Back to Journals » International Journal of General Medicine » Volume 14
The Correlation Between MYO9B Gene Polymorphism and Inflammatory Bowel Disease in the Guangxi Zhuang Population
Authors Zeng RZ, Lv XD, Liu GF, Gu GL, Li SQ, Chen L, Fan JH, Liang ZL, Wang HQ, Lu F, Zhan LL, Lv XP
Received 10 September 2021
Accepted for publication 27 October 2021
Published 1 December 2021 Volume 2021:14 Pages 9163—9172
DOI https://doi.org/10.2147/IJGM.S338142
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Scott Fraser
Rui-zhi Zeng,1 Xiao-dan Lv,2 Geng-feng Liu,1 Guang-li Gu,1 Shi-quan Li,1 Lan Chen,1 Jun-hua Fan,1 Zhao-liang Liang,1 Hui-qin Wang,1 Fei Lu,1 Ling-ling Zhan,2 Xiao-ping Lv1
1Department of Gastroenterology, The First Affiliated Hospital of Guangxi Medical University, Nanning, 530021, Guangxi Zhuang Autonomous Region, People’s Republic of China; 2Department of Clinical Laboratory, The First Affiliated Hospital of Guangxi Medical University, Nanning, 530021, Guangxi Zhuang Autonomous Region, People’s Republic of China
Correspondence: Xiao-ping Lv
Department of Gastroenterology, The First Affiliated Hospital of Guangxi Medical University, No. 6 of Shuangyong Street, Qingxiu District, Nanning, 530021, Guangxi Zhuang Autonomous Region, People’s Republic of China
Tel +86 135 1788 8390
Email [email protected]
Objective: To analyze the correlation between site rs962917 of the MYO9B gene and inflammatory bowel disease (IBD) in the Guangxi Zhuang nationality population.
Methods: The intestinal mucosa tissue of 153 IBD subjects (Han and Zhuang patients only) in the Guangxi Zhuang autonomous region comprised the case group, and the intestinal mucosa tissue of 155 healthy subjects (Han and Zhuang patients only) in the same region represented the control group. Deoxyribonucleic acid was extracted from the intestinal mucosa tissue of each experimental group, and the MYO9B gene-target fragment containing the single nucleotide polymorphism (SNP) site rs962917 was designed. Finally, polymerase chain reaction products were obtained by amplification, analyzed, and compared using the sequencing results.
Results: The results indicated that the genotype frequency of the MYO9B SNP site rs962917 between Crohn’s disease (CD) and control groups of Zhuang and Han participants differed significantly (P < 0.05). Furthermore, the genotype frequency of MYO9B site rs962917 differed significantly between the Zhuang and Han population groups (P < 0.05).
Conclusion: Site rs962917 of the MYO9B gene is related to CD susceptibility and incidence among the Guangxi Zhuang population.
Keywords: inflammatory bowel disease, IBD, ulcerative colitis, UC, Crohn’s disease, CD, MYO9B gene, gene polymorphism
Introduction
Inflammatory bowel disease (IBD) is chronic and recurrent intestinal inflammation of unknown etiology. Epidemiological data indicate that the incidence of IBD is high in Western regions, such as the Americas and Europe, and low in Asian regions. However, IBD has become increasingly common in Asia in recent years with the number of cases increasing annually.1–3
The etiology and pathogenesis of IBD include several factors, such as genetic susceptibility, the effect of environmental factors on the intestinal flora, chronic, repeated alternating with easy bowel and an abnormal immune response, intestinal mucosa barrier damage, aggravating inflammation, and deferment and recurrence of pathological change.4 Research showed that intestinal mucosal permeability and the structure and function of the mesenteric barrier are mainly related to the tight junction (TJ) between intestinal mucosal epithelial cells; The intestinal mucosa of IBD patients shows the expression and function of TJs between intestinal epithelial cells are destroyed, that causes damage to the intestinal mucosal barrier structure and function.5–7
TJs are affected by myosin and various genes regulate its formation, function, metabolism, and other physiological processes. Myosin IXB, which includes a Ras homolog family member (Rho) GTPase binding site, and its role is related to TJs.8 GWAS studies have shown that in addition to MYO9B, multiple genes may affect the protein structure and function of gene loci (eg, rs962917). The locus mutation or loss in these genes may cause damage to the structure and function of the intestinal mucosa barrier, increase the permeability of intestinal mucosa, and promote the onset and development of IBD, which will affect the susceptibility of patients to IBD.9,10
Based on the research focused on these SNPs, the effect of MYO9B gene variation on the structure and function of human intestinal myosin IXB and TJs, as well as the incidence and development of IBD, was clarified.11 However, only a small number of studies have investigated the correlation between the IBD population and the MYO9B gene in China, and none have to date analyzed the correlation between the IBD population and the MYO9B gene within the Guangxi Zhuang population. Accordingly, we aimed to investigate the correlation between the IBD population and MYO9B gene polymorphism among the Guangxi Zhuang population.
