Back to Journals » Infection and Drug Resistance » Volume 17
The Emerging Pathogen Candida metapsilosis: Biological Aspects, Virulence Factors, Diagnosis, and Treatment
Authors Gómez-Gaviria M, García-Carnero LC, Baruch-Martínez DA , Mora-Montes HM
Received 4 November 2023
Accepted for publication 16 January 2024
Published 20 January 2024 Volume 2024:17 Pages 171—185
DOI https://doi.org/10.2147/IDR.S448213
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Manuela Gómez-Gaviria, Laura C García-Carnero, Dario A Baruch-Martínez, Héctor M Mora-Montes
Departamento de Biología, División de Ciencias Naturales y Exactas, Universidad de Guanajuato, Guanajuato, Gto., México
Correspondence: Héctor M Mora-Montes, Departamento de Biología, División de Ciencias Naturales y Exactas, Campus Guanajuato, Universidad de Guanajuato, Noria Alta s/n, col. Noria Alta, C.P. 36050, Guanajuato, Gto., México, Tel +52 473-7320006 Ext. 8193, Fax +52 473-7320006 Ext. 8153, Email [email protected]
Abstract: Fungal infections represent a constant and growing menace to public health. This concern is due to the emergence of new fungal species and the increase in antifungal drug resistance. Mycoses caused by Candida species are among the most common nosocomial infections and are associated with high mortality rates when the infection affects deep-seated organs. Candida metapsilosis is part of the Candida parapsilosis complex and has been described as part of the oral microbiota of healthy individuals. Within the complex, this species is considered the least virulent; however, the prevalence has been increasing in recent years, as well as an increment in the resistance to some antifungal drugs. One of the main concerns of candidiasis caused by this species is the wide range of clinical manifestations, ranging from tissue colonization to superficial infections, and in more severe cases it can spread, which makes diagnosis and treatment difficult. The study of virulence factors of this species is limited, however, proteomic comparisons between species indicate that virulence factors in this species could be similar to those already described for C. albicans. However, differences may exist, taking into account changes in the lifestyle of the species. Here, we provide a detailed review of the current literature about this organism, the caused disease, and some sharing aspects with other members of the complex, focusing on its biology, virulence factors, the host-fungus interaction, the identification, diagnosis, and treatment of infection.
Keywords: antifungal resistance, candidiasis, Candida parapsilosis complex, emerging pathogens, virulence factors
Introduction
The Candida genus groups fungal species of yeast-like organisms, and from the approximately 200 species belonging to it, only 17 species are the most frequently isolated from clinical specimens.1 Collectively, these species are the etiological agents of candidiasis, a medically relevant fungal infection that can affect superficial or deep-seated organs. Candida albicans is the most common organism that causes candidiasis in humans and thus is the species whose pathogenesis, virulence, epidemiology, and susceptibility to antifungal drugs have been thoroughly studied. However, nowadays the emergence of new species in the clinical setting is widely recognized and other Candida species, such as Candida parapsilosis, Candida orthopsilosis, Candida metapsilosis, Candida krusei, Candida lusitaniae, Candida glabrata, and Candida tropicalis, have recently attracted attention.1–6
Candidiasis, colloquially known as thrush, shows a wide range of infections throughout the body and is responsible for about 96% of all mycoses in immunocompromised patients.7,8 With no doubt, the highest mortality toll is associated with candidemia,1,4,6 which implies the fungal dispersion to different organs by the bloodstream. On the contrary, superficial candidiasis is often an infection of mucosal (buccal, vulvovaginal, esophageal) or cutaneous (skin and nails) tissues, with benign or mild symptoms that can affect both immunocompetent and immunocompromised patients.9 It is estimated that up to 75% of women suffer from vulvovaginal candidiasis caused by Candida spp. at least once in their life.10
C. parapsilosis sensu lato is the second most found Candida isolate in bloodstream samples from Latin America and Asia, and the third overall Candida species isolated worldwide.11 From those isolates, it has been reported that around 90% were identified as Candida parapsilosis sensu stricto; while the rest as C. orthopsilosis and C. metapsilosis.12
C. metapsilosis is ubiquitous and has been reported as part of the oral mycobiome of healthy individuals.13,14 This species is reported as the least common and virulent species of the complex, given that just around 1.1 to 8.4% of the infections caused by C. parapsilosis sensu lato are attributed to C. metapsilosis.15,16 Even though it is considered the least important species of the complex, its prevalence has increased over the years and has also been linked to challenging clinical failures.17,18 Although many aspects of this organism, are still unclear, here we offer a literature revision of the known information, which we consider will be helpful for the understanding of this emerging pathogen.
Biological Aspects
Morphology
The members of the C. parapsilosis complex are phenotypically indistinguishable from each other but genetically different, with differences in epidemiology, virulence, and antifungal susceptibility.19 The C. parapsilosis species have the selective advantage of growing in high glucose concentration media, and in these rich growing conditions, the fungus grows like white creamy colonies of subglobose cells, with a size of 4×3-6 µm.20 It was previously thought that, like C. parapsilosis, C. metapsilosis was capable of forming pseudohyphae,21 but a study of 93 different C. parapsilosis sensu lato isolates demonstrated that C. metapsilosis does not produce this morphology, being killed more efficiently by the host macrophages.16 This aspect remains contradictory though, since several studies report the formation of pseudohyphae in C. metapsilosis.22 The discrepancies shown in the literature could be related to the fungal strain analyzed, since genetic variations between strains may affect phenotypic traits, including morphological switch. Another explanation could be related to the culture conditions, factors such as temperature, pH and the composition of the culture medium could also affect dimorphism. However, most of the studies indicate that C. metapsilosis can form pseudohyphae.
