Back to Journals » OncoTargets and Therapy » Volume 14
The Biological Roles of Exosomal Long Non-Coding RNAs in Cancers
Authors Da M , Jiang H, Xie Y, Jin W, Han S
Received 8 October 2020
Accepted for publication 22 December 2020
Published 12 January 2021 Volume 2021:14 Pages 271—287
DOI https://doi.org/10.2147/OTT.S281175
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Federico Perche
Miao Da,1 Hao Jiang,1 Yangyang Xie,2 Weili Jin,3 Shuwen Han4
1Department of Nursing, Huzhou Third Municipal Hospital, Huzhou, Zhejiang, People’s Republic of China; 2Key Laboratory of Diagnosis and Treatment of Digestive System Cancers of Zhejiang Province, Ningbo 315000, Zhejiang, People’s Republic of China; 3Department of Gastroenterology, Nanxun District People’s Hospital, Huzhou, Zhejiang 313009, People’s Republic of China; 4Department of Oncology, Huzhou Central Hospital, Affiliated Central Hospital HuZhou University, Huzhou, Zhejiang, People’s Republic of China
Correspondence: Shuwen Han
Department of Oncology, Huzhou Central Hospital, Affiliated Central Hospital HuZhou University, No.1558, Sanhuan North Road, Wuxing District, Huzhou, Zhejiang, People’s Republic of China
Tel +86 5722555750
Email [email protected]
Abstract: Although it has many treatment strategies, cancer is still one of the most common causes of morbidity and mortality in the world. Exosomes are small extracellular vesicles (EVs) that can be secreted by almost all cells. Exosomes can encapsulate various types of molecules, including lipids, proteins, DNA, messenger RNAs, and non-coding RNAs [microRNAs (miRNAs) and long non-coding RNAs (lncRNAs)]. Exosome release is a way of communication between cells. They act as powerful signaling molecules between cancer cells and the surrounding cells that make up the cancer microenvironment. lncRNAs are a class of non-coding P, with a length of more than 200 bp, which are differentially expressed in many cancers. lncRNAs have been widely regarded as a new medium for cancer behavior. The presence of lncRNAs in circulation can be acellular or encapsulated in exosomal bodies released by cancer cells. Exosomal lncRNAs are functional and can transmit different phenotypic patterns to neighboring cells. Here, we reviewed the molecular mechanism of exosomal lncRNAs in regulating cancer progression, angiogenesis, and chemotherapy resistance, as well as the prospective applications of exosomal lncRNAs in cancer diagnosis, treatment and prognosis. These findings potentially promote the current understanding of exosomal lncRNAs and provide a new research direction for exosomal lncRNAs in cancer prevention, diagnosis, and treatment.
Keywords: exosome, long non-coding RNAs, cancer
Introduction
The American Cancer Society has estimated that there were more than 1.7 million new cancer cases and 160 million cancer-related deaths in the United States in 2018.1 At present, surgery, radiotherapy, and chemotherapy are the main strategies for the treatment of cancer. Surgery is usually recommended in the early stages of cancers, and is most effective when all cancer cells can be removed. It is also used in advanced stages, but it is mainly used to remove cancers and improve the quality of life. Therefore, chemotherapy and radiotherapy are the most widely used interventions in the treatment of cancer. Compared with surgery, chemotherapy and radiotherapy mostly kill only a small number of cancer cells in each treatment cycle, and usually never completely cures the cancer.1,2 Therefore, there is an urgent need to find at least one effective biological target that can improve the quality of life of cancer patients.
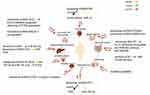
Exosomes are a kind of vesicle with a bilayer membrane structure, and can be secreted by many kinds of cells. It mainly transports nucleic acids (DNA, mRNAs, lncRNAs and miRNAs), it contains a variety of the biological characteristics of their source cells, and it participates in many important physiological and pathological processes.3 Exosomes were found in a variety of body fluids, such as plasma, serum, urea, saliva and tears.4 Exosomes act on receptor cells in three steps: (1) the transmembrane proteins of the exosomal bodies directly interact with the signal receptors of the target cells, (2) the plasma membranes of the exosomes fuse with the receptor cells and transfer their contents to the cytoplasm, and finally (3) the exosomes are directly endocytosed by the receptor cells or ingested by the receptor cells through phagocytosis.5,6 In recent years, exosomes gained increasing attention as an important biomarker of many diseases and a medium of intercellular communication in the progression, invasion, metastasis and recurrence of many cancers.4 Several features of exosomes have emerged, especially in cancer biology. First, exosomes contain a various biological information and functions. Exosome is a key medium for cell-to-cell communication. Substances secreted by exosomes, including proteins and mRNA, lncRNA, can regulate cell function.7 Second, exosomes have a strong influence on the progression and metastasis of cancers. They can predict the metastatic site and establish a niche before metastasis, depending on their interaction with the stromal cells.8 Third, exosomes can induce effective pro-cancer and anti-cancer immune responses, but exosomes derived from advanced cancer cells showed a stronger immunosuppressive effect than immune activation.9
The progress of whole genome and whole exome sequencing has promoted the study of the DNA regions of non-coding proteins. Only 3% of the genome encodes proteins, and the non-coding regions are considered “junk DNA”. However, recent exome sequencing found that 66% of RNA transcripts came from non-coding regions. These non-coding RNAs include lncRNAs, miRNAs, circular RNAs, etc.10 lncRNAs refer to long non-protein-coded transcripts with lengths of more than 200 bp.11 The common modes of lncRNAs in regulating cell physiology are as follows: lncRNAs regulate target genes by affecting epigenetics (such as promoter methylation), and lncRNAs compete with miRNAs for their target RNAs, so that miRNAs cannot affect the function of target mRNAs.12 lncRNAs can be used as 1) chromatin modified scaffolds, 2) bait-induced transcriptional inhibitors, 3) RNA processing guides, or 4) signals of gene activation.13 Recent evidence suggested that lncRNAs play a key role in many biological processes, such as inflammation, cell growth, cell differentiation, and tumorigenesis.14,15 lncRNAs have a regulatory function in human diseases, especially cancer. lncRNA dysfunction can lead to changes in cell behavior, including proliferation, invasion, and migration, as well as inhibition of apoptosis.16,17 lncRNA not only was detected in cancer tissues, but also was in various body fluids, including, urine, blood and saliva.13
Exosomes have been proven to be a bridge for the exchange of important information between cells, carrying nucleic acids, proteins, and lipids to recipient cells.18 Interestingly, exosomes also carry a wide range of lncRNAs. lncRNAs regulate gene expression through translation inhibition or as a competitive endogenous RNA (ceRNA).14,16 Exosomal lncRNAs play a key role in cancer progression and drug resistance. For example, the lncRNAs Exo1-4 and RMRP were preferentially packaged and transmitted by exosomes, thus improving the viability of receptor cells.19
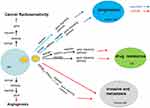
Exosomal lncRNAs are functional and can transmit different phenotypic patterns to neighboring cells, however, the exact role of exosomal lncRNAs have not been fully revealed. This article reviews the role of exosomal lncRNAs in cancers. We reviewed the molecular mechanism of exosomal lncRNAs in regulating cancer progression, angiogenesis, and chemotherapy resistance, and the potential applications of exosomal lncRNAs from three aspects, including cancer diagnosis, treatment and prognosis. The molecular mechanism of exosomal lncRNAs in cancers is shown in Figure 1.