Materials and Methods
Research Subjects
The case group comprised 153 IBD patients from Guangxi who had been treated in the Gastroenterology Department of the First Affiliated Hospital of Guangxi Medical University (China) from August 2016 to December 2018. These patients included 79 cases of Han and 74 of Zhuang nationality. Among them, 41 were male and 33 were female of Zhuang nationality, while those of Han nationality comprised 39 males and 40 females. Furthermore, the series included 39 cases of UC and 35 cases of CD among those of Zhuang nationality and 43 cases of UC, and 36 cases of CD among those of Han nationality. The inclusion criteria for patients in the IBD group were as follows: (1) consensus reached on the diagnosis and treatment of IBD in 2012; (2) participants of Zhuang nationality has been a resident of Guangxi for a long time and their immediate relatives of the three preceding generations were all of Zhuang nationality and had not intermarried with other nationalities. The study’s exclusion criteria were as follows: combined with other infectious and non-infectious colitis conditions; IBD patients with other autoimmune diseases. The control group comprised 155 healthy individuals without blood relationships in the Guangxi region. The sampling sites were corresponding to the case group This group included 40 males and 30 females of Zhuang nationality and 45 males and 40 females of Han nationality. In addition, 82 patients with UC showed lesions in the rectum (33 patients), the left-half colon (22 patients), and the entire colon (27 patients). Among 71 cases of CD, 50 lesions were located in the ileum, 11 in the colon, and 10 in the ileocolon. The control group comprised 155 patients.
Genomic Deoxyribonucleic Acid Collection
Intestinal Mucosa Specimen Collection
After informing the patient and receiving their consent, and following the approval of the ethics committee of our hospital, 20–40 mg of fresh intestinal mucosal tissue were removed under an endoscope by biopsy forceps from the participants in the case and control groups and stored at –20°C.
Deoxyribonucleic Acid Extraction from the Intestinal Mucosa
After the samples were washed and broken into cell suspension, DNA was extracted from the intestinal tissue using a TIANamp Genomic DNA Kit (DP304; Tiangen Biotechnology Co., Ltd., Beijing, China). Firstly, Proteinase K was added and mixed, placed at 56°C until tissue was dissolved; 200 μL anhydrous ethanol was added in the clear solution and mixed well. The solution in the previous step were added to a spin columns, buffer was added to wash the DNA product. Then elution buffer was added, and the solution in the spin columns was into a new collection tube. Finally, DNA product was stored in at −20°C and DNA degradation should be avoid.
Deoxyribonucleic Acid Purity Determination
The purity of DNA (1ul per DNA sample) was determined using a nucleic acid-protein analyzer, and A260/A280 was recorded. A ratio closer to 1.80 indicated high purity. After that, the quantity and quality of DNA was analysis by gel electrophoresis, the qualified DNA was collected for the next step.