Cell Wall
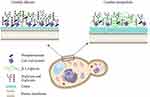
Regarding cellular structures, thus far the cell wall is the most studied organelle and currently, there are well-described models of the C. albicans wall,23–25 but limited information about this structure in other Candida species. A comparative analysis of the cell wall composition of C. albicans, C. parapsilosis, C. orthopsilosis, and C. metapsilosis revealed that the arrangement of this component is different among the species, although with a similar wall composition.26 When compared to C. albicans, the species from the C. parapsilosis complex showed lower mannan content, similar protein levels, and an increase in chitin and β-1,3-glucan exposure on the cell wall (Figure 1). 26 Indeed, it has been showed that C. parapsilosis has shorter N-linked mannans than C. albicans,27 an observation likely extrapolated to C. metapsilosis, and that could explain the overexposure of chitin and β-1,3-glucans in C. parapsilosis sensu lato cell surface, as a compensatory mechanism. In addition, the C. metapsilosis cell wall has a higher porosity and reduced phosphomannan content, which can also be explained by shorter mannan chains (Figure 1). 26 It was also found that there are subtle differences in the distribution of structural polysaccharides among the C. parapsilosis complex species, being the most remarkable that C. orthopsilosis exposes all of its β-1,3-glucan and chitin at the cell wall surface, which might suggest that cell wall proteins have even shorter mannans than C. parapsilosis and C. metapsilosis, or that these proteins contain fewer glycosylation sites, making the inner wall polysaccharides more accessible.26
Genome
To determine the C. metapsilosis evolution and the genetic and molecular basis of its pathogenesis, the genome of 11 clinical isolates from different locations was sequenced and compared to each other, and against C. parapsilosis and C. orthopsilosis. The C. metapsilosis genome assembly revealed a total size of 13.3 Mbp distributed in 7 putative chromosomes, with 5973 protein-coding genes, different from what has been reported for C. parapsilosis (5752 protein-coding genes) and C. orthopsilosis (5784 protein-coding genes), a GC percentage of 38.1, and 17 pseudogenes.16 These predicted chromosomes presented ends enriched in telomere repeats (GGTTAGGATGTCCAAAGTATTGA) that correspond to the telomerase template domain.16 The analysis of the C. metapsilosis genome revealed that this organism is highly heterozygous, with a divergence between alleles in heterozygous regions of 4.5%, which facilitated the delimitation of heterozygous and homozygous regions. It was found that around 50 and 60% of the genome from the different strains are heterozygous regions, much higher than that reported for C. orthopsilosis (17%).16 This finding could mean two things: (i) the C. metapsilosis hybridization is a more recent event, or (ii) C. orthopsilosis lost its heterozygosity faster.16 The heterozygous regions’ sizes had identical boundaries, which might suggest that all 11 C. metapsilosis strains derived from the same primary hybridization event.16 Due to the lack of a homozygous C. metapsilosis strain and a clear parental mapping, an indirect approach was used to obtain an idea of this organism’s parental lineage, based on the identification of two haplotypes in a determined homozygous region. The first haplotype represented the reference assembly, while the second the remaining strains. This procedure provided an estimate of 50% for the first haplotype and 40% for the second haplotype in homozygous regions, which cannot be assigned to a specific parental genome but that might suggest a balanced presence of both parents in these regions from the C. parapsilosis genome and no preference for either parental strain.16
It was also shown that the 11 C. metapsilosis strains showed diploidy, which suggests that mating between two haploid cells not very divergent is the possible hybridization mechanism for this organism. In addition, most of the C. metapsilosis strains contain the MTLa and MTLα idiophorms, which supports the mating event in C. metapsilosis, species that was thought to be asexual.16 Also, it was found that the 11 strains, isolated from different locations, had quite different evolutionary relationships, which suggests a lineage relatively ancient that spread worldwide a long time ago, unlike what has been seen with C. orthopsilosis, which shows stable similarities among isolates from different locations.16
Finally, the sequencing of the C. metapsilosis genome allowed a comparison among the C. parapsilosis sensu lato, which showed a conserved synteny between the three species, being C. parapsilosis and C. orthopsilosis closer to each other, with 98% of conserved synteny. In addition, phylogeny analysis revealed that C. metapsilosis has a basal position in this complex.16
The C. parapsilosis mitochondrial genome has also been assembled, resulting in a single 21 kb contig,28 and although the species from the C. parapsilosis complex usually have this genome as linear, some C. metapsilosis and C. orthopsilosis isolates have circular mitochondrial DNA (mtDNA).29–31 In addition, the C. metapsilosis mtDNA is compact and encodes the same number of genes, arranged in the same order as C. parapsilosis and C. orthopsilosis, with the only difference that C. metapsilosis mtDNA has one intronic sequence, while the other two species have seven and two introns, respectively.32 Although the mtDNA architecture is not considered a taxonomic criterion,29 these differences might suggest biodiversity.20,30,31
The C. parapsilosis complex is a member of the CTG clade, which means that the CTG codon codes for serine instead of leucine,33 and includes other clinically relevant species, like C. albicans and C. tropicalis. However, unlike these two species, C. parapsilosis sensu lato independently evolved their virulence.16
General Aspects
As mentioned, the C. parapsilosis complex species cannot be differentiated by phenotypical or biochemical analyses, since they all share similar characteristics, such as (i) macro and microscopic cellular morphology, presenting colonies of different colorations that vary from white to pink, and oval, round or cylindrical yeast cells (Figure 2), 30 (ii) biochemical markers, like their ability to grow on high glucose concentrations,34 and a strong activity for proteinase,30 and (iii) fermentation profiles, like their ability to ferment maltose, D-ribose, and D-gluconate.20,34 However, a recent study found subtle differences in the growth kinetics among the three species when grown under different salinity and pH levels, being C. orthopsilosis the species with a lower number of viable cells when grown at 10 and 15% of NaCl and at pH 7.35
 |
Figure 2 Microscopic morphology of Candida metapsilosis. Yeast cells growing at 28 °C in Sabouraud medium, with the typical round or cylindrical cells. |
Some studies have shown that C. metapsilosis can ferment some sugars such as glucose, maltose, D-ribose, D-gluconate, galactose, sucrose, trehalose, N-acetyl-glucosamine, 2-K-D-gluconate, and glycerol.36 In addition, this organism can assimilate melezitose, ribitol, glucose, galactose, D-xylose, and D-mannitol.29,36 Recent work found that C. metapsilosis can significantly reduce the content of alkaloids and increase the content of aromatic components in tobacco leaves.37
Putative Virulence Factors in Candida metapsilosis
Although C. albicans and C. metapsilosis are two organisms found within the same genus, both species show notable differences concerning biological aspects and virulence. The study of C. metapsilosis virulence is limited if compared with the large amount of information available for C. albicans and its closest species C. parapsilosis sensu stricto.38 This section will provide information about C. metapsilosis virulence factors and a proteomic comparison to predict possible orthologs already described in C. albicans.
Cell adhesion plays an important role in numerous biological processes. Many fungi, such as those of the Candida genus, have cell wall glycoproteins known as adhesins, which provide distinctive adhesion properties.39 Adhesion is the first step during Candida infection. The ability of Candida spp. to bind to host tissues or abiotic surfaces, such as catheters or medical devices is achieved through the biological products of genes that encode several adhesin families that are found in the cell wall.38 One of the groups involved in this process is the Als family (agglutinin-like sequence). Although its importance as a virulence factor in C. albicans is known, little information is available on C. metapsilosis.38,40 A comparative genomics analysis identified the presence of several ALS genes in C. orthopsilosis (3 genes), C. parapsilosis (5 genes), and C. metapsilosis (4 genes).41–43 The proteins encoded by these genes are similar to the C. albicans Als proteins, and have a signal peptide, an N-terminal domain with a peptide-binding cavity, an amyloid-forming region, repeat sequences, and a C-terminal site for the addition of the glycosylphosphatidylinositol anchor.42 Molecular modeling revealed the presence of unique features, including differences in shape and charge in different peptide-binding cavities.43 Previous reports have indicated that C. metapsilosis invades reconstituted human oral and epidermal epithelial tissues; however, unlike C. parapsilosis and C. orthopsilosis, this species does not cause significant cell damage.38 Furthermore, tests carried out with human bronchial epithelial cells showed that C. metapsilosis has a reduced adherence capacity.41,44 This low adhesion phenotype could be correlated with the low virulence of this species, as has been reported in different studies.17,26 Members of the ALS gene family, EAP1, ECM33, IFF4, INT1, MP65, and PHR1, encode the main C. albicans adhesins,45 and although little is known about these in C. metapsilosis, putative orthologs could be identified within its genome, indicating that adhesion may occur through these adhesins (Table 1).
 |
Table 1 Prediction of Some Important Virulence Factors in Candida metapsilosis |
Biofilm development has also been described as an important virulence factor in Candida species.46 It is known that biofilm formation can contribute to resistance to antifungal drugs and immune evasion.47 Biofilms formed by the C. parapsilosis complex members have gained important attention because isolates have been obtained from a wide variety of both biotic and abiotic surfaces.46,48 To date, there have been few investigations of these properties in clinical isolates of the C. parapsilosis complex, especially in C. metapsilosis. Some studies have shown that clinical isolates of this species are classified as weak and moderate biofilm producers.48 The isolates that were considered strong biofilm producers were C. parapsilosis sensu stricto.48 Analysis of biofilm production and scanning electron microscopy revealed that 65 clinical isolates obtained from complex species were able to form biofilms. However, C. parapsilosis and C. orthopsilosis exhibited greater metabolic activity at 24 hours, compared to C. metapsilosis.49 Taking into account the microscopic observations, C. metapsilosis biofilms showed different surface topography when compared to other species of the complex.49–51 Other essays have shown that C. metapsilosis is the lowest biofilm producer when compared with other Candida species.22,52,53 Although C. metapsilosis biofilms exhibit different characteristics than C. parapsilosis, it is still concerning this virulence trait, in particular in the clinical setting. C. metapsilosis can be isolated from hospital environments, representing a potential risk of infection/reinfection for patients.46 Taking into account the bioinformatic analysis, it is possible to predict that C. metapsilosis has seven genes involved in biofilm formation (Table 1). However, more research is required to assess the function of these genes in this organism.