The Molecular Mechanism of Exosomal lncRNAs in Cancers
Exosomal lncRNAs Regulate Cancer Cells Proliferation
Exosomal lncRNAs not only promote cancers, but they can also inhibit cancer growth. As shown in Figure 2, a network diagram has been constructed to illustrate exosomal lncRNAs regulate cancer cells proliferation. The exosomal lncRNA SENP3-EIF4A1 inhibited the progression of hepatocellular cancer (HCC) and the exosomal lncRNA SENP3-EIF4A1 sponging miR-9-5p protected ZFP36, inhibited the biological malignant behavior of HCC.20 The lncRNA TCONS_00027227 activated by APC (TCNS_00027227), that is, lncRNA APC1, adenomatous polyposis coli (APC)-activated lncRNA APC1, in colorectal adenomatous polyposis inhibited the production of exosomes and reduced its stability through direct binding to Rab5b mRNA, inhibiting the growth, metastasis, and angiogenesis of colorectal cancer (CRC) cells.21 The exosome secreted by SKOV3 cells inhibited the PTEN/AKT signal pathway by transduction of the lncRNA FAL1, inhibiting the metastasis of ovarian cancer cells in vivo and in vitro.22 The exosome secreted by Huc-MSC cells inhibited the growth of glioma cells through lncRNA PTENP1/miR-10a-5p/PTEN pathway.23 Exosomal lncRNA PTENP1 could reduce the malignant phenotype of bladder cancer cells in vivo and in vitro. In addition, exosomal lncRNA PTENP1 might be used as a bait for miR-17 to regulate PTEN and inhibit the progression of bladder cancer.24
Exosomal lncRNA SENP3-EIF4A1, exosomal lncRNA TCONS_00027227, and exosomal lncRNA FAL1 inhibited cancer cells proliferation of HCC, CRC and ovarian cancer, respectively. Exosomal lncRNA PTENP1 also inhibited cancer cells proliferation in glioma and bladder cancer.
Exosomal lncRNAs Regulate Cancer Invasion and Metastasis
The basic process of cancer metastasis includes: primary cancer growth, cancer cells falling off and invading the matrix, angiogenesis, cancer cells leaking into the vascular system, thrombus formation, localized growth in secondary tissues and organs, and finally, metastatic cancer continues to spread.25 As shown in Figure 3, a network diagram has been constructed to illustrate exosomal lncRNAs regulate cancer invasion and metastasis. The lncRNA PCGEM1 was highly expressed in gastric cancer (GC) cells, and a part of PCGEM1 could be encapsulated in exosomes. The exosomal lncRNA PCGEM1 promoted invasion and metastasis of GC by stabilizing SNAIL.26 Exosomal lncRNA CEBPA-AS1 secreted by GC cells could promote cell proliferation, inhibiting apoptosis and induces GC progression.27 Exosomal lncRNA metastasis-associated lung adenocancer transcript 1 (MALAT-1) sponging miR-26a/26b promoted the invasion and metastasis of CRC by regulating FUT4 and activating the PI3K/Akt/mTOR pathway.28 Exosomal lncRNA UCA1 in the serum regulated the expression of MYO6 by acting on sponge miR-143, as a ceRNA, affecting the proliferation and apoptosis of CRC cells.29 A newly-discovered lncRNA, RP11-85G21.1 (Lnc85), promoted the proliferation and migration of HCC cells through targeted binding and regulation with miR-324-5p.30 Exosomal lncRNA FAL1 promoted cell proliferation and metastasis through competitive binding with miR-1236. In addition, the expression of lncRNA FAL1 in the sera of patients with HCC was also upregulated, and lncRNA FAL1 could be transferred to hepatoma cells, which enhanced their proliferation and migration ability.31 Exosomes derived from non-small cell lung cancer (NSCLC) cells could promote the proliferation, migration, and invasion of NSCLC cells through UFC1 transfer. In addition, exosomal lncRNA UFC1 inhibited PTEN expression and activated the Akt pathway through EZH2-mediated epigenetic silencing, promoting the progression of NSCLC.32 In NSCLC, the expression of lncRNA MALAT-1 in the serum was upregulated after being protected by exosomes, promoting cell proliferation and migration.33 Bioactive lncRNA PCa-specific expression and EZH2-associated transcript (PCSEAT) was bound to the exosome and passed on to neighboring cells, promoting cell proliferation and movement in prostate cancer. It also promoted cell proliferation, at least in part, through the regulation of EZH2 mediated by a competitive “sponge” miR-143-3p and miR-24-2-5p.34 During hypoxia, exosomes promoted the growth and development of bladder cancer by transferring lncRNA UCA1.35 Overexpression of the lncRNA TIRY in cancer-associated fibroblasts activated the wnt/β-catenin signaling pathway and promoted the invasion and metastasis of oral squamous cell cancer cells through the sponge action of miR-14 secreted by cancer-related exosomes.36 Melanoma-derived exosomes reprogrammed fibroblasts into cancer-associated fibroblasts that promoted cancer by transferring lncRNA Gm26809, thereby promoting melanoma cell proliferation and migration.37 lncRNA PVT1 wrapped in exosomes derived from bone marrow mesenchymal stem cells promoted the growth and metastasis of osteosarcoma by stabilizing ERG and sponging miR-183-5p.38
To sum up, exosomal lncRNA PCGEM1 and exosomal lncRNA CEBPA-AS1 promoted invasion and metastasis in GC. Exosomal lncRNA MALAT-1 and exosomal lncRNA UCA1 promoted invasion and metastasis in CRC. Exosomal lncRNA Lnc85 and exosomal lncRNA FAL1 promoted invasion and metastasis in HCC. Exosomal lncRNA UFC1 and exosomal lncRNA MALAT promoted invasion and metastasis in NSCLC. Exosomal lncRNA PCSEAT, exosomal lncRNA UCA1, exosomal lncRNA TIRY, exosomal lncRNA Gm26809, and exosomal lncRNA PVT1 promoted invasion and metastasis in prostate cancer, bladder cancer, oral squamous cell cancer, melanoma and osteosarcoma, respectively.