DNA Extraction and Genotyping of the MYO9B Polymorphisms
The PCR primers that were employed in this study are listed in Table 1 (primers were designed and synthesized by Takara Biotechnology Co., Ltd., Dalian, China).
 |
Table 1 Polymerase Chain Reaction Primers |
The PCR was a 50-μL reaction comprising 25 μL Premier Taq and a 2μL DNA template (RR001A, Takara Biotechnology Co., Ltd.), 21 μL enzyme-free water (9012; Takara Biotechnology Co., Ltd.), 1 μL rs962917-f, and 1 μL rs962917-r. The reaction conditions were as follows: pre-denaturation for 0 s; 30 cycles: denaturation at 98°C for 30 s, annealing at 55°C for 30 s, extension: 72°C for 30 s. The amplified products were analyzed by agarose gel electrophoresis in a 1×Tris - boric acid buffer, and images were captured under ultraviolet illumination (Bio-Rad Gel Doc-2000; Hercules, CA, United States). Then, the PCR products were excised from the gel and stored at –20°C, until sequencing by Takara Biotechnology Co., Ltd (Using ddNTP labeled with four-color fluorescent dye, ABI3730XL automatic sequencer detected four fluorophore groups on DNA fragments of different lengths by capillary electrophoresis and fluorescence labeling technology, and directly translated DNA fragments into DNA sequences).
Statistical Analysis
The DNA sequencing data were classified and processed, and the genotype and allele frequencies of SNP site rs962917 of the MYO9B gene were calculated using the gene-counting method. A Hardy–Weinberg balanced test and the SPSS Statistics 20.0 software package (chi-square (χ2) and Fisher’s exact probability tests) were used to compare the genotype and allele frequencies between groups; P < 0.05 indicated a statistically significant difference between each group and among individual groups.
Results
Electrophoresis of the Single Nucleotide Polymorphism Site rs962917 of the MYO9B Gene
The results for PCR gel electrophoresis of the extended gene fragment in the case and control groups were compared (Figure 1). The electrophoresis bands of the product were consistent with the size of the target fragment (the molecular weight range of DNA marker 100–500 bp).
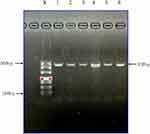 |
Figure 1 Gel electrophoresis of PCR extended gene fragments of MY09B SNP locus rs962917 in each group. |
Nucleotide Sequencing of the Polymerase Chain Reaction Product
The sequencing results showed three genotypes, and homozygous; Figures 2–4 show the sequencing maps of the rs962917 SNP site of the MYO9B gene.
 |
Figure 2 Homozygous CC sequencing diagram, blue single peak indicated by arrow (location 149). |
 |
Figure 3 Homozygous TT sequencing map, arrow pointing to red single peak (location 144). |
 |
Figure 4 Heterozygote CT sequencing map, blue and red double peaks indicated by arrow (location 148). |
Genotype and Allele Frequency
A Comparison of Genotypes and Allele Frequencies of the MYO9B Gene SNP Site rs962917 Between the Case and Control Groups
As shown in Tables 2 and 3, the genotype frequency of rs962917 in the CD group comprising participants of Zhuang nationality was compared to that in the control group; the P-value was <0.05 (P = 0.033), indicating a statistically significant difference between the groups. However, when comparing the genotype and allele frequencies between the Zhuang UC and control group, the P-value was not statistically significant (the P-values were 0.885 and 0.782, respectively).
 |
Table 2 Genotype and Allele Frequency of rs962917 in the Zhuang IBD Case Group and the Control Group |
 |
Table 3 Genotype and Allele Frequency of rs962917 in the Han IBD Case Group and the Control Group |
Compared to the control group, the genotype frequency of rs962917 in the CD participant group of Han nationality was P < 0.05 (P = 0.015), and the difference was statistically significant. However, when comparing the genotype and allele frequencies between the UC and control groups of Han nationality, the P-values were not statistically significant (P = 0.791 and 0.063, respectively). In summary, the rs962917 site of the MYO9B gene was not related to Guangxi Han or Zhuang UC but was positively related to cases of Guangxi Han and Zhuang CD.
A Comparison of the rs962917 Site Genotype and the Allele Frequency of the MYO9B Gene in the IBD Groups of Zhuang and Han Nationality
As shown in Table 4, when comparing the genotype frequency between the IBD groups of Zhuang and Han nationality, a P-value of >0.05 was not statistically significant (the P-values were 0.479 and 0.286, respectively). When comparing the genotype and allele frequency between the UC groups of Zhuang and Han nationality, the P-values were >0.05 with no statistical significance (the P-values were 0.536 and 0.429, respectively).
 |
Table 4 Genotype and Allele Frequency of MYO9B Site rs962917 in the IBD Group of Zhuang Nationality and IBD Group of Han Nationality |
When comparing the genotype frequency between the CD groups of Zhuang and Han nationality, a P-value < 0.05 indicated a statistically significant difference (P = 0.013). A P-value > 0.05 was not statistically significant (P = 0.723). In conclusion, MYO9B gene site rs962917 is related to CD incidence in Zhuang and Han nationalities but not related to UC in Zhuang and Han nationalities.