Biofilms are associated with the dimorphism. As reported, C. metapsilosis cannot form true hyphae, only pseudohyphae, which implies changes in the expression of some virulence factors that are morphology-specific.54,55 The yeast-to-hypha transition in Candida is a key virulence determinant that allows the organism to invade host tissues and evade the host immune response.56 Dimorphism in C. metapsilosis is poorly studied; however, work carried out with the Galleria mellonella model indicates that C. metapsilosis has a reduced ability to produce pseudohyphae. After 48 hours of incubation, none of the C. metapsilosis isolates managed to produce hyphae or pseudohyphae, which could be correlated with the reduced virulence of this species.17 In the case of infection in the invertebrate Caenorhabditis elegans, the appearance of C. metapsilosis pseudohyphae was demonstrated after 24 and 48 hours, and distention of the nematode intestine was observed after ingesting fungal cells.57 Although the death of some individuals was evident, this was lower than that observed in animals infected with C. parapsilosis.57 Taking into account the bioinformatic analysis (Table 1), C. metapsilosis has putative functional orthologs of these genes coding for dimorphism regulators; however, the process that controls the dimorphism could be regulated differently in C. metapsilosis and C. albicans.
The secretion of hydrolytic enzymes is considered an important virulence factor in Candida spp. These enzymes fulfill important functions such as facilitating adhesion, internalization into tissues, and, therefore, invasion into host cells.58 Among these enzymes, the most studied and best characterized are phospholipases and proteinases; however, esterase activity has also been described in Candida species.20,59 In non-albicans species, such as C. metapsilosis these virulence factors have not been widely described. Enzyme activity assays of 12 C. metapsilosis clinical isolates obtained from cutaneous and mucocutaneous samples demonstrated the presence of phospholipase activity.60 Furthermore, 10 of these isolates were positive for proteinase, and 9 out of the 10 showed to be strong producers of this enzyme.60 When the esterase activity was evaluated in C. metapsilosis, it was found that 2 of the clinical isolates were producers of this enzyme, after an incubation period of 10 days.60 These findings correlate with other reports working with isolates from different geographical origins.61 Recent work has shown that C. metapsilosis is a proteinase producer and only some isolates synthesize phospholipases, which is of great interest since the production of these enzymes is not common in this species.48,62 In vivo tests with G. mellonella demonstrated that C. metapsilosis has a reduced capacity to produce hydrolytic enzymes, however, strains of this species showed esterase activity.17
These results indicate that both phospholipases and proteinases prevail in C. metapsilosis, despite being the least virulent species of the complex. Taking into account that it is a species that is frequently isolated from abiotic surfaces, these enzymes can facilitate the survival and adaptation of the fungus to different niches. Furthermore, although it is considered a non-invasive species, it can also be lethal, since it has also been isolated from patient blood cultures.12,60,63,64 Table 1 shows the putative orthologs of LIP, SAP, and PLB found in C. metapsilosis, genes encoding for esterase, proteinase, and phospholipase activities, respectively, although their function remains poorly understood.
In Candida spp., other virulence factors are known to play important functions, such as immune evasion and thermotolerance. Thermotolerance contributes to growth, dimorphism, and tissue colonization. This virulence factor plays an important role in fungal adaptation processes to stressful environments.56,65 Bioinformatic analysis suggests that the C. metapsilosis genome contains putative orthologs of the thermotolerance genes HSP60, HSP104, and SSA1 (Table 1). Immune evasion is considered a mechanism that involves other cellular processes, such as the formation of biofilms, morphological changes, secretion of proteases, and synthesis of proteins that contribute to evading oxidative stress.65 Some of these processes have already been mentioned earlier in this review. The C. albicans genes involved in immune evasion, HGT1, and MSB2, were found in C. metapsilosis (Table 1). However, no ortholog of the PRA1 gene was found. This finding is consistent with other non-albicans species, such as Candida lusitaniae.65
Based on the bioinformatic analysis carried out, it is possible to hypothesize that, despite the genomic similarities between C. metapsilosis and the virulence factors previously described in C. albicans, there could be a differential expression of these factors in C. metapsilosis. This could be related to the lifestyle of this species since several authors suggest that it can also behave as an environmental pathogen.20
Candida metapsilosis-Host Interaction
The C. parapsilosis complex members have become as important as C.albicans in the clinical setting.26 To better understand the clinical relevance of these species, it is necessary to elucidate how they interact with host immune cells. Currently, the best described and studied interaction with the host immunity is that of C. albicans; however, although there are specie-specific variations, the elemental recognition process of Candida cells is mediated by pattern recognition receptors (PRRs) and pathogen-associated molecular patterns (PAMPs).66 Furthermore, both innate and adaptive immune responses play an important role in the control of these pathogens.56,66–68
The fungal cell wall is a relevant structure during immune sensing. This has very specific functions, among which are structural support, signal transduction, and molecular scaffold to display virulence factors. It is also involved in the first events of interaction with the host cells, containing most of the PAMPs recognized by the PRRs in innate immune cells.23,24,69
A comparative study of C. parapsilosis complex species with C. albicans revealed that the cell wall composition of C. parapsilosis, C. metapsilosis, and C. orthopsilosis is similar, however, the organization of wall components may have some differences, which impacts on the ability to activate human peripheral blood mononuclear cells (PBMCs)26 In contrast to C. albicans, C. metapsilosis stimulated higher and similar levels of the cytokines TNFα, IL-1β, IL-6, and IL-10. When yeast cells were subjected to different cell treatments, such as heat inactivation and β-elimination, C. metapsilosis was found to stimulate higher levels of TNFα, IL-6, and IL-10 in both types of treatments, contrasting with C. albicans that only stimulated higher cytokine levels upon heat inactivation.26
The cytokine stimulation by C. metapsilosis has a strong dependence on the dectin-1, at the difference of C. albicans. This observation is in line with the higher β-1,3-glucan content at the C. metapsilosis cells surface.26 In the case of the TLR4 receptor, it is known to be the only receptor that recognizes O-linked mannan in C. albicans,70 and probably also plays an important role in the recognition of C. metapsilosis wall components. Blockade of the TLR4 receptor resulted in reduced TNFα stimulation by C. metapsilosis, but IL-1β, IL-6, and IL-10 production was not affected, suggesting a modest contribution of this receptor for cytokine stimulation.26 Contrary to these observations, the mannose receptor, the main PRR for N-linked mannans,70 plays an important role during the stimulation of proinflammatory cytokines by C. metapsilosis.26 These observations are relevant, as they allow proposing the costimulation hypothesis, which has already been observed in other fungal species.67,71,72