Exosomal lncRNAs Regulate Cancer Angiogenesis
Cancer angiogenesis includes several steps: enzymatic hydrolysis of the vascular basement membrane, cell proliferation, migration, germination, branching, and tubular formation of endothelial cells. In the cancer microenvironment, exosomes released by different cell types have been proven to be important mediators in the process of cancer angiogenesis, such as mesenchymal stem cells, stromal cells, and endothelial cells. lncRNAs secreted by cancer-derived exosomes could stimulate the pro-angiogenic potential of circulating angiogenic cells by increasing the expression of membrane molecules and soluble factors.19 The exosomal lncRNA RAMP2-AS1 derived from chondrosarcoma cells promoted angiogenesis through the miR-2355-5p/VEGFR2 axis.39 Epithelial ovarian cancer cells transferred the lncRNA MALAT1 to recipient human umbilical vein endothelial cells (HUVECs) through exosomal bodies, and then stimulated the expression of angiogenesis-related genes to affect HUVECs, eventually promoting angiogenesis.40 The exosomal lncRNA H19 released by cancer stem cell-like (CSC) could promote the angiogenic phenotype of endothelial cells.41
Exosomal lncRNAs Regulate Chemotherapy Drug Resistance
Chemotherapy is one of the most commonly used and most important modes of treatment for malignant cancers. Cancer cells often develop resistance to chemotherapeutic drugs. The drug resistance of cancer cells can be divided into two types, including primary drug resistance and acquired drug resistance. Primary drug resistance is congenital. Acquired drug resistance is caused by the mutation of cancer cells after drug treatment. As shown in Figure 4, a network diagram has been constructed to illustrate exosomal lncRNAs regulate chemotherapy drug resistance. In breast cancer, exosomal lncRNA SNHG14 inhibited the expression of apoptotic proteins and apoptosis by targeting the Bcl-2/Bax signaling pathway, promoting drug resistance of trastuzumab.42 In breast cancer, exosomal lncRNA AFAP1-AS1 promoted trastuzumab resistance by binding to AUF1 and activating ERBB2 translation.43 Exosomal lncRNA H19 induced adriamycin resistance in metastatic breast cancer.44 lncRNA UCA1 promoted gefitinib resistance as a ceRNA to target FOSL2 by sponging miR-143 in NSCLC.45 Exosomal lncRNA RP11-838N2.4 transfection promoted the resistance of NSCLC to erlotinib.46 Exosomal lncRNA H19 promoted the drug resistance of NSCLC to erlotinib through the miR-615-3p/ATG7 axis, which provided a potential target for the diagnosis and treatment of NSCLC.47 lncRNA ARSR could be secreted from drug-resistant cells through exosomes. lncRNA ARSR promoted drug resistance to sunitinib by upregulating axl/c-met, and activating the STAT3, AKT and ERK signal pathways through competitive combination of miR-34 and miR-449. In turn, activated AKT promoted the expression of lncRNA ARSR by inhibiting transcription factors FOXO1 and FOXO3a, forming a positive feedback loop in sunitinib-resistant renal cancer cells.48 Exosomal lncRNA PART1 induced gefitinib resistance in esophageal squamous cell cancer through competition for endogenous RNA. Downregulation of the lncRNA PART1 could effectively promote gefitinib-induced cell death, while upregulation of PART1 promoted gefitinib resistance by promoting Bcl-2 expression through competitive binding with miR-129. In addition, the lncRNA PART1 could be integrated into the exosomal body and transmitted to sensitive cells, thus spreading resistance to gefitinib.49
To sum up, in breast cancer, exosomal lncRNA SNHG14 and exosomal lncRNA AFAP1-AS1 promoted drug resistance of trastuzumab. Exosomal lncRNA H19 induced adriamycin resistance in breast cancer. Exosomal lncRNA UCA1 and exosomal lncRNA RP11-838N2.4 promoted resistance of gefitinib and erlotinib in NSCLC, respectively. Exosomal lncRNA H19 also promoted the erlotinib resistance in NSCLC. Exosomal lncRNA ARSR promoted drug resistance to sunitinib in renal cancer. Exosomal lncRNA PART1 induced gefitinib resistance in esophageal squamous cell cancer.
Exosomal lncRNAs Regulate Radiosensitivity
Radiotherapy is considered to be an important choice for cancer treatment, not only because it can cause irreversible DNA damage to cancer cells, but also because it can promote anti-cancer immune response.50 The exosomal lncRNA HOTAIR reduced the radiosensitivity of laryngeal cancer by regulating the microRNA-454-3p/E2F2 axis.51
lncRNAs Regulate Exosome Secretion in Cancers
In cancers, lncRNAs can regulate related proteins to promote cancer cells to secrete exosomes. In pancreatic cancer, the lncRNA PVT1 promoted exosome secretion through YKT6v-SNARE homologue (YKT6), Ras-associated protein RAB-7 (RAB7), and vesicle-associated membrane protein 3 (VAMP3).52 lncRNA HOTAIR promoted exosome secretion by regulating RAB35 and SNAP23 in HCC.53 Highly upregulated in liver cancer (HULC) and the lncRNA HULC/miR-372-3p/Rab11a axis promoted the secretion of exosomes in HCC cells.54
Clinical Studies of lncRNAs Mediated by Exosomes in Cancers
Circulating lncRNAs are widely distributed in the peripheral blood and other biological body fluids, such as urine and saliva. Exosomal lncRNAs are related to the occurrence and development of cancers. It can be as promising biomarkers for cancers diagnosis and prognosis. A number of studies have confirmed the diagnostic, therapeutic, and prognostic value of circulating exosomal lncRNAs in a variety of cancers, including HCC, NSCLC, GC, bladder cancer, CRC, and breast cancer. As shown in Table 1, a table has been constructed to illustrate exosomal lncRNAs in body fluids of human cancers. Therefore, an in-depth understanding of the basic biology involved in circulating exosomal lncRNAs is essential for the development of new biomarkers, especially for clinical applications.
 | 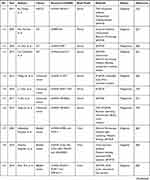 | 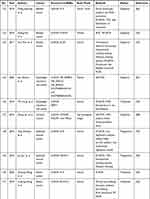 |
Table 1 Exosomal lncRNAs in Body Fluids of Human Cancers |
Diagnostic Markers
Exosomal lncRNA ENSG00000248932.1, ENST00000440688.1 and ENST00000457302.2 were significantly increased in patients with HCC.55
These lncRNAs were stable enough in human plasma. Circulating exosomal lncRNA ENSG00000248932.1, ENST00000440688.1 and ENST00000457302.2 could be used as potential biomarkers to predict tumorigenesis in patients with HCC.55 A new DE-lncRNA, RP11-85G21.1 (Lnc85), promoted the proliferation and migration of HCC cells through targeted binding with and regulation of miR-324-5p. The level of Lnc85 in serum was higher in both alpha-fetoprotein-positive and alpha-fetoprotein-negative patients with HCC, suggesting that lnc85 might be a potential biomarker of HCC.30 Multivariate analysis showed that high miRNA-21 and high lncRNA ATB expression were independent predictors of mortality and disease progression. The overall survival time and progression-free survival time were significantly decreased in patients with high levels of circulating exosomal miRNA21 and lncRNA ATB, indicating that circulating exosomal lncRNA ATB could be used as a new prognostic biomarker and therapeutic target for HCC.56 LINC00161 was significantly expressed in the serum and exosomes of patients with HCC. The upregulated expression of LINC00161 in the serum of patients with HCC comes from the exosome into the serum and could at least be partially metabolized through the urine. Exosomal LINC00161 in serum could be used as a potential non-invasive biomarker to predict the diagnosis of HCC.57 The expression of lncRNA HEIH in the serum and exosomal body increased in patients with HCV-related HCC, while the ratio of lncRNA HEIH expression in serum to exosome decreased. lncRNA HEIH in serum and exosomes could be used as a predictive molecule in hepatitis C virus (HCV)-associated HCC.58 XIST activator (Jpx) could be transported from HCC cells to extracellular bodies and activate the expression of lncRNA X–inactive-specific transcript (Xist) in blood cells by inhibiting the trans regulation of CCCTC-binding factor (CTCF).59