The Odds Ratio Values of Genotype and Allele Frequencies of Site rs962917 of the MYO9B Gene: A Comparison Between Inflammatory Bowel Disease Cases and Control Groups
As shown in Tables 5 and 6, combined with the above conclusions, the odds ratio (OR) values of genotype and allele frequencies were compared between the Zhuang case group (IBD and CD) and the Zhuang control group. The TT genotype and T-allele may increase the risk of IBD OR CD and CT genotype, while the C-allele may reduce this risk; currently, the role of the CC-genotype remains unclear. In addition, when comparing the OR value of the genotype and allele frequencies between the Han nationality case group (IBD and CD) and the Han nationality control group, the CT genotype and T-allele were shown to increase the risk of developing IBD OR CD, while the CC genotype and C-allele may reduce these risks; however, the role of the TT genotype remains unclear.
 |
Table 5 Comparison Between the Zhuang Case Group (IBD, UC, and CD) and the Zhuang Control Group (MYO9B Locus rs962917 Genotype and Allele Frequency Dominance Ratio and 95% CI) |
 |
Table 6 Comparison Between the Han Case Group (IBD, UC, and CD) and the Han Control Group (MYO9B Locus rs962917 Genotype and Allele Frequency Dominance Ratio and 95% Confidence Interval) |
Discussion
Intestinal mucosal barrier injury and changes in intestinal mucosal permeability, as well as characteristic pathological changes in chronic inflammatory intestinal diseases (IBD, such as UC and CD) are related to the TJ contraction of intestinal epithelial cells and myosin.12 Studies have shown that in experimental colitis models, the expression of TJ protein isoforms (including claudin, occludin, and ZOs) was decreased and often accompanied by an increase in intestinal epithelial permeability.13,14 Bacteria, toxins, and other substances in the intestinal cavity can disrupt and break through the intestinal epithelial cells’ TJs, penetrate the intestinal mucosa, and enter other tissue, organs, and the circulatory system, causing bacterial and toxin translocation. Concurrently, intestinal inflammation causes the release of a large number of inflammatory factors that affect epithelial barrier function and can lead to the development of IBD.15 The TJ intestinal epithelial cells can serve as a signaling center that transfers cell information between the inside and outside; contrastingly, the structure and function are regulated by various materials, such as silk that cracks the original mitogen-activated protein kinase, myosin light-chain kinase, Rho/Rho-kinase, signal transducers, and the activation of transcription factor 3. Furthermore, the inhibition of Rho activity decreases the expression of TJ protein isoforms, such as closed small-loop protein ZOs and occludin proteins, whereas the activation of Rho activity increases the expression of Z0s and occludin proteins, thereby maintaining TJ function.16
Several genes that can affect the intestinal barrier have been identified including MAGI2, PARD3, and MYO9B.17 The MYO9B gene exists in the mammalian body as myosin IX, its tail includes a Rho GTPase activation domain, which negatively regulates the RhoA signal. The encoding gene, myosin IXB, is involved in the formation of TJs between the intracellular skeleton and adjacent cells, thereby participating in the regulation of the morphology and differentiation of epithelial cells in the body.18 In MYO9B knockout (KO) studies of mice, fecal occult blood (indicators) reflected gastrointestinal bleeding. Intestinal epithelial cell apoptosis was able to spread along the entire intestine, and pathological intestinal biopsy showed intestinal mucosa damage in the ileum. The most common pathological features include surface ulcers and neutrophil infiltration; ultrastructural examination confirmed epithelial discontinuous and deposition of extracellular matrix. The present study showed the percutaneous resistance of KO ileum to wild-type (WT) ileum and found that the percutaneous resistance of KO ileum was three times smaller compared with WT ileum, and the intestinal mucosa was also permeable to high-molecular-weight glucan, suggesting that the impaired mucosal barrier function and the change in intestinal permeability of KO mice may have been due to the presence of mucosal surface ulcers.19 Interestingly, when MYO9B gene expression was deficient or the C-terminal dominant-negative tail expression was lacking, damaged intestinal epithelial cell line Caco2 (BBe) was unable to migrate to the wound surface, indicating the destruction of TJ proteins, as well as the decreased torsional motor ability and connection permeability.20 This suggested that MYO9B played a crucial role in epithelial wound healing and TJ integrity maintenance, key functions that may be altered in related IBD patients. The MYO9B gene is also involved in the pathogenesis and development of IBD, and its genetic variation may affect the tight connection and assembly of epithelial cells and remodeling of the cytoskeleton.21