An in vitro infection model using microglial cells was employed to investigate the pathogenic potential of clinical isolates of the C. parapsilosis complex species.73 C. metapsilosis isolates were found to be more susceptible to microglia-mediated antifungal activity compared to the other two species. Isolates of this species are less phagocytosed, however, phagosomes internalizing C. metapsilosis yeasts showed a higher degree of acidification compared to phagosomes containing other species of the C. parapsilosis complex.73 This may indicate that this species is less susceptible to phagocytosis, but when phagocytosed, it is less effective in counteracting the host’s intracellular antimicrobial defenses.73 When the secretory response of microglia to infection was assessed, comparable levels of macrophage inflammatory protein-1 (MIP-1α) were observed in all species of the complex. On the other hand, C. metapsilosis did not cause any apparent damage to microglial cells, unlike C. parapsilosis and C. orthopsilosis.73 Cytotoxicity was measured using the enzyme lactate dehydrogenase, and it was found that microglial cells infected with C. metapsilosis released low levels of the enzyme, compared to the other species of the complex.73 All these results suggested that C. metapsilosis is the least virulent species of the complex. Tests on female mice confirmed C. metapsilosis reduced virulence. When they were infected with cells of this species, the fungal load found during the first stages of infection was lower than that found with C. parapsilosis and C. orthopsilosis.44 Tests with human buccal epithelial cells showed that C. metapsilosis strains have reduced adhesion capacity to these cells.44 When the virulence, hemocyte density, and phagocytic activity were evaluated in Galleria mellonella larvae, it was found that the larvae managed to survive infection by this pathogen, compared to the other species of the complex, where greater mortality was documented.17 During the hemocyte counting, two hours after the yeast infection, a significantly greater increase was observed in the larvae infected by C. metapsilosis.17 Work carried out with the invertebrate model C. elegans demonstrated that unlike G. mellonella, all species of the complex can kill C. elegans. However, the time to death when infected with C. metapsilosis was longer (6 days), compared to C. parapsilosis (4 days).57 To assess the defense mechanisms that are involved during C. metapsilosis infection, five antimicrobial peptides were evaluated, which are thought to have antifungal activity in vivo. 57 After infection, the expression of cnc-4, cnc-7, and fipr22/23 was demonstrated, showing that C. elegans has a specific defense response against C. metapsilosis.57
Candidiasis Associated with Candida metapsilosis
The C. parapsilosis complex members play an important role in human infections. These species can cause a wide range of clinical manifestations, ranging from superficial to disseminated infections.74,75 These species are highly prevalent in neonates, especially C. parapsilosis.18
In Brazil, there are few reports on the prevalence of the three species of the C. parapsilosis complex. In 2014, a study was conducted to determine whether candidemia in neonates and children was related to these species.76 A total of 3 cases of fungemia identified in the Brazilian Pediatric Hospital, in São Paulo, Brazil, were reported, the persistent symptomatology in these patients was high fever, chills, rapid breathing, and tachycardia.76 The treatment was based on fluconazole and amphotericin B, and at the time of distinguishing between species, the isolates were sequenced, resulting in the species C. metapsilosis and C. orthopsilosis.76 For the case of C. metapsilosis, the sample was isolated from a single child, and those of C. orthopsilosis were isolated from the blood of a newborn and a child.76 Even though the patients were treated immediately, infection by both pathogens was lethal, as these three strains were resistant to fluconazole, which was used as a first-line treatment.76
The incidence of C. metapsilosis and C. orthopsilosis infections has increased since 2004, with prevalence rates ranging from 0.9 and 6.9% to 2.3 and 9%, respectively.12 However, there are still poorly identified risk factors to learn more about these species, along with limited data about their prevalence in hospitals.
In Spain, during the years 2009 to 2010, 1356 cases of fungemia were analyzed, and C. parapsilosis was isolated in 400 of these cases, which corresponds to an incidence of 29%.15 Although this species was found in a higher proportion of men, statistical analysis did not determine significant differences between genders. Of the 400 isolates, 330 corresponded to C. parapsilosis sensu stricto, 30 to C. orthopsilosis, and 4 to C. metapsilosis.15 Infections caused by these fungi were mainly observed in patients older than 15 years; candidemia in newborns was not related to C. orthopsilosis and C. metapsilosis, as reported in Brazil.15 It was also found that these species were associated with different hospital wards, with C. parapsilosis being found more frequently in the hematology, pediatrics, and neonatology departments, while C. metapsilosis was found in the adult and elderly hospitalization areas.15
In Asia, the frequency of C. metapsilosis is 2.6% of the total fungal isolates, but in China, this species represents 10.7% of candidiasis cases.60 The reason for this is not clear yet; however, when reviewing the clinical records, it was hypothesized that a high incidence of this species could be related to a unique distribution pattern in China. However, this hypothesis has not yet been validated.60
Identification and Diagnosis of Candida metapsilosis
Accurate identification of Candida species in the clinical setting has become essential, given the increasing frequency of candidiasis in recent years. Over time, various strategies have been developed for Candida species identification. However, these strategies may vary considering the similarity that exists among species.
For C. metapsilosis, it is not possible to make its specific identification with biochemical and morphological techniques, due to its similarity with the other members of the C. parapsilosis complex.77 When clinical samples are inoculated on CHROMagar Candida medium for 24 and 48 hours, the colonies show a light brown color, which does not provide relevant information to distinguish them from the species of the complex.78 When Sabouraud dextrose agar is used, all cultures are identified as C. parapsilosis, without discriminating between C. metapsilosis and C. orthopsilosis,78 ie, this medium does not provide informative results for species identification.29,74,79 API 20C AUX carbohydrate assimilation test has been used most frequently for C. parapsilosis identification, with 100% efficiency.80
Considering that biochemical and morphological approaches do not yield much information, molecular and biophysical methods have been proposed to significantly accelerate C. metapsilosis identification in laboratories.29,61,81,82 Molecular strategies are based on the amplification of the SADH gene by PCR, with oligonucleotides that amplify a specific region of this locus.29 The amplicons are digested with BanI and the digestion products are separated by electrophoresis.74 For C. metapsilosis, there are three cleavage sites at positions 96, 469, and 529, while for C. parapsilosis and C. orthopsilosis, the cleavage is at position 196, demonstrating that by this analysis it is possible to differentiate between the complex species.29
To differentiate between species, 32 isolates were sequenced and divided into groups (I, II and III), using this technique and sequence polymorphism data for ACPL, ACPR, COX3, GAL1, L1A1, LIP1, SADH, SAPP3, SYA1, TOP2 and URA3 genes, it was found that C. metapsilosis had no amplification product for ACPL, ACPR, L1A1, LIP2, TOP2, and URA3 genes, unlike C. parapsilosis and C. orthopsilosis. In addition, for GAL1 and SAPP3 genes, higher amplicons were observed than those obtained in C. parapsilosis.29 The analysis of the rRNA and ITS regions is another of the tools that have been improved for the C. metapsilosis identification. It has been found that the ITS1 sequence similarity between C. metapsilosis and C. parapsilosis is 82.5% and with C. orthopsilosis 86.1%.29 Thus, there is a significative dissimilarity between them.