The expression of the lncRNA UFC1 was upregulated in cancer tissue, serum and exosome of NSCLC patients, and high levels of lncRNA UFC1 were related to cancer invasion. Exosomal lncRNA UFC1 promoted the progression of NSCLC through EZH2-mediated epigenetic silencing of PTEN expression, which provided a potential marker for the diagnosis of NSCLC.32 The blood plasma levels of exosomal lncRNA SOX2-OT in post-operative patients with squamous cell lung cancer was significantly decreased, and exosomal lncRNA SOX2-OT was expected to become a non-invasive plasma marker of squamous cell lung cancer.60 Exosomal lncRNA MALAT-1 in the serum promoted the proliferation and migration of NSCLC cells, which laid a foundation for the use of exosomal lncRNA MALAT-1 as a non-invasive serum-based biomarker for the diagnosis and prognosis of NSCLC.33
The level of lncRNA CEBPA-AS1 in the tissue and plasma of patients with GC was also significantly increased. Most of the lncRNA CEBPA-AS1 in plasma was stable. The diagnostic accuracy of lncRNA CEBPAAS1 was higher than that of other traditional cancer markers. Exosomal lncRNA CEBPA-AS1 was a promising new biomarker for the clinical diagnosis of GC.27 Compared with the healthy control group, the expression level of exosomal lncRNA PCSK2-2:1 in the serum of patients with GC was significantly downregulated. In addition, the expression level of lncRNA PCSK2-2:1 was correlated with cancer size, cancer stage and venous invasion. Exosomal lncRNA pcsk2-2:1 in serum could be used as a potential marker for the diagnosis of GC.61
The expression level of colon cancer-associated transcript 2 (CCAT2) in cancer tissues was significantly higher than that in paracancerous tissues. The high expression of CCAT2 was associated with advanced CRC. The expression level of CCAT2 in the serum of patients with CRC was significantly higher than that of normal controls. The level of CCAT2 after operation was significantly lower than that before operation. Circulating lncRNA CCAT2 might be protected by exosomes and could be used as a potential predictor of CRC.62
The expression of lncRNA ANRIL and PCAT-1 in patients with bladder cancer was significantly higher than that in normal subjects. The transcriptional levels of lncRNA ANRIL and PCAT-1 in urinary exosomes were potential biomarkers for the diagnosis of bladder cancer.63 lncRNAs UCA1-201, UCA1-203, MALAT1, and LINC00355 had 92% sensitivity and 91.7% specificity in the diagnosis of normal bladder cancer.64 Exosomal lncRNAs UCA1-201, UCA1-203, MALAT1 and LINC00355 in urine could be used as potential biomarkers for the diagnosis of bladder cancer.64 The high expression of lncRNA PCAT-1 and MALAT-1 was related to poor recurrence-free survival (RFS) of non-muscle invasive bladder cancer. The overexpression of exosomal lncRNA PCAT-1 was an independent prognostic factor for RFS of non-muscle invasive bladder cancer.65 Urinary exosomal lncRNAs could therefore be used as a new non-invasive biomarker for the diagnosis and recurrence prediction of bladder cancer.65
Exosomal lncRNA H19 in serum showed a higher AUC value and diagnostic sensitivity, while urine cytology showed higher diagnostic specificity. The detection of exosomal lncRNA H19 in the serum was helpful for future use of exosomal lncRNAs as a non-invasive biomarker in the diagnosis and prognosis in patients with bladder cancer.66 Exosomal lncRNA PTENP1 inhibited the progression of bladder cancer and played an intermediary role in the intercellular communication during the occurrence of bladder cancer. Exosomal lncRNA PTENP1 was a promising new biomarker for bladder cancer diagnosis.24 Exosomal lncRNA UCA1 secreted by hypoxic bladder cancer cells promoted the growth and development of bladder cancer. Exosomal lncRNA UCA1 in human serum could be used as a biomarker for the diagnosis of bladder cancer.35
The expression levels of exosomal lncRNA NR_039819, NR_036133, NR_003353, ENST00000442416.1, and ENST00000416100.1 in the plasma of esophageal cancer patients were significantly higher than those of non-cancer controls, which might be an efficient and non-invasive biomarker for the diagnosis of esophageal cancer.67
Exosomal lncRNA HOTAIR, MALAT1, and MEG3 of cervical cancer were found in cervicovaginal lavage fluids. The expression level of lncRNAs in patients with cervical cancer was significantly different from that in normal controls. Exosomal lncRNAs had great potential in the early detection and diagnosis of cervical cancer; it was a convenient and non-invasive biomarker.68
Treatment Markers
The expression level of exosomal lncRNA H19 in the serum was high in NSCLC patients who could not benefit after erlotinib treatment.47 The exosomal lncRNA H19 in the serum was stable and could be used as a potential biomarker of erlotinib in the treatment of NSCLC.47 The expression level of lncRNA RP11-838N2.4 in the serum of patients resistant to erlotinib was upregulated, and exosomal lncRNA RP11-838N2.4 might be a therapeutic target for patients with NSCLC.46
In patients with esophageal squamous cell cancer, high levels of lncRNA Prostate Androgen-Regulated Transcript 1 (PART1) in serum exosomes were associated with poor response to gefitinib treatment. lncRNA PART1 could promote gefitinib resistance by regulating the miR-129/Bcl-2 pathway, which could be used as a therapeutic target for patients with esophageal squamous cell cancer.49
The level of exosomal lncRNA H19 in patients resistant to doxorubicin was higher than that in patients who were sensitive to doxorubicin, therefore exosomal lncRNA H19 might be used as a non-invasive biomarker of doxorubicin resistance in breast cancer.44 High expression of exosomal lncRNA SNHG14 was correlated with distant metastasis and lymph node metastasis of breast cancer.42 It was suggested that exosomal lncRNA SNHG14 in serum could be used as a potential diagnostic marker of breast cancer.42
Prognostic Markers
The overall survival time and progression-free survival time were significantly decreased in patients with high levels of circulating exosomal miRNA21 and lncRNA ATB0.09, indicating that circulating exosomal lncRNA ATB could be used as a new prognostic biomarker and therapeutic target for HCC.56 Compared with that before treatment, the level of serum exosomal lncRNA myocardial infarction associated transcript (MIAT) decreased significantly after treatment, but increased significantly in recurrent cases.69 Exosomal lncRNA MIAT in serum might be a new biomarker for monitoring the progression of GC.69
The high expression of lncRNA SPINT1-AS1 was associated with poor clinical prognosis. Serum exosomal lncRNA SPINT1-AS1 in patients with CRC could be used as a biomarker to predict the prognosis and monitor the curative effect of CRC treatment.70 High exosomal lncRNA CRNDE-p and low miR-217 levels in the serum were associated with advanced clinical stage (III/IV), cancer classification (T3/T4), lymph node metastasis, or distant metastasis.71 A combined evaluation of exosomal lncRNA CRNDE-p and miR-217 levels in serum had diagnostic and prognostic potential in patients with CRC.71 Furthermore, the level of exosomal lncRNA CRNDE-h in the serum was significantly increased in patients with CRC. Exosomal lncRNA CRNDE-h in serum could be used as a non-invasive cancer marker for diagnosis and prognosis of CRC.72
The expression of serum exosomal lncRNA antisense hypoxia inducible factor (aHIF) was higher in patients with epithelial ovarian cancer, and it was positively correlated with the expression level of lncRNA aHIF in epithelial ovarian cancer tissues.73 The overall survival rate of patients with ovarian cancer with a high expression of exosomal lncRNA aHIF in serum was lower.73 Exosomal lncRNA aHIF in serum may therefore be an important non-invasive predictive index for the prognosis of epithelial ovarian cancer.73
The increase of exosomal lncRNA MALAT1 in serum was highly correlated with advanced and metastatic phenotypes of ovarian cancer, and was an independent predictor of the overall survival (OS) from ovarian cancer. Circulating exosomal lncRNA MALAT1 could therefore be used as a promising serum-based and non-invasive biomarker for predicting the prognosis of ovarian cancer.40
Finding the molecular target of prognosis was to evaluate the therapeutic effect of cancer. Exosomal lncRNA ATB was a new prognostic markers of HCC. Exosomal lncRNA MIAT was a new prognostic markers of GC. Exosomal lncRNA SPINT1-AS1 and exosomal lncRNA CRNDE-p were a new prognostic markers of CRC. Exosomal lncRNA aHIF and exosomal lncRNA MALAT1 were a new prognostic markers of ovarian cancer. These markers provided molecular targets for cancers prognosis.