Currently, studies on susceptible genes use modified signal transduction pathways as therapeutic targets for IBD, and MYO9B has proven to be related to IBD.22 Nowadays, many new drug therapies have been studied to participate in the regulation of intestinal mucosal barrier. MYO9B, as an important expression gene involved in intestinal mucosal barrier, should be paid more attention to its expression in patients.23,24 Van Bodegraven et al studied eight SNPs in the MYO9B gene including six sites related to digestive diseases and found the gene to be closely related to IBD.25 Latiano et al demonstrated that the MYO9B gene was closely linked to IBD in the Italian population, indicating that the rs962917 genotype may increase susceptibility to IBD.26 A meta-analysis conducted by Wang et al revealed that rs962917 in MY09B increased the risk of IBD, UC, and CD.27 The number of MYO9B IBD patients in our country is small; the study of genetic polymorphisms, such as rs962917 in the UC clinical subgroups, did not differ, while the frequency of rs962917 distribution differed significantly in the CD and phenotype cases. This phenomenon could be attributed to the different locations; however, the correlation between these MYO9B polymorphisms and Chinese Han patients with IBD intestinal permeability has not yet been identified.28
We explored the correlation between MYO9B and IBD, UC, and CD populations of Guangxi Zhuang nationality in China. The genotype frequency of rs962917 in the CD groups of Zhuang and Han nationality differed significantly compared to that of the control group; however, the difference in allele frequency was not statistically significant. Furthermore, no significant difference was detected in the genotype and allele frequencies between the UC and control groups. Therefore, MYO9B gene rs962917 may be related to cases of CD in the Zhuang and Han populations in Guangxi. In addition, the genotype frequency of MYO9B gene site rs962917 in the Zhuang population differed significantly from that of the Han nationality CD group. The genotypes and alleles for the MYO9B gene site rs962917 in the Zhuang group and the IBD and UC groups of Han nationality did not show any statistical significance, which further supported the above conclusion. However, many similarities and differences were detected in the conclusions of this study, as well as in studies conducted worldwide. This may be ascribed to the different races and regions related to IBD patients, who exhibit variable clinical characteristics, as well as the different effects of MYO9B gene polymorphism.
Conclusion
In conclusion, IBD is a genetically susceptible disease. The transcriptional regulation of the susceptibility genes in IBD, the processing and modification of ribosomal ribonucleic acid (RNA) and messenger RNA, translation control and DNA rearrangement, the pathogenesis of IBD, and the distribution of susceptibility genes must be investigated further in this regard. Early preventive treatment for individuals with a genetic risk by screening for common IBD- susceptible genes is important for providing new methods for the accurate diagnosis, treatment, and prevention of IBD and to improve the therapeutic effect on the disease.29–31 In particular, individual or group differences must be noted in the incidence of the disease; additionally, accurate classification of the disease and its symptomatic treatment via a nutritional diet, indications of drug and surgical treatment, effective treatments, cause evaluation of side effects and adverse reactions, and long-term monitoring of disease outcome and recurrence requires further research.32
Ethics Approval and Consent to Participate
This study was conducted in accordance with the declaration of Helsinki. This study was conducted with approval from the Ethics Committee of The First Affiliated Hospital of Guangxi Medical University [2021(KY-E-194)]. A written informed consent was obtained from all participants.