Multiplex PCR assays have been performed using primers targeting genes such as HWP1 and ITS regions (ITS1, ITS2) of different Candida species, including C. metapsilosis.82,83 These assays, together with techniques such as matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) and DNA sequencing have been able to reproducibly differentiate between Candida species, including C. metapsilosis.83 MALDI-TOF-MS, a technique that can be useful for the correct identification of C. metapsilosis, can be performed in a few minutes and is mainly based on the analysis of the proteins contained in a crude cell extract of the different isolates. The instrument’s database provides the references to compare the spectra obtained and thus conclude the specific identity of the sample. This technique discriminates between the three species of the C. parapsilosis complex.81,84 Among all the proposed techniques, this has proved to be the most effective for timely diagnosis.81
Real-time PCR techniques based on mtDNA, using as a target a polymorphic region of the NADH5 gene, have contributed to a precise identification of each member of the C. parapsilosis complex, due to the species-specific variability of this gene.85
Antifungal Resistance and Therapeutic Options
The treatment for different types of candidiasis is based on azoles, polyenes, echinocandins, and pyrimidine analogs, such as flucytosine.3 These drugs are effective against a wide range of Candida spp; however, several studies have demonstrated that non-albicans Candida species may become resistant or less susceptible to different antifungal classes, such as echinocandins.29,82,86
C. metapsilosis is known to show higher minimal inhibitory concentrations (MICs) for echinocandins, and although no significant clinical failures have been reported, there is concern that strains of this species may be predisposed to higher levels of resistance to these drugs.87 Echinocandins inhibit β-1,3-glucan synthases, which are important for cell wall glucan biosynthesis, by targeting the Fks1 catalytic subunit.88,89 When the in vitro activity of different echinocandins, such as caspofungin, micafungin, and anidulafungin was evaluated on C. parapsilosis, C. metapsilosis, and C. orthopsilosis, it was found that the MIC of anidulafungin and micafungin was 20 to 50 times higher than the control strains. The antifungal caspofungin showed a minor 3.6-to-7.8-fold increase, however, this was still statistically significant.82 Echinocandin resistance has been linked to amino acid substitutions in two conserved regions of Fks1 or Fks2.90 In C. metapsilosis, and the other species of the complex, a variation in alanine at position 660 in Fks1 has been observed, replacing proline. This substitution has not been observed in other species; however, other types of substitutions have been identified in species such as C. guilliermondii, which are also related to reduced susceptibility to echinocandins.82,91 These data further suggest that mutations within the Fks1 regions may alter the in vitro sensitivity of the glucan synthase enzyme and alter its catalytic capacity.
Previous reports have revealed that the complex species are susceptible to azoles and polyenes, and the MIC of caspofungin for C. metapsilosis is lower than that found for C. parapsilosis.92 Caspofungin is the most active echinocandin against these species and is known to generate significant therapeutic effects for the treatment of systemic infections. On the other hand, micafungin is more efficient against C. metapsilosis.92 A multicenter study in Spain showed that all C. orthopsilosis and C. metapsilosis isolates were susceptible to nine antifungal agents tested, including amphotericin B, flucytosine, fluconazole, itraconazole, voriconazole, posaconazole, anidulafungin, caspofungin, and micafungin.15 Other studies have indicated that C. orthopsilosis and C. metapsilosis isolates can be inhibited by concentrations >1mg/l of amphotericin B.93 Considering the results showed in different studies, it is possible to observe variation concerning antifungal susceptibility in the different species of the complex. This could be related to the clinical isolate and the methodology used to determine MIC values.15
Tests in neutropenic mice have shown that when intraperitoneal treatment with caspofungin is initiated 24 hours after infection with C. metapsilosis, the antifungal exerts a fungistatic response of ≥2-8 µg/mL in vitro against this fungus. This indicates that at clinically achievable concentrations this antifungal is effective against C. metapsilosis.94 For C. metapsilosis, the available information about antifungal treatment is scarce, however, there is no consensus regarding treatment, so it is normal to empirically administer amphotericin B, azoles, flucytosine, and echinocandins.20 Although amphotericin B is the most commonly used therapy, it is usually associated with some side effects, such as renal affectations.20 Fluconazole is often used in patients with candidiasis associated with members of the C. parapsilosis complex; however, it has been observed that prolonged exposure to it often leads to drug resistance.95,96 The choice of antifungal and the duration of treatment may vary according to the severity of the infection and the area affected.
Considering that C. metapsilosis is a poorly studied but frequently occurring species, it is necessary to determine which are the appropriate antifungal therapies that contribute to treating infection caused by this species.
Concluding Remarks
In recent years, there has been increasing interest in research on candidiasis caused by non-albicans species. Most studies have focused on understanding the biological, epidemiological, and clinical aspects of species such as C. parapsilosis, C. orthopsilosis, C. lusitaniae, and C. krusei. C. metapsilosis has been largely overlooked due to its low frequency of isolation in medical settings, and because its diagnosis is often confused with its closest species C. orthopsilosis and C. parapsilosis. However, it is important to note that infections caused by this species can be fatal when spread to the bloodstream. Additionally, C. metapsilosis can develop resistance to some antifungal drugs, making it a difficult species to treat.87
The findings of this work highlight the lack of substantial information about this species, which allows carrying out more detailed research that will provide the scientific community with information that may help to clarify fundamental aspects of the biology of C. metapsilosis. Although there are already phenotypic and molecular strategies for the identification of C. metapsilosis, the implementation of options that are more efficient and economical is sought.
The bioinformatics analyses included here may increase the information to understand basic aspects of this species. Sequence analyses allow the generation of genetic predictions, which facilitate the creation of working models to identify general and species-specific virulence factors.
Acknowledgments
This work was supported by Consejo Nacional de Ciencia y Tecnología (ref. Ciencia de Frontera 2019-6380). The funding source that supported this work did not have any involvement in the design, acquisition, and analysis of data and writing of the manuscript.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis, and interpretation, or all these areas; took part in drafting, revising, or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors declare no conflict of interest.