Discussion and Future Perspectives
Cancer is undoubtedly a serious public health problem worldwide. According to the latest data, the incidence of cancer will continue to increase over a long period of time, especially among younger people, with new cases increasing every year. Therefore, the occurrence and treatment of cancer is a complicated problem. Over time, new therapeutic targets should be discovered, with current and emerging techniques in biotechnology. Exosomes are very small EVs secreted by cells to local and distant tissues. Cancer cells abnormally secrete a large number of exosomes, which contain different cargo molecules from normal cells, depending on the phenotype of malignant cancers, that is, cancer cells progress by changing the quality and quantity of exosomes. Exosomes can encapsulate lncRNAs, and these functional exosomal lncRNAs can transmit different phenotypic patterns to neighboring cells. This article summarized the molecular mechanisms of exosomal lncRNAs in regulating cancer progression, angiogenesis, and chemotherapy resistance, as well as the prospective clinical applications of exosomal lncRNAs in cancer diagnosis, treatment, and prognosis.
Exosomal lncRNAs Play a Role Through a ceRNA Mechanism
lncRNAs regulate gene expression through several different mechanisms, including acting as a co-regulator, regulating the activity of basic transcription factors, epigenetic modification, and post-transcriptional regulation. More studies have shown that lncRNAs participate in the occurrence, development, and drug resistance of cancers through ceRNAs.16,17 Competitive endogenous RNA (ceRNA) is an important regulatory mechanism mediated by lncRNAs.74 Based on the ceRNA hypothesis, exosomal lncRNA compete with miRNA each other to regulate downstream target genes. The target genes play a regulatory role in the occurrence and development of multiple cancers. For example, exosomal lncRNA UCA1 in serum regulated the expression of MYO6 by acting on sponge miR-143, as a ceRNA, affecting the proliferation and apoptosis of CRC cells.29 Exosomal lncRNA SENP3-EIF4A1 sponging miR-9-5p protected ZFP36 from inhibiting the biological malignant behavior of HCC.20 Exosomal lncRNA UCA1 of NSCLC promoted drug resistance to gefitinib by targeting FOSL2 through sponging miR-143.45
Exosomal lncRNAs can participate in tumorigenesis and development, chemotherapy resistance, angiogenesis, and radiosensitivity through a variety of pathways, especially through the ceRNA mechanism. On the basis of existing work, the network map of exosomal lncRNAs action mechanisms was constructed to further explore the role of exosomal lncRNAs in cancers.
Exosomal lncRNAs Can Induce Drug Resistance to Chemotherapy
The key requirements of an ideal drug delivery system (DDS) are safety, non-toxicity, high efficiency, non-immunogenicity, bioavailability, and targeting.75 Although many DDS such as liposomes, micelles, nanoparticles, and hydrogels have been developed for effective drug delivery,75 most of them face two key problems: high systemic toxicity and low bioavailability. Recently, nanoscale EVs, such as exosomes and microvesicles, which have organic tendency, good bioavailability, and low toxicity and immunogenicity, have been added to the field of DDS.76,77 Compared with traditional targeted carriers, exosomes have better safety and bioavailability, and show lower systemic toxicity and immunogenicity. In addition, compared with chemotherapies, current studies have shown that exosomal lncRNAs can induce drug resistance, which provides a new direction for the use of exosomal lncRNAs in the treatment of chemotherapeutic drug resistance.
Although many studies have focused on the role and mechanism of exosomal lncRNAs in cancer drug resistance, a large number of them have not been revealed, and are therefore worthy of further study. Future cancer chemotherapy drugs may be a combination of existing anti-cancer drugs and drugs targeting exosomal lncRNAs associated with drug resistance. Some studies can be conducted to determine the basic mechanisms of drug resistance associated with exosomal lncRNAs, which may provide some common targets for cancer treatment. The pathway through which exosomal lncRNAs enter the cells is also unclear. Exosomes can transfer bioactive molecules between cancer cells and their microenvironment, but the mechanism by which exosomes accurately transport lncRNAs to specific target cells needs to be further elucidated.
Early Detection of Cancers is a Long-Term Hot Issue in Cancer Research
Using exosomal lncRNAs as a biomarker has three advantages: 1) lncRNAs in vitro and in vivo are not affected by RNase, so it can be inferred that the integrity and function of lncRNAs will not change; 2) compared with other types of EVs, the number of exosomal lncRNAs are higher; and 3) the expression of many lncRNAs are tissue-specific.78 Therefore, the analysis of exosomal lncRNAs is helpful to develop valuable diagnostic markers. Recent research has turned to the development of minimally invasive biomarkers in the form of liquid biopsies, which sample and analyze various types of cells and molecules collected from biological fluids. These fluids can be blood, plasma, cerebrospinal fluid, bronchoalveolar lavage fluid, pleural effusion, saliva, or urine. In the current clinical environment, routine biopsies may not reflect cancer heterogeneity and cancer dynamics. In contrast, liquid biopsies detect circulating cancer cells, circulating DNAs, circulating RNAs, and exosomes, providing a minimally invasive alternative therapy because it can provide a wealth of information for cancers. Although the exact potential mechanism of nucleic acid release into the extracellular environment is not yet fully understood, recent observations have shown that microbubbles, especially exosomes, can act as shuttle particles to protect their functional cargo from the effects of endogenous ribonucleases. The exosomes transfer a large amount of its contents (including mRNAs, miRNAs, lncRNAs, and proteins) to the recipient cells, which may lead to changes in phenotypic characteristics. In particular, exosomal lncRNA markers have been studied and may become representative biomarkers to distinguish different cancers and contribute to the diagnosis and treatment of cancer. Exosomal lncRNAs are promising biomarkers for cancer diagnosis, prediction, and prognosis as well as providing real-time monitoring of treatment response. However, these studies usually have small samples and poor repeatability, so it is necessary to conduct large multicenter studies to improve the effectiveness of liquid biopsies.
Limitations
Exosomal lncRNAs have a good prospect, but they also have some limitations. At present, a large number of exosomal lncRNAs have not been discovered, and their mechanism has not been fully revealed. Exosomal lncRNAs with more dysfunctional expression have been confirmed at the cellular level in animal experiments and a large number of clinical data, and so these lncRNAs may be transformed into a clinical diagnostic method, but the specificity and sensitivity of exosomal lncRNAs as cancer markers still need to be further improved. In view of the fact that there are many lncRNA targets, different lncRNAs can also act on the same target, so it will be difficult to construct a complete network of lncRNA regulation mechanisms. Finally, although there are many methods of exosomal extraction at present, such as gradient centrifugation, filtration centrifugation, immunomagnetic bead method, and chromatography, there is still no unified protocol for extracting exosomes. At present, there is no method to ensure the content, purity, and biological activity of the exosomes simultaneously.
Conclusion
The successful treatment of cancer depends on understanding the complex mechanisms involved in the cancer microenvironment. Exosomes are a new means of information exchange between cells, which play an important role in tumorigenesis, development, and drug resistance. Cancer-derived exosomes contain specific proteins and RNA profiles. As nanoscale biological vesicles, the contents of exosomes are well protected and will not be degraded. Targeted lncRNA delivery can promote the communication and exchange of genetic material and have a significant impact on cell behavior. We focused on the molecular mechanism of exosomal lncRNAs in regulating cancer progression, angiogenesis, chemotherapy resistance, and metastasis, to help understand and manipulate the mechanisms of exosomal lncRNAs in intercellular information exchange. In addition, it is necessary to further explore how the cell uptake of exosomal lncRNAs leads to proliferation, invasion, metastasis, drug resistance, and other behavioral changes, and whether exosomes can be used as early liquid biopsy specimens for cancer diagnosis.
