNAs) in Health and Disease. Genes. 2017;8(12):353.
41. Ji W, Qiu C, Wang M, et al. Hsa_circ_0001649: a circular RNA and potential novel biomarker for colorectal cancer. Biochem Biophys Res Commun. 2018;497(1):122126.
42. Qin M, Liu G, Huo X, et al. Hsa_circ_0001649: a circular RNA and potential novel biomarker for hepatocellular carcinoma. Cancer Biomarkers. 2016;16(1):161–169.
43. Liu T, Song Z, Gai Y. Circular RNA circ_0001649 acts as a prognostic biomarker and inhibits NSCLC progression via sponging miR-331-3p and miR-338-5p. Biochem Biophys Res Commun. 2018;503(3):1503–1509.
44. Ebbesen KK, Hansen TB, Kjems J. Insights into circular RNA biology. RNA Biol. 2017;14(8):1035–1045.
45. Sun H, Wang Q, Yuan G, et al. Hsa_circ_0001649 restrains gastric carcinoma growth and metastasis by downregulation of miR-20a. J Clin Lab Anal. 2020;34:
46. Yang Y, Fan X, Mao M, et al. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res. 2016;27(5):626–641.
47. Legnini I, Di Timoteo G, Rossi F, et al. Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Mol Cell. 2017;66(1):22–37.e9.
48. Begum S, Yiu A, Stebbing J, et al. Novel tumour suppressive protein encoded by circular RNA, circ-SHPRH, in glioblastomas. Oncogene. 2018;37(30):4055–4057.
49. Biswas A, Chowdhury N, Bagchi A. Structural characterization of the hidden peptide SHPRH-146aa encoded by non-coding circ-SHPRH to act as tumor suppressor. Appl Biochem Biotechnol. 2021. doi:10.1007/s12010-021-03520-0
50. Motegi A, Sood R, Moinova H, et al. Human SHPRH suppresses genomic instability through proliferating cell nuclear antigen polyubiquitination. J Cell Biol. 2006;175(5):703–708.
51. Bray F, Ferlay J, Soerjomataram I, et al. y. CA Cancer J Clin. 2018;68(6):394–424.
52. Ko KL, Mak LY, Cheung KS, et al. Hepatocellular carcinoma: recent advances and emerging medical therapies. F1000Res. 2020;9:F1000 Faculty Rev–620.
53. Ye Q, Ling S, Zheng S, et al. Liquid biopsy in hepatocellular carcinoma: circulating tumor cells and circulating tumor DNA. Mol Cancer. 2019;18(1):114.
54. Zhang X, Qiu S, Luo P, et al. Down-regulation of hsa_circ_0001649 in hepatocellular carcinoma predicts a poor prognosis. Cancer Biomarkers. 2018;22(1):135–142.
55. Su Y, Xu C, Liu Y, et al. Circular RNA hsa_circ_0001649 inhibits hepatocellular carcinoma progression via multiple miRNAs sponge. Aging. 2019;11(10):3362–3375.
56. Sze KM, Chu GK, Lee JM, et al. C-terminal truncated hepatitis B virus x protein is associated with metastasis and enhances invasiveness by C-Jun/matrix metalloproteinase protein 10 activation in hepatocellular carcinoma. Hepatology. 2013;57(1):131–139.
57. Chen X, Bo L, Zhao X, et al. MicroRNA-133a inhibits cell proliferation, colony formation ability, migration and invasion by targeting matrix metallopeptidase 9 in hepatocellular carcinoma. Mol Med Rep. 2015;11(5):3900–3907.
58. Wang C, Jin G, Jin H, et al. Clusterin facilitates metastasis by EIF3I/Akt/MMP13 signaling in hepatocellular carcinoma. Oncotarget. 2015;6(5):2903–2916.
59. Smyth EC, Nilsson M, Grabsch HI, et al. Gastric cancer. Lancet. 2020;396(10251):635–648.
60. Choi YJ, Kim N. Gastric cancer and family history. Korean J Intern Med. 2016;31(6):1042–1053.
61. Song Z, Wu Y, Yang J, et al. Progress in the treatment of Advanced gastric cancer. Tumour Biol. 2017;39(7):1010428317714626.
62. Gu W, Sun Y, Zheng X, et al. Identification of gastric cancer-related circular RNA through microarray analysis and bioinformatics analysis. Biomed Res Int. 2018;2381680.
63. Tang W, Fu K, Sun H, et al. CircRNA microarray profiling identifies a novel circulating biomarker for detection of gastric cancer. Mol Cancer. 2018;17(1):137.
64. Sun HD, Xu ZP, Sun ZQ, et al. Down-regulation of circPVRL3 promotes the proliferation and migration of gastric cancer cells. Sci Rep. 2018;8(1):10111.
65. Huang Z, Zhu D, Wu L, et al. Six serum-based miRNAs as potential diagnostic biomarkers for gastric cancer. Cancer Epidemiol Biomarkers Prev. 2017;26(2):188–196.
66. Yang R, Fu Y, Zeng Y, et al. Serum miR-20a is a promising biomarker for gastric cancer. Biomedical Rep. 2017;6(4):429–434.
67. Pourhanifeh MH, Vosough M, Mahjoubin-Tehran M, et al. Autophagy-related microRNAs: possible regulatory roles and therapeutic potential in and gastrointestinal cancers. Pharmacol Res. 2020;161:105133.
68. Xin L, Zhou LQ, Liu L, et al. METase promotes cell autophagy via promoting SNHG5 and suppressing miR-20a in gastric cancer. Int J Biol Macromol. 2019;122:1046–1052.
69. Li WH, Song YC, Zhang H, et al. Decreased expression of Hsa_circ_00001649 in gastric cancer and its clinical significance. Dis Markers. 2017;4587698.