Funding
The National Natural Science Foundation of China (No. 81860104, 81860120, 81460114); The Natural Science Foundation of Guangxi Zhuang Autonomous Region (No. 2017GXNSFBA1981-34, No. 2017GXNSFAA- 198299); the Development and Application of Medical and Health Appropriate Technology Project in Guangxi Zhuang Autonomous Region (No. S2018049); The Youth Science Foundation of Guangxi Medical University (No. GXMUYSF201913, GXMUYSF201908); The Self-financing Project of Health- Commission of Guangxi Zhuang Autonomous Region (No. Z20200398).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Liu TC, Stappenbeck TS. Genetics and pathogenesis of inflammatory bowel disease. Annu Rev Pathol. 2016;11:127–148. doi:10.1146/annurev-pathol-012615-044152
2. Hu S, Vich Vila A, Gacesa R, et al. Whole exome sequencing analyses reveal gene-microbiota interactions in the context of IBD. Gut. 2021;70:285–296.
3. Graham DB, Xavier RJ. Pathway paradigms revealed from the genetics of inflammatory bowel disease. Nature. 2020;578:527–539. doi:10.1038/s41586-020-2025-2
4. Xavier RJ, Podolsky DK. Unravelling the pathogenesis of inflammatory bowel disease. Nature. 2007;448(7152):427–434. doi:10.1038/nature06005
5. He W-Q, Wang J, Sheng J-Y, et al. Contributions of myosin light chain kinase to regulation of epithelial paracellular permeability and mucosal homeostasis. Int J Mol Sci. 2020;21(3):993. doi:10.3390/ijms21030993
6. Allam-Ndoul B, Castonguay-Paradis S, Veilleux A. Gut microbiota and intestinal trans-epithelial permeability. Int J Mol Sci. 2020;21(17):6402. doi:10.3390/ijms21176402
7. Schulzke JD, Bojarski C, Zeissig S, et al. Disrupted barrier function through epithelial cell apoptosis. Ann N Y Acad Sci. 2006;1072:288–299. doi:10.1196/annals.1326.027
8. Liping S, Shen L, Clayburgh DR, et al. Targeted epithelial tight junction dysfunction causes immune activation and contributes to development of experimental colitis. Gastroenterology. 2009;136(2):551–563. doi:10.1053/j.gastro.2008.10.081
9. Sánchez de Medina F, Romero-Calvo I, Mascaraque C, Martínez-Augustin O. Intestinal inflammation and mucosal barrier function. Inflamm Bowel Dis. 2014;20(12):2394–2404. doi:10.1097/MIB.0000000000000204
10. Kurashima Y, Kiyono H. Mucosal ecological network of epithelium and immune cells for gut homeostasis and tissue healing. Annu Rev Immunol. 2017;35:119–147. doi:10.1146/annurev-immunol-051116-052424
11. Frenkel S, Bernstein CN, Sargent M, et al. Genome-wide analysis identifies rare copy number variations associated with inflammatory bowel disease. PLoS One. 2019;14:e0217846. doi:10.1371/journal.pone.0217846
12. Martini E, Krug SM, Siegmund B, Neurath MF, Becker C. Mend your fences: the epithelial barrier and its relationship with mucosal immunity in inflammatory bowel disease. Cell Mol Gastroenterol Hepatol. 2017;4(1):33–46. doi:10.1016/j.jcmgh.2017.03.007
13. Shigetomi K, Ikenouchi J. Regulation of the epithelial barrier by post- translational modifications of tight junction membrane proteins. J Biochem. 2018;163:265–272. doi:10.1093/jb/mvx077
14. Foerster E, Mukherjee T, Cabral-Fernandes L, et al. How autophagy controls the intestinal epithelial barrier. Autophagy. 2021:1–18. doi:10.1080/15548627.2021.1909406
15. Lu RY, Yang WX, Hu YJ. The role of epithelial tight junctions involved in pathogen infections. Mol Biol Rep. 2014;41:6591–6610. doi:10.1007/s11033-014-3543-5
16. Fries W, Belvedere A, Vetrano S. Sealing the broken barrier in IBD: intestinal permeability, epithelial cells and junctions. Curr Drug Targets. 2013;14(12):1460–1470. doi:10.2174/1389450111314120011