Abbreviations
AUC, area under curve; AUF1, AU-binding factor 1; AFAP1-AS1, actin filament associated protein 1 antisense RNA 1; ATGs, Autophagy-related proteins; APC, adenomatous polyposis coli; aHIF, antisense hypoxia inducible factor; CCAT2, colon cancer-associated transcript 2; CRC, colorectal cancer; ceRNA, competitive endogenous RNA; CTCF, CCCTC-binding factor; CSC, cell-like cells; RFS, recurrence-free survival (); DDS, drug delivery system; EZH2, enhancer of zeste homolog 2; EV, extracellular vesicles; FAL1, focally amplified lncRNA on chromosome 1; GC, gastric cancer; HOTAIR, HOX transcript antisense intergenic RNA; lncRNA, long non-coding RNA; Lnc85, ncRNA, RP11-85G21.1; MALAT-1, metastasis-associated lung adenocancer transcript 1; HUVECs, human umbilical vein endothelial cell; HCC, hepatocellular cancer; HULC, highly upregulated in liver cancer; Jpx, XIST activator; MIAT, myocardial infarction associated transcript; NEAT2, nuclear-enriched abundant transcript 2; NSCLC, non-small cell lung cancer; PCSEAT, PCa specifific expression and EZH2-associated transcript; PART1, Prostate Androgen-Regulated Transcript 1; qRT-PCR, quantitative real-time reverse transcription-polymerase chain reaction; PI3K, phosphatidylinositol 3-kinase; PTEN, phosphatase and tensin homologue; RAB7, ras-associated protein RAB-7; ROC, receiver operating characteristic curve; SPINT1-AS1, kunitz type 1 antisense RNA1; TAMs, tumor-associated macrophages; UCA1, urothelial cancer-associated; VAMP3, vesicle-associated membrane protein 3; Xist, X-inactive-specific transcript; YKT6, YKT6v-snare homologue.
Consent to Participate
We did not recruit any human subjects for this study; therefore, consent is not applicable.
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Funding
This work was supported by the Public Welfare Technology Application Research Program of Huzhou (No. 2017GYB18), and the Zhejiang Medical and Health Technology Projects (No.2020KY311, No.LGC20H160002, No.2019KY595).
Disclosure
The authors declare that they have no conflicts of interest for this work.
References
1. Morales-Cruz M, Delgado Y, Castillo B, et al. Smart targeting to improve cancer therapeutics. Drug Des Devel Ther. 2019;13:3753–3772. doi:10.2147/DDDT.S219489
2. Akhter MH, Rizwanullah M, Ahmad J, et al. Nanocarriers in advanced drug targeting: setting novel paradigm in cancer therapeutics. Artif Cells, Nanomed Biotechnol. 2018;46(5):873–884. doi:10.1080/21691401.2017.1366333
3. Cai J, Qiao B, Gao N, et al. Oral squamous cell cancer-derived exosomes promote M2 subtype macrophage polarization mediated by exosome-enclosed miR-29a-3p. Am J Physiol Cell Physiol. 2019;316(5):C731–C740. doi:10.1152/ajpcell.00366.2018.
4. Inubushi S, Kawaguchi H, Mizumoto S, et al. Oncogenic miRNAs identified in tear exosomes from metastatic breast cancer patients. Anticancer Res. 2020;40(6):3091–3096. doi:10.21873/anticanres.14290
5. Lim W, Kim H-S. Exosomes as therapeutic vehicles for cancer. Tissue Eng Regen Med. 2019;16(3):213–223. doi:10.1007/s13770-019-00190-2
6. Weston WW, Ganey T, Temple HT The relationship between exosomes and cancer: implications for diagnostics and therapeutics. BioDrugs. 2019;33(2):137–158.
7. Hannafon BN, Gin AL, Xu Y-F, et al. Metastasis-associated protein 1 (MTA1) is transferred by exosomes and contributes to the regulation of hypoxia and estrogen signaling in breast cancer cells. Cell Commun Signal. 2019;17(1):13. doi:10.1186/s12964-019-0325-7
8. Zhang L, Yu D. Exosomes in cancer development, metastasis, and immunity. Biochimica Et Biophysica Acta (BBA) - Reviews on Cancer. 2019;1871(2):455–468. doi:10.1016/j.bbcan.2019.04.004
9. Wang X, Shen H, He Q, et al. Exosomes derived from exhausted CD8+ T cells impaired the anticancer function of normal CD8+ T cells. J Med Genet. 2019;56(1):29–31. doi:10.1136/jmedgenet-2018-105439.
10. Ai Y, Wu S, Zou C, et al. LINC00941 promotes oral squamous cell cancer progression via activating CAPRIN2 and canonical WNT/β-catenin signaling pathway. J Cell Mol Med. 2020;24:10512–10524.
11. Zhang M, Wang N, Song P, et al. lncRNA GATA3-AS1 facilitates tumour progression and immune escape in triple-negative breast cancer through destabilization of GATA3 but stabilization of PD-L1. Cell Prolif. 2020;53:e12855.
12. Lin Z, Ni X, Dai S, et al. Screening and verification of long noncoding RNA promoter methylation sites in hepatocellular cancer. Cancer Cell Int. 20;2020:311. doi:10.1186/s12935-020-01407-4
13. Zeuschner P, Linxweiler J, Junker K, Non-coding RNAs as biomarkers in liquid biopsies with a special emphasis on extracellular vesicles in urological malignancies. Expert Rev Mol Diagn. 2020;20(2):151–167. doi:10.1080/14737159.2019.1665998
14. Chang H, Zhang X, Li B, et al. MAGI2-AS3 suppresses MYC signaling to inhibit cell proliferation and migration in ovarian cancer through targeting miR-525-5p/MXD1 axis. Cancer Med. 2020;9:6377–6386.
15. Liu J, Xu R, Mai S-J, et al. lncRNA CSMD1-1 promotes the progression of Hepatocellular cancer by activating MYC signaling. Theranostics. 2020;10(17):7527–7544. doi:10.7150/thno.45989.
16. Kong S, Xue H, Li Y, et al. The long noncoding RNA OTUD6B-AS1 enhances cell proliferation and the invasion of hepatocellular cancer cells through modulating GSKIP/Wnt/β-catenin signalling via the sequestration of miR-664b-3p. Exp Cell Res. 2020;395:112180.
17. Du C, Wang Y, Zhang Y, et al. lncRNA DLX6-AS1 contributes to epithelial-mesenchymal transition and cisplatin resistance in triple-negative breast cancer via modulating mir-199b-5p/paxillin axis. Cell Transplant. 2020;29:963689720929983. doi:10.1177/0963689720929983
18. Hikita T, Miyata M, Watanabe R, Oneyama C, Sensitive and rapid quantification of exosomes by fusing luciferase to exosome marker proteins. Sci Rep. 2018;8(1):14035. doi:10.1038/s41598-018-32535-7
19. Sun Z, Yang S, Zhou Q, Emerging role of exosome-derived long non-coding RNAs in cancermicroenvironment. Mol Cancer. 2018;17(1):82. doi:10.1186/s12943-018-0831-z
20. Wang J, Pu J, Zhang Y, et al. SENP3-EIF4A1Exosome-transmitted long non-coding RNA suppresses the progression of hepatocellular cancer. Aging. 2020;12(12):11550–11567. doi:10.18632/aging.103302.