70. Bade BC, Dela Cruz CS. Lung cancer 2020: epidemiology, etiology, and prevention. Clin Chest Med. 2020;41(1):1–24.
71. World Health Organization, Cancer fact sheets, lung cancer. 2018. Cancer Today.2020. Available from: http://gco.iarc.fr/today/fact-sheets-cancers.
72. Gelatti ACZ, Drilon A, Santini FC. Optimizing the sequencing of tyrosine kinase inhibitors (TKIs) in epidermal growth factor receptor (EGFR) mutation-positive non-small cell lung cancer (NSCLC). Lung Cancer. 2019;137:113–122.
73. Harrison DJ, Geller DS, Gill JD, et al. Current and future therapeutic approaches for osteosarcoma. Expert Rev Anticancer Ther. 2018;18(1):39–50.
74. Simpson E, Brown HL. Understanding osteosarcomas. JAAPA. 2018;31(8):15–19.
75. Sun D, Zhu D. Circular RNA hsa_circ_0001649 suppresses the growth of osteosarcoma cells via sponging multiple miRNAs. Cell Cycle. 2020;19(20):2631–2643.
76. Jamali Z, Taheri-Anganeh M, Shabaninejad Z, et al. Autophagy regulation by microRNAs: novel insights into osteosarcoma therapy. IUBMB Life. 2020;72(7):1306–1321.
77. Owen KL, Brockwell NK, Parker BS. JAK-STAT signaling: a double- edged sword of immune regulation and cancer progression. Cancers. 2019;11(12):2002.
78. Thomas SJ, Snowden JA, Zeidler MP, et al. The role of JAK/STAT signalling in the pathogenesis, prognosis and treatment of solid tumours. Br J Cancer. 2015;113(3):365–371.
79. Dekker E, Tanis PJ, Vleugels J, et al. Colorectal cancer. Lancet. 2019;394(10207):1467–1480.
80. Arnold M, Sierra MS, Laversanne M, et al. Global patterns and trends in colorectal cancer incidence and mortality. Gut. 2017;66(4):683–691.
81. Banales JM, Marin JJG, Lamarca A, et al. Cholangiocarcinoma 2020: the next horizon in mechanisms and management. Nat Rev Gastroenterol Hepatol. 2020;17(9):557–588.
82. Koch C, Franzke C, Bechstein WO, et al. Poor prognosis of advanced cholangiocarcinoma: real-world data from a tertiary referral center. Digestion. 2020;101(4):458–465.
83. Rodrigues PM, Olaizola P, Paiva NA, et al. Pathogenesis of cholangiocarcinoma. Annu Rev Pathol. 2021;16:433–463.
84. Xu Y, Yao Y, Zhong X, et al. Downregulated circular RNA hsa_circ_0001649 regulates proliferation, migration and invasion in cholangiocarcinoma cells. Biochem Biophys Res Commun. 2018;496(2):455–461.
85. Storz P, Crawford HC. Carcinogenesis of pancreatic ductal adenocarcinoma. Gastroenterology. 2020;158(8):2072–2081.
86. Chandana S, Babiker HM, Mahadevan D. Therapeutic trends in pancreatic ductal adenocarcinoma (PDAC). Expert Opin Investig Drugs. 2019;28(2):161–177.
87. Jiang Y, Wang T, Yan L, et al. A novel prognostic biomarker for pancreatic ductal adenocarcinoma: hsa_circ_0001649. Gene. 2018;675:88–93.
88. Li P, Zhou L, Zhao T, et al. Caspase −9: structure, mechanisms and clinical application. Oncotarget. 2017;8(14):23996–24008.
89. Lossi L, Castagna C, Merighi A. Caspase −3 medi ated cell death in the normal development of the mammalian cerebellum. Int J Mol Sci. 2018;19(12):3999.
90. Rao R, Honavar SG. Retinoblastoma. Indian J Pediatr. 2017;84(12):937–944.
91. Xing L, Zhang L, Feng Y, et al. Downregulation of circular RNA hsa_circ_0001649 indicates poor prognosis for retinoblastoma and regulates cell proliferation and apoptosis via AKT/mTOR signaling pathway. Biomedicine Pharmacother. 2018;105:326–333.
92. Xu Z, Han X, Ou D, et al. Targeting PI3K/AKT/mTOR-mediated autophagy for tumor therapy. Appl Microbiol Biotechnol. 2020;104(2):575–587.
93. Zhang Y, Hu B, Li Y, et al. Binding of Avibirnavirus VP3 to the PIK3C3-PDPK1 complex inhibits autophagy by activating the AKT-MTOR pathway. Autophagy. 2020;16(9):1697–1710.
94. Lim M, Xia Y, Bettegowda C, et al. Current state of immunotherapy for glioblastoma. Nat Rev Clin Oncol. 2018;15(7):422–442.
95. Khani P, Nasri F, Khani Chamani F, et al. Genetic and epigenetic contribution to astrocytic gliomas pathogenesis. J Neurochem. 2019;148(2):188–203.
96. Chen A, Zhong L, Ju K, et al. Plasmatic circRNA predictingthe occurrence of human glioblastoma. Cancer Manag Res. 2020;12:2917–2923.
97. Wang Y, Sui X, Zhao H, et al. Decreased circular RNA hsa_circ_0001649 predicts unfavorable prognosis in glioma and exerts oncogenic properties in vitro and in vivo. Gene. 2018;676:117–122.
98. Bagherian A, Mardani R, Roudi B, et al. Combination therapy with nanomicellar-curcumin and temozolomide for in vitro therapy of glioblastoma multiforme via wnt signaling pathways. J Mol Neurosci. 2020;70(10):1471–1483.
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.