References
1. Pfaller MA, Diekema DJ, Procop GW, Rinaldi MG. Multicenter comparison of the VITEK 2 antifungal susceptibility test with the CLSI broth microdilution reference method for testing amphotericin B, flucytosine, and voriconazole against Candida spp. J Clin Microbiol. 2007;45(11):3522–3528. doi:10.1128/jcm.00403-07
2. Hani U, Shivakumar HG, Vaghela R, Osmani RA, Shrivastava A. Candidiasis: a fungal infection-current challenges and progress in prevention and treatment. Infect Disord Drug Targets. 2015;15(1):42–52. doi:10.2174/1871526515666150320162036
3. Arendrup M, Horn T, Frimodt-Møller N. In vivo pathogenicity of eight medically relevant Candida species in an animal model. Infection. 2002;30(5):286–291. doi:10.1007/s15010-002-2131-0
4. Pfaller M, Neofytos D, Diekema D, et al. Epidemiology and outcomes of candidemia in 3648 patients: data from the prospective antifungal therapy (PATH Alliance®) registry, 2004–2008. Diagn Microbiol Infect Dis. 2012;74(4):323–331. doi:10.1016/j.diagmicrobio.2012.10.003
5. Pfaller MA, Diekema DJ, Mendez M, et al. Candida guilliermondii, an opportunistic fungal pathogen with decreased susceptibility to fluconazole: geographic and temporal trends from the ARTEMIS DISK antifungal surveillance program. J Clin Microbiol. 2006;44(10):3551–3556. doi:10.1128/JCM.00865-06
6. Pfaller MA, Jones RN, Messer SA, Edmond MB, Wenzel RP. National surveillance of nosocomial blood stream infection due to species of Candida other than Candida albicans: frequency of occurrence and antifungal susceptibility in the SCOPE Program. Diagn Microbiol Infect Dis. 1998;30(2):121–129. doi:10.1016/S0732-8893(97)00192-2
7. Wisplinghoff H, Bischoff T, Tallent SM, Seifert H, Wenzel RP, Edmond MB. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin Infect Dis. 2004;39(3):309–317. doi:10.1086/421946
8. Marchetti O, Bille J, Fluckiger U, et al. Epidemiology of candidemia in Swiss tertiary care hospitals: secular trends, 1991–2000. Clin Infect Dis. 2004;38(3):311–320. doi:10.1086/380637
9. Bhattacharya S, Sae-Tia S, Fries BC. Candidiasis and mechanisms of antifungal resistance. Antibiotics. 2020;9(6):312. doi:10.3390/antibiotics9060312
10. Sobel JD. Vulvovaginal candidosis. Lancet. 2007;369(9577):1961–1971. doi:10.1016/S0140-6736(07)60917-9
11. Pfaller MA, Jones RN, Doern GV, et al. Bloodstream infections due to Candida species: SENTRY antimicrobial surveillance program in North America and Latin America, 1997–1998. Antimicrob Agents Chemother. 2000;44(3):747–751. doi:10.1128/aac.44.3.747-751.2000
12. Lockhart SR, Messer SA, Pfaller MA, Diekema DJ. Geographic distribution and antifungal susceptibility of the newly described species Candida orthopsilosis and Candida metapsilosis in comparison to the closely related species Candida parapsilosis. J Clin Microbiol. 2008;46(8):2659–2664. doi:10.1128/JCM.00803-08
13. Tosun I, Akyuz Z, Guler NC, et al. Distribution, virulence attributes and antifungal susceptibility patterns of Candida parapsilosis complex strains isolated from clinical samples. Med Mycol. 2013;51(5):483–492. doi:10.3109/13693786.2012.745953
14. Ghannoum MA, Jurevic RJ, Mukherjee PK, et al. Characterization of the oral fungal microbiome (mycobiome) in healthy individuals. PLoS Pathog. 2010;6(1):e1000713. doi:10.1371/journal.ppat.1000713
15. Canton E, Peman J, Quindos G, et al. Prospective multicenter study of the epidemiology, molecular identification, and antifungal susceptibility of Candida parapsilosis, Candida orthopsilosis, and Candida metapsilosis isolated from patients with candidemia. Antimicrob Agents Chemother. 2011;55(12):5590–5596. doi:10.1128/AAC.00466-11
16. Pryszcz LP, Nemeth T, Saus E, et al. The genomic aftermath of hybridization in the opportunistic pathogen Candida metapsilosis. PLoS Genet. 2015;11(10):e1005626. doi:10.1371/journal.pgen.1005626
17. Gago S, García-Rodas R, Cuesta I, Mellado E, Alastruey-Izquierdo A. Candida parapsilosis, Candida orthopsilosis, and Candida metapsilosis virulence in the non-conventional host Galleria mellonella. Virulence. 2014;5(2):278–285. doi:10.4161/viru.26973
18. Arastehfar A, Khodavaisy S, Daneshnia F, et al. Molecular identification, genotypic diversity, antifungal susceptibility, and clinical outcomes of infections caused by clinically underrated yeasts, Candida orthopsilosis, and Candida metapsilosis: an Iranian multicenter study (2014–2019) Original Research. Front Cell Infect Microbiol. 2019;9:264. doi:10.3389/fcimb.2019.00264
19. Feng X, Wu Z, Ling B, et al. Identification and differentiation of Candida parapsilosis complex species by use of exon-primed intron-crossing PCR. J Clin Microbiol. 2014;52(5):1758–1761. doi:10.1128/JCM.00105-14
20. Trofa D, Gacser A, Nosanchuk JD. Candida parapsilosis, an emerging fungal pathogen. Clin Microbiol Rev. 2008;21(4):606–625. doi:10.1128/CMR.00013-08
21. Lackey E, Vipulanandan G, Childers DS, Kadosh D. Comparative evolution of morphological regulatory functions in Candida species. Eukaryot Cell. 2013;12(10):1356–1368. doi:10.1128/EC.00164-13
22. Melo AS, Bizerra FC, Freymüller E, Arthington-Skaggs BA, Colombo AL. Biofilm production and evaluation of antifungal susceptibility amongst clinical Candida spp. isolates, including strains of the Candida parapsilosis complex. Med Mycol. 2011;49(3):253–262. doi:10.3109/13693786.2010.530032
23. Gómez-Gaviria M, García-Carnero LC, Tamez-Castrellón AK, Mora-Montes HM. The cell wall of medically relevant yeasts and molds. In: Zaragoza Ó, Casadevall A, editors. Encyclopedia of Mycology. Elsevier. Elsevier; 2021:12–22.
24. Díaz-Jiménez DF, Pérez-García LA, Martínez-álvarez JA, Mora-Montes HM. Role of the fungal cell wall in pathogenesis and antifungal resistance. Curr Fungal Infect Rep. 2012;6(4):275–282. doi:10.1007/s12281-012-0109-7
25. Klis FM, de Groot P, Hellingwerf K. Molecular organization of the cell wall of Candida albicans. Med Mycol. 2001;39(Suppl 1):1–8. doi:10.1080/mmy.39.1.1.8-0
26. Estrada-Mata E, Navarro-Arias MJ, Perez-Garcia LA, et al. Members of the Candida parapsilosis complex and Candida albicans are differentially recognized by human peripheral blood mononuclear cells. Front Microbiol. 2016;6:1527. doi:10.3389/fmicb.2015.01527
27. Shibata N, Ikuta K, Imai T, et al. Existence of branched side chains in the cell wall mannan of pathogenic yeast, Candida albicans. Structure-antigenicity relationship between the cell wall mannans of Candida albicans and Candida parapsilosis. J Biol Chem. 1995;270(3):1113–1122. doi:10.1074/jbc.270.3.1113
28. Valach M, Pryszcz LP, Tomaska L, Gacser A, Gabaldón T, Nosek J. Mitochondrial genome variability within the Candida parapsilosis species complex. Mitochondrion. 2012;12(5):514–519. doi:10.1016/j.mito.2012.07.109
29. Tavanti A, Davidson AD, Gow NA, Maiden MC, Odds FC. Candida orthopsilosis and Candida metapsilosis spp. nov. to replace Candida parapsilosis groups II and III. J Clin Microbiol. 2005;43(1):284–292. doi:10.1128/JCM.43.1.284-292.2005
30. Toth R, Nosek J, Mora-Montes HM, et al. Candida parapsilosis: from genes to the bedside. Clin Microbiol Rev. 2019;32(2). doi:10.1128/CMR.00111-18