21. Wang W, Cao C-H, Han K, et al. APC-activated long noncoding RNA inhibits colorectal cancer pathogenesis through reduction of exosome production. J Clin Invest. 2019;129(2):727–743. doi:10.1172/JCI122478
22. Zhang Q, Len T-Y, Zhang S-X, et al. Exosomes transferring long non-coding RNA FAL1 to regulate ovarian cancer metastasis through the PTEN/AKT signaling pathway. Eur Rev Med Pharmacol Sci. 2020;24(1):43–54. doi:10.26355/eurrev_202001_19894.
23. Hao S-C, Ma H, Niu Z-F, et al. hUC-MSCs secreted exosomes inhibit the glioma cell progression through PTENP1/miR-10a-5p/PTEN pathway. Eur Rev Med Pharmacol Sci. 2019;23(22):10013–10023. doi:10.26355/eurrev_201911_19568
24. Zheng R, Du M, Wang X, et al. Exosome-transmitted long non-coding RNA PTENP1 suppresses bladder cancer progression. Mol Cancer. 2018;17(1):143. doi:10.1186/s12943-018-0880-3.
25. Yilmaz M, Christofori G, Lehembre F, et al. Distinct mechanisms of cancerinvasion and metastasis. Trends Mol Med. 2007;13(12):535–541. doi:10.1016/j.molmed.2007.10.004
26. Piao HY, Guo S, Wang Y, et al. Exosome-transmitted lncRNA PCGEM1 promotes invasive and metastasis in gastric cancer by maintaining the stability of SNAI1. Clin Translat Oncol. 2020.
27. Piao H-Y, Guo S, Wang Y, et al. Exosomal long non-coding RNA CEBPA-AS1 inhibits cancerApoptosis and functions as a non-invasive biomarker for diagnosis of gastric cancer. Onco Targets Ther. 2020;13:1365–1374. doi:10.2147/OTT.S238706
28. Xu J, Xiao Y, Liu B, et al. exosomal MALAT1 sponges miR-26a/26b to promote the invasion and metastasis of CRC via FUT4 enhanced fucosylation and PI3K/Akt pathway. BioDrugs. 2020;39(1):54. doi:10.1186/s13046-020-01562-6.
29. Luan Y, Li X, Luan Y, et al. Circulating lncRNA UCA1 promotes malignancy of CRC via the miR-143/MYO6 axis. Mol Ther Nucleic Acids. 19;2020:790–803. doi:10.1016/j.omtn.2019.12.009
30. Huang X, Sun L, Wen S, et al. RNA sequencing of plasma exosomes revealed novel functional long noncoding RNAs in hepatocellular cancer. Cancer Sci. 2020;111:3338.
31. Li B, Mao R, Liu C, et al. lncRNA FAL1 promotes cell proliferation and migration by acting as a CeRNA of miR-1236 in hepatocellular cancer cells. Life Sci. 197;2018:122–129. doi:10.1016/j.lfs.2018.02.006
32. Zang X, Gu J, Zhang J, et al. Exosome-transmitted lncRNA UFC1 promotes non-small-cell lung cancer progression by EZH2-mediated epigenetic silencing of PTEN expression. Cell Death Dis. 2020;11(4):215. doi:10.1038/s41419-020-2409-0.
33. Zhang R, Xia Y, Wang Z, et al. Serum long non coding RNA MALAT-1 protected by exosomes is upregulated and promotes cell proliferation and migration in non-small cell lung cancer. Biochem Biophys Res Commun. 2017;490(2):406–414. doi:10.1016/j.bbrc.2017.06.055.
34. Yang X, Wang L, Li R, et al. The long non-coding RNA PCSEAT exhibits an oncogenic property in prostate cancer and functions as a competing endogenous RNA that associates with EZH2. Biochem Biophys Res Commun. 2018;502(2):262–268. doi:10.1016/j.bbrc.2018.05.157.
35. Xue M, Chen W, Xiang A, et al. Hypoxic exosomes facilitate bladder cancergrowth and development through transferring long non-coding RNA-UCA1. Mol Cancer. 2017;16(1):143. doi:10.1186/s12943-017-0714-8.
36. Jin N, Jin N, Bu W, et al. Long non-coding RNA TIRY promotes cancermetastasis by enhancing epithelial-to-mesenchymal transition in oral cancer. Exp Biol Med. 2020;245(7):585–596. doi:10.1177/1535370220903673
37. Hu T, Hu J. Melanoma-derived exosomes induce reprogramming fibroblasts into cancer-associated fibroblasts via Gm26809 delivery. Cell Cycle. 2019;18(22):3085–3094. doi:10.1080/15384101.2019.1669380
38. Zhao W, Qin P, Zhang D, et al. Long non-coding RNA PVT1 encapsulated in bone marrow mesenchymal stem cell-derived exosomes promotes osteosarcoma growth and metastasis by stabilizing ERG and sponging miR-183-5p. Aging. 2019;11(21):9581–9596. doi:10.18632/aging.102406.
39. Cheng C, Zhang Z, Cheng F, et al. exosomal lncRNA RAMP2-AS1 derived from chondrosarcoma cells promotes angiogenesis through miR-2355-5p/VEGFR2 axis. Onco Targets Ther. 13;2020:3291–3301. doi:10.2147/OTT.S244652
40. Qiu JJ, Lin X-J, Tang X-Y, et al. exosomal metastasis-associated lung adenocancer transcript 1 promotes angiogenesis and predicts poor prognosis in epithelial ovarian cancer. Int J Biol Sci. 2018;14(14):1960–1973. doi:10.7150/ijbs.28048
41. Conigliaro A, Costa V, Lo Dico A, et al. CD90+ liver cancer cells modulate endothelial cell phenotype through the release of exosomes containing H19 lncRNA. Mol Cancer. 14;2015:155. doi:10.1186/s12943-015-0426-x
42. Dong H, Wang W, Chen R, et al. exosomal transfer of lncRNA‑SNHG14 promotes trastuzumab chemoresistance in breast cancer. Int J Oncol. 2018;53(3):1013–1026. doi:10.3892/ijo.2018.4467.
43. Han M, Gu Y, Lu P, et al. exosomal lncRNA AFAP1-AS1 promotes trastuzumab resistance through binding with AUF1 and activating ERBB2 translation. Mol Cancer. 2020;19(1):26. doi:10.1186/s12943-020-1145-5.
44. Wang X, Pei X, Guo G, et al. exosomal transfer of long noncoding RNA H19 induces doxorubicin resistance in breast cancer. J Cell Physiol. 2020;235(10):6896–6904. doi:10.1002/jcp.29585.
45. Chen X, Wang Z, Tong F, et al. lncRNA UCA1 promotes gefitinib resistance as a ceRNA to target FOSL2 by sponging miR-143 in non-small cell lung cancer. Mol Ther Nucleic Acids. 19;2020:643–653. doi:10.1016/j.omtn.2019.10.047
46. Zhang W, Cai X, Yu J, et al. exosomal transfer of lncRNA RP11‑838N2.4 promotes erlotinib resistance in non-small cell lung cancer. Int J Oncol. 2018;53(2):527–538. doi:10.3892/ijo.2018.4412.
47. Pan R, Zhou H. Exosomal transfer of lncRNA H19 promotes erlotinib resistance in non-small cell lung cancer via miR-615-3p/ATG7 axis. Cancer Manag Res. 2020;12:4283–4297. doi:10.2147/CMAR.S241095
48. Qu L, Ding J, Chen C, et al. Exosome-transmitted lncARSR promotes sunitinib resistance in renal cancer by acting as a competing endogenous RNA. Cancer Cell. 2016;29(5):653–668. doi:10.1016/j.ccell.2016.03.004
49. Kang M, Ren M, Li Y, et al. exosomal transfer of lncRNA PART1 induces gefitinib resistance in esophageal squamous cell cancer via functioning as a competing endogenous RNA. BioDrugs. 2018;37(1):171. doi:10.1186/s13046-018-0845-9.