17. McGovern Dermot PB, Kugathasan S, Cho JH. Genetics of inflammatory bowel diseases. Gastroenterology. 2015;149(5):1163. doi:10.1053/j.gastro.2015.08.001
18. Wolters VM, Xu W, Zhao X, et al. Replication of genetic variation in the MYO9B gene in crohn’s disease. Hum Immunol. 2011;72(7):592–597. doi:10.1016/j.humimm.2011.03.025
19. Garg A, Zhao A, Erickson SL, et al. Pregnane X receptor activation attenuates inflammation-associated intestinal epithelial barrier dysfunction by inhibiting cytokine-induced myosin light-chain kinase expression and c-Jun N-terminal kinase 1/2 activation. J Pharmacol Exp Ther. 2016;359(1):91–101. doi:10.1124/jpet.116.234096
20. Hemkemeyer SA, Vollmer V, Schwarz V, et al. Local Myo9b RhoGAP activity regulates cell motility. J Biol Chem. 2021;296:100136. doi:10.1074/jbc.RA120.013623
21. Monsuur AJ, De Bakker PIW, Alizadeh BZ, et al. Myosin IXB variant increases the risk of celiac disease and points toward a primary intestinal barrier defect. Nat Genet. 2005;37(12):1341–1344. doi:10.1038/ng1680
22. Chandhoke SK, Mooseker MS. A role for myosin IXb, a motor-RhoGAP chimera, in epithelial wound healing and tight junction regulation. Mol Biol Cell. 2012;23:2468–2480. doi:10.1091/mbc.e11-09-0803
23. Hegan PS, Chandhoke SK, Barone C, et al. Mice lacking myosin IXb, an inflammatory bowel disease susceptibility gene, have impaired intestinal barrier function and superficial ulceration in the ileum. Cytoskeleton. 2016;73(4):163–179. doi:10.1002/cm.21292
24. Sanchez E, Alizadeh BZ, Valdigem G, et al. MYO9B gene polymorphisms are associated with autoimmune diseases in Spanish population. Hum Immunol. 2007;68:610–615. doi:10.1016/j.humimm.2007.03.006
25. Van Bodegraven AA, Curley CR, Hunt KA, et al. Genetic variation in myosin IXB is associated with ulcerative colitis. Gastroenterology. 2006;131(6):1768–1774. doi:10.1053/j.gastro.2006.09.011
26. Latiano A, Palmieri O, Valvano MR, et al. The association of MYO9B gene in Italian patients with inflammatory bowel diseases. Aliment Pharmacol Ther. 2008;27(3):241–248. doi:10.1111/j.1365-2036.2007.03551.x
27. Wang MJ, Xu XL, Yao GL, et al. MYO9B gene polymorphisms are associated with the risk of inflammatory bowel diseases. Oncotarget. 2016;7(37):58862–58875. doi:10.18632/oncotarget.11186
28. Hu J, Mei Q, Huang J, et al. Association of MYO9B gene polymorphisms with inflammatory bowel disease in Chinese Han population. World J Gastroenterol. 2014;20(23):7466–7472. doi:10.3748/wjg.v20.i23.7466
29. Torres J, Ellul P, Langhorst J, et al. European crohn’s and colitis organisation topical review on complementary medicine and psychotherapy in inflammatory bowel disease. J Crohns Colitis. 2019;13(6):673–85e. doi:10.1093/ecco-jcc/jjz051
30. Loy L, Roda G, Fiorino G, et al. Detection and management of early stage inflammatory bowel disease: an update for clinicians. Expert Rev Gastroenterol Hepatol. 2019;13(6):547–555. doi:10.1080/17474124.2019.1605291
31. Powell JJ, Cook WB, Hutchinson C, et al. Dietary fortificant iron intake is negatively associated with the quality of life in patients with mildly active inflammatory bowel diseases. Nutr Metab. 2013;10:9. doi:10.1186/1743-7075-10-9
32. Queiroz NSF, Regueiro M. Safety considerations with biologics and new inflammatory bowel disease therapies. Curr Opin Gastroenterol. 2020;36:257–264. doi:10.1097/MOG.0000000000000607
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