31. Rycovska A, Valach M, Tomaska L, Bolotin-Fukuhara M, Nosek J. Linear versus circular mitochondrial genomes: intraspecies variability of mitochondrial genome architecture in Candida parapsilosis. Microbiology. 2004;150(Pt 5):1571–1580. doi:10.1099/mic.0.26988-0
32. Kosa P, Valach M, Tomaska L, Wolfe KH, Nosek J. Complete DNA sequences of the mitochondrial genomes of the pathogenic yeasts Candida orthopsilosis and Candida metapsilosis: insight into the evolution of linear DNA genomes from mitochondrial telomere mutants. Nucleic Acids Res. 2006;34(8):2472–2481. doi:10.1093/nar/gkl327
33. Sai S, Holland LM, McGee CF, Lynch DB, Butler G. Evolution of mating within the Candida parapsilosis species group. Eukaryot Cell. 2011;10(4):578–587. doi:10.1128/EC.00276-10
34. Guérin M, Camougrand N. The alternative oxidase of Candida parapsilosis. Eur J Biochem. 1986;159(3):519–524. doi:10.1111/j.1432-1033.1986.tb09917.x
35. Cordeiro RA, Sales JA, Ponte YB, et al. Phenotype-driven strategies for screening Candida parapsilosis complex for molecular identification. Braz J Microbiol. 2018;49(Suppl 1):193–198. doi:10.1016/j.bjm.2017.11.004
36. Bednarek M, Szwengiel A, Flórez AB, Czarnecki Z, Mayo B. Effect of different starter cultures on chemical and microbial parameters of buckwheat honey fermentation. Food Microbiol. 2019;82:294–302. doi:10.1016/j.fm.2019.03.006
37. Jia Y, Zhou W, Yang Z, et al. A critical assessment of the Candida strains isolated from cigar tobacco leaves. Front Bioeng Biotechnol. 2023;11:1201957. doi:10.3389/fbioe.2023.1201957
38. Gacser A. Adhesins in Candida parapsilosis: understudied players in virulence. Virulence. 2016;7(2):65–67. doi:10.1080/21505594.2015.1135288
39. de Groot PW, Bader O, de Boer AD, Weig M, Chauhan N. Adhesins in human fungal pathogens: glue with plenty of stick. Eukaryot Cell. 2013;12(4):470–481. doi:10.1128/ec.00364-12
40. Cota E, Hoyer LL. The Candida albicans agglutinin-like sequence family of adhesins: functional insights gained from structural analysis. Future Microbiol. 2015;10(10):1635. doi:10.2217/fmb.15.79
41. Oh SH, Smith B, Miller AN, et al. Agglutinin-like sequence (ALS) genes in the Candida parapsilosis species complex: blurring the boundaries between gene families that encode cell-wall proteins. Front Microbiol. 2019;10:781. doi:10.3389/fmicb.2019.00781
42. Lombardi L, Zoppo M, Rizzato C, et al. Characterization of the Candida orthopsilosis agglutinin-like sequence (ALS) genes. PLoS One. 2019;14(4):e0215912. doi:10.1371/journal.pone.0215912
43. Essen LO, Vogt MS, Mösch HU. Diversity of GPI-anchored fungal adhesins. Biol Chem. 2020;401(12):1389–1405. doi:10.1515/hsz-2020-0199
44. Bertini A, De Bernardis F, Hensgens LA, Sandini S, Senesi S, Tavanti A. Comparison of Candida parapsilosis, Candida orthopsilosis, and Candida metapsilosis adhesive properties and pathogenicity. Int J Med Microbiol. 2013;303(2):98–103. doi:10.1016/j.ijmm.2012.12.006
45. Talapko J, Juzbašić M, Matijević T, et al. Candida albicans-the virulence factors and clinical manifestations of infection. J Fungi. 2021;7(2):79. doi:10.3390/jof7020079
46. Araújo D, Henriques M, Silva S. Portrait of Candida species biofilm regulatory network genes. Trends Microbiol. 2017;25(1):62–75. doi:10.1016/j.tim.2016.09.004
47. Pulcrano G, Panellis D, De Domenico G, Rossano F, Catania MR, Calderone R. Ambroxol influences voriconazole resistance of Candida parapsilosis biofilm. FEMS Yeast Res. 2012;12(4):430–438. doi:10.1111/j.1567-1364.2012.00792.x
48. Paula-Mattiello S, Oliveira SD, Medina-Silva R. In vitro evaluation of hydrolytic enzyme activity and biofilm formation of Candida parapsilosis species complex from a nosocomial environment. Rev Soc Bras Med Trop. 2017;50(4):558–561. doi:10.1590/0037-8682-0032-2017
49. Modiri M, Khodavaisy S, Barac A, et al. Comparison of biofilm-producing ability of clinical isolates of Candida parapsilosis species complex. J Mycol Med. 2019;29(2):140–146. doi:10.1016/j.mycmed.2019.02.003
50. Lattif AA, Mukherjee PK, Chandra J, et al. Characterization of biofilms formed by Candida parapsilosis, C. metapsilosis, and C. orthopsilosis. Int J Med Microbiol. 2010;300(4):265–270. doi:10.1016/j.ijmm.2009.09.001
51. de Toro M, Torres MJ, Maite R, Aznar J. Characterization of Candida parapsilosis complex isolates. Clin Microbiol Infect. 2011;17(3):418–424. doi:10.1111/j.1469-0691.2010.03302.x
52. Melo AS, Colombo AL, Arthington-Skaggs BA. Paradoxical growth effect of caspofungin observed on biofilms and planktonic cells of five different Candida species. Antimicrob Agents Chemother. 2007;51(9):3081–3088. doi:10.1128/aac.00676-07
53. Treviño-Rangel Rde J, Rodríguez-Sánchez IP, Rosas-Taraco AG, Hernández-Bello R, González JG, González GM. Biofilm formation and genetic variability of BCR1 gene in the Candida parapsilosis complex. Rev Iberoam Micol. 2015;32(3):180–184. doi:10.1016/j.riam.2014.11.001
54. Gácser A, Schäfer W, Nosanchuk JS, Salomon S, Nosanchuk JD. Virulence of Candida parapsilosis, Candida orthopsilosis, and Candida metapsilosis in reconstituted human tissue models. Fungal Genet Biol. 2007;44(12):1336–1341. doi:10.1016/j.fgb.2007.02.002
55. Treviño-Rangel R, González-González JG, Garza-González E, González GM. Candida parapsilosis, una amenaza desafiante. Medicina Universitaria. 2012;14(56):157–165.
56. Gómez-Gaviria M, Mora-Montes HM. Current aspects in the biology, pathogeny, and treatment of Candida krusei, a neglected fungal pathogen. Infect Drug Resist. 2020;13:1673–1689. doi:10.2147/idr.S247944
57. Souza ACR, Fuchs BB, Alves VS, Jayamani E, Colombo AL, Mylonakis E. Pathogenesis of the Candida parapsilosis complex in the model host Caenorhabditis elegans. Genes. 2018;9(8):401. doi:10.3390/genes9080401
58. Schaller M, Borelli C, Korting HC, Hube B. Hydrolytic enzymes as virulence factors of Candida albicans. Mycoses. 2005;48(6):365–377. doi:10.1111/j.1439-0507.2005.01165.x
59. Slifkin M. Tween 80 opacity test responses of various Candida species. J Clin Microbiol. 2000;38(12):4626–4628. doi:10.1128/jcm.38.12.4626-4628.2000
60. Ge YP, Lu GX, Shen YN, Liu WD. In vitro evaluation of phospholipase, proteinase, and esterase activities of Candida parapsilosis and Candida metapsilosis. Mycopathologia. 2011;172(6):429–438. doi:10.1007/s11046-011-9440-8
61. Hensgens LA, Tavanti A, Mogavero S, Ghelardi E, Senesi S. AFLP genotyping of Candida metapsilosis clinical isolates: evidence for recombination. Fungal Genet Biol. 2009;46(10):750–758. doi:10.1016/j.fgb.2009.06.006
62. da Silva BV, Silva LB, de Oliveira DB, et al. Species distribution, virulence factors, and antifungal susceptibility among Candida parapsilosis complex isolates recovered from clinical specimens. Mycopathologia. 2015;180(5–6):333–343. doi:10.1007/s11046-015-9916-z
63. Tay ST, Na SL, Chong J. Molecular differentiation and antifungal susceptibilities of Candida parapsilosis isolated from patients with bloodstream infections. J Med Microbiol. 2009;58(Pt 2):185–191. doi:10.1099/jmm.0.004242-0
64. Mirhendi H, Bruun B, Schønheyder HC, et al. Molecular screening for Candida orthopsilosis and Candida metapsilosis among Danish Candida parapsilosis group blood culture isolates: proposal of a new RFLP profile for differentiation. J Med Microbiol. 2010;59(Pt 4):414–420. doi:10.1099/jmm.0.017293-0