50. Gupta A, Sharma A, von Boehmer L, et al. Radiotherapy supports protective tumor-specific immunity. Oncoimmunology. 2012;1(9):1610–1611. doi:10.4161/onci.21478.
51. Cui X, Xiao D, Cui Y, et al. Exosomes-derived long non-coding RNA HOTAIR reduces laryngeal cancer radiosensitivity by regulating microRNA-454-3p/E2F2 axis. Onco Targets Ther. 12;2019:10827–10839. doi:10.2147/OTT.S224881
52. Sun C, Wang P, Dong W, et al. lncRNA PVT1 promotes exosome secretion through YKT6, RAB7, and VAMP3 in pancreatic cancer. Aging. 2020;12(11):10427–10440. doi:10.18632/aging.103268.
53. Yang L, Peng X, Li Y, et al. Long non-coding RNA HOTAIR promotes exosome secretion by regulating RAB35 and SNAP23 in hepatocellular cancer. Mol Cancer. 2019;18(1):78. doi:10.1186/s12943-019-0990-6.
54. Cao S-Q, Zheng H, Sun B-C, et al. Long non-coding RNA highly upregulated in liver cancer promotes exosome secretion. World J Gastroenterol. 2019;25(35):5283–5299. doi:10.3748/wjg.v25.i35.5283
55. Lu Y, Duan Y, Xu Q, et al. Circulating exosome-derived bona fide long non-coding RNAs predicting the occurrence and metastasis of hepatocellular cancer. J Cell Mol Med. 2020;24(2):1311–1318. doi:10.1111/jcmm.14783
56. Lee YR, Kim G, Tak WY, et al. Circulating exosomal noncoding RNAs as prognostic biomarkers in human hepatocellular cancer. Int J Cancer. 2019;144(6):1444–1452. doi:10.1002/ijc.31931
57. Sun L, Su Y, Liu X, et al. Serum and exosome long non coding RNAs as potential biomarkers for hepatocellular cancer. J Cancer. 2018;9(15):2631–2639. doi:10.7150/jca.24978
58. Zhang C, Yang X, Qi Q, et al. lncRNA-HEIH in serum and exosomes as a potential biomarker in the HCV-related hepatocellular cancer. Cancer Biomarkers. 2018;21(3):651–659. doi:10.3233/CBM-170727
59. Ma X, Yuan T, Yang C, et al. X-inactive-specific transcript of peripheral blood cells is regulated by exosomal Jpx and acts as a biomarker for female patients with hepatocellular cancer. Ther Adv Med Oncol. 2017;9(11):665–677. doi:10.1177/1758834017731052
60. Teng Y, Kang H, Chu Y, et al. Identification of an exosomal long noncoding RNA SOX2-OT in plasma as a promising biomarker for lung squamous cell cancer. Genet Test Mol Biomarkers. 2019;23(4):235–240. doi:10.1089/gtmb.2018.0103
61. Cai C, Zhang H, Zhu Y, et al. Serum exosomal long noncoding RNA pcsk2-2:1 as a potential novel diagnostic biomarker for gastric cancer. Onco Targets Ther. 12;2019:10035–10041. doi:10.2147/OTT.S229033
62. Wang L, Duan W, Yan S, et al. Circulating long non-coding RNA colon cancer-associated transcript 2 protected by exosome as a potential biomarker for CRC. Biomed Pharmacother. 113;2019:108758. doi:10.1016/j.biopha.2019.108758
63. Abbastabar M, Sarfi M, Golestani A, et al. Tumor-derived urinary exosomal long non-coding RNAs as diagnostic biomarkers for bladder cancer. EXCLI J. 19;2020:301–310. doi:10.17179/excli2019-1683
64. Yazarlou F, Modarressi MH, Mowla SJ, et al. Urinary exosomal expression of long non-coding RNAs as diagnostic marker in bladder cancer. Cancer Manag Res. 10;2018:6357–6365. doi:10.2147/CMAR.S186108
65. Zhan Y, Du L, Wang L, et al. Expression signatures of exosomal long non-coding RNAs in urine serve as novel non-invasive biomarkers for diagnosis and recurrence prediction of bladder cancer. Mol Cancer. 2018;17(1):142. doi:10.1186/s12943-018-0893-y
66. Wang J, Yang K, Yuan W, et al. Determination of serum exosomal H19 as a noninvasive biomarker for bladder cancer diagnosis and prognosis. Med Sci Mon. 24;2018:9307–9316. doi:10.12659/MSM.912018
67. Jiao Z, Yu A, Rong W, et al. Five-lncRNA signature in plasma exosomes serves as diagnostic biomarker for esophageal squamous cell cancer. Aging. 2020;12(14):15002–15010. doi:10.18632/aging.103559
68. Zhang J, Liu S-C, Luo X-H, et al. Exosomal long noncoding RNAs are differentially expressed in the cervicovaginal lavage samples of cervical cancer patients. J Clin Lab Anal. 2016;30(6):1116–1121. doi:10.1002/jcla.21990
69. Xu H, Zhou J, Tang J, et al. Identification of serum exosomal lncRNA MIAT as a novel diagnostic and prognostic biomarker for gastric cancer. J Clin Lab Anal. 2020;34(8):e23323.
70. Li C, Li W, Zhang Y, et al. SPINT1-AS1Increased expression of antisense lncRNA predicts a poor prognosis in CRC and is negatively correlated with its sense transcript. Onco Targets Ther. 11;2018:3969–3978. doi:10.2147/OTT.S163883
71. Yu B, Du Q, Li H, et al. Diagnostic potential of serum exosomal colorectal neoplasia differentially expressed long non-coding RNA (CRNDE-p) and microRNA-217 expression in colorectal cancer. Oncotarget. 2017;8(48):83745–83753. doi:10.18632/oncotarget.19407.
72. Liu T, Zhang X, Gao S, et al. exosomal long noncoding RNA CRNDE-h as a novel serum-based biomarker for diagnosis and prognosis of CRC. Oncotarget. 2016;7(51):85551–85563. doi:10.18632/oncotarget.13465
73. Tang X, Liu S, Liu Y, et al. Circulating serum exosomal aHIF is a novel prognostic predictor for epithelial ovarian cancer. Onco Targets Ther. 12;2019:7699–7711. doi:10.2147/OTT.S220533
74. Gonzalez-Villasana V, Rashed MH, Gonzalez-Cantú Y, et al. Presence of circulating miR-145, miR-155, and miR-382 in exosomes isolated from serum of breast cancer patients and healthy donors. Dis Markers. 2019;2019:6852917.
75. Seo JY, Lee B, Kang TW, et al. Electrostatically interactive injectable hydrogels for drug delivery. Tissue Eng Regen Med. 2018;15(5):513–520. doi:10.1007/s13770-018-0146-6
76. Antimisiaris SG, Mourtas S, Marazioti A. Exosomes and exosome-inspired vesicles for targeted drug delivery. Pharmaceutics. 2018;10(4):218.
77. Lee JY, Kim H-S. Extracellular vesicles in neurodegenerative diseases: a double-edged sword. Tissue Eng Regen Med. 2017;14(6):667–678. doi:10.1007/s13770-017-0090-x
78. Dragomir M, Chen B, Calin GA. Exosomal lncRNAs as new players in cell-to-cell communication. Transl Cancer Res. 2018;7:S243–S252. doi:10.21037/tcr.2017.10.46
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.