65. Mendoza-Reyes DF, Gómez-Gaviria M, Mora-Montes HM. Candida lusitaniae: biology, pathogenicity, virulence factors, diagnosis, and treatment. Infect Drug Resist. 2022;15:5121–5135. doi:10.2147/idr.S383785
66. Martinez-Alvarez JA, Perez-Garcia LA, Flores-Carreon A, et al. The immune response against Candida spp. and Sporothrix schenckii. Rev Iberoam Micol. 2014;31(1):62–66. doi:10.1016/j.riam.2013.09.015
67. Netea MG, Joosten LA, van der Meer JW, Kullberg BJ, van de Veerdonk FL. Immune defence against Candida fungal infections. Nat Rev Immunol. 2015;15(10):630–642. doi:10.1038/nri3897
68. Hernandez-Chavez MJ, Perez-Garcia LA, Nino-Vega GA, Mora-Montes HM. Fungal strategies to evade the host immune recognition. J Fungi. 2017;3(4):51. doi:10.3390/jof3040051
69. Gómez-Gaviria M, Vargas-Macías AP, García-Carnero LC, Martínez-Duncker I, Mora-Montes HM. Role of protein glycosylation in interactions of medically relevant fungi with the host. J Fungi. 2021;7(10):875. doi:10.3390/jof7100875
70. Netea MG, Gow NA, Munro CA, et al. Immune sensing of Candida albicans requires cooperative recognition of mannans and glucans by lectin and Toll-like receptors. J Clin Invest. 2006;116(6):1642–1650. doi:10.1172/JCI27114
71. Dennehy KM, Ferwerda G, Faro-Trindade I, et al. Syk kinase is required for collaborative cytokine production induced through Dectin-1 and Toll-like receptors. Eur J Immunol. 2008;38(2):500–506. doi:10.1002/eji.200737741
72. Yadav B, Mora-Montes HM, Wagener J, et al. Differences in fungal immune recognition by monocytes and macrophages: n-mannan can be a shield or activator of immune recognition. Cell Surf. 2020;6:100042. doi:10.1016/j.tcsw.2020.100042
73. Orsi CF, Colombari B, Blasi E. Candida metapsilosis as the least virulent member of the ‘C. parapsilosis’ complex. Med Mycol. 2010;48(8):1024–1033. doi:10.3109/13693786.2010.489233
74. Feng X, Ling B, Yang G, Yu X, Ren D, Yao Z. Prevalence and distribution profiles of Candida parapsilosis, Candida orthopsilosis and Candida metapsilosis responsible for superficial candidiasis in a Chinese university hospital. Mycopathologia. 2012;173(4):229–234. doi:10.1007/s11046-011-9496-5
75. Barbedo LS, Vaz C, Pais C, et al. Different scenarios for Candida parapsilosis fungaemia reveal high numbers of mixed C. parapsilosis and Candida orthopsilosis infections. J Med Microbiol. 2015;64(Pt 1):7–17. doi:10.1099/jmm.0.080655-0
76. Oliveira VK, Paula CR, Colombo AL, et al. Candidemia and death by Candida orthopsilosis and Candida metapsilosis in neonates and children. Pediatr Neonatol. 2014;55(1):75–76. doi:10.1016/j.pedneo.2013.07.006
77. De Carolis E, Hensgens LA, Vella A, et al. Identification and typing of the Candida parapsilosis complex: MALDI-TOF MS vs AFLP. Med Mycol. 2014;52(2):123–130. doi:10.1093/mmy/myt009
78. Ghelardi E, Pichierri G, Castagna B, Barnini S, Tavanti A, Campa M. Efficacy of Chromogenic Candida Agar for isolation and presumptive identification of pathogenic yeast species. Clin Microbiol Infect. 2008;14(2):141–147. doi:10.1111/j.1469-0691.2007.01872.x
79. Criseo G, Scordino F, Romeo O. Current methods for identifying clinically important cryptic Candida species. J Microbiol Methods. 2015;111:50–56. doi:10.1016/j.mimet.2015.02.004
80. Willemsen M, Breynaert J, Lauwers S. Comparison of Auxacolor with API 20 C Aux in yeast identification. Clin Microbiol Infect. 1997;3(3):369–375. doi:10.1111/j.1469-0691.1997.tb00628.x
81. Bader O, Weig M, Taverne-Ghadwal L, Lugert R, Gross U, Kuhns M. Improved clinical laboratory identification of human pathogenic yeasts by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin Microbiol Infect. 2011;17(9):1359–1365. doi:10.1111/j.1469-0691.2010.03398.x
82. Garcia-Effron G, Canton E, Pemán J, Dilger A, Romá E, Perlin DS. Assessment of two new molecular methods for identification of Candida parapsilosis sensu lato species. J Clin Microbiol. 2011;49(9):3257–3261. doi:10.1128/jcm.00508-11
83. Arastehfar A, Fang W, Pan W, Liao W, Yan L, Boekhout T. Identification of nine cryptic species of Candida albicans, C. glabrata, and C. parapsilosis complexes using one-step multiplex PCR. BMC Infect Dis. 2018;18(1):480. doi:10.1186/s12879-018-3381-5
84. Quiles-Melero I, García-Rodríguez J, Gómez-López A, Mingorance J. Evaluation of matrix-assisted laser desorption/ionisation time-of-flight (MALDI-TOF) mass spectrometry for identification of Candida parapsilosis, C. orthopsilosis and C. metapsilosis. Eur J Clin Microbiol Infect Dis. 2012;31(1):67–71. doi:10.1007/s10096-011-1277-z
85. Souza AC, Ferreira RC, Gonçalves SS, et al. Accurate identification of Candida parapsilosis (sensu lato) by use of mitochondrial DNA and real-time PCR. J Clin Microbiol. 2012;50(7):2310–2314. doi:10.1128/jcm.00303-12
86. Gomez-Lopez A, Alastruey-Izquierdo A, Rodriguez D, et al. Prevalence and susceptibility profile of Candida metapsilosis and Candida orthopsilosis: results from population-based surveillance of candidemia in Spain. Antimicrob Agents Chemother. 2008;52(4):1506–1509. doi:10.1128/aac.01595-07
87. Bennett JE. Echinocandins for candidemia in adults without neutropenia. N Engl J Med. 2006;355(11):1154–1159. doi:10.1056/NEJMct060052
88. Douglas CM, Foor F, Marrinan JA, et al. The Saccharomyces cerevisiae FKS1 (ETG1) gene encodes an integral membrane protein which is a subunit of 1,3-beta-D-glucan synthase. Proc Natl Acad Sci U S A. 1994;91(26):12907–12911. doi:10.1073/pnas.91.26.12907
89. Douglas CM, D’Ippolito JA, Shei GJ, et al. Identification of the FKS1 gene of Candida albicans as the essential target of 1,3-beta-D-glucan synthase inhibitors. Antimicrob Agents Chemother. 1997;41(11):2471–2479. doi:10.1128/aac.41.11.2471
90. Perlin DS. Resistance to echinocandin-class antifungal drugs. Drug Resist Updat. 2007;10(3):121–130. doi:10.1016/j.drup.2007.04.002
91. Katiyar S, Pfaller M, Edlind T. Candida albicans and Candida glabrata clinical isolates exhibiting reduced echinocandin susceptibility. Antimicrob Agents Chemother. 2006;50(8):2892–2894. doi:10.1128/aac.00349-06
92. Spreghini E, Orlando F, Tavanti A, et al. In vitro and in vivo effects of echinocandins against Candida parapsilosis sensu stricto, Candida orthopsilosis and Candida metapsilosis. J Antimicrob Chemother. 2012;67(9):2195–2202. doi:10.1093/jac/dks180
93. Diekema DJ, Messer SA, Boyken LB, et al. In vitro activity of seven systemically active antifungal agents against a large global collection of rare Candida species as determined by CLSI broth microdilution methods. J Clin Microbiol. 2009;47(10):3170–3177. doi:10.1128/jcm.00942-09
94. Földi R, Kovács R, Gesztelyi R, et al. Comparison of in vitro and vivo efficacy of caspofungin against Candida parapsilosis, C. orthopsilosis, C. metapsilosis and C. albicans. Mycopathologia. 2012;174(4):311–318. doi:10.1007/s11046-012-9554-7
95. Sarvikivi E, Lyytikäinen O, Soll DR, et al. Emergence of fluconazole resistance in a Candida parapsilosis strain that caused infections in a neonatal intensive care unit. J Clin Microbiol. 2005;43(6):2729–2735. doi:10.1128/jcm.43.6.2729-2735.2005
96. Fleck R, Dietz A, Hof H. In vitro susceptibility of Candida species to five antifungal agents in a German university hospital assessed by the reference broth microdilution method and Etest. J Antimicrob Chemother. 2007;59(4):767–771. doi:10.1093/jac/dkl555
 © 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.