Back to Journals » Cancer Management and Research » Volume 12
The Biogenesis, Functions, and Roles of circRNAs in Bladder Cancer
Authors Li C, Fu X, He H , Chen C, Wang Y, He L
Received 11 January 2020
Accepted for publication 16 April 2020
Published 19 May 2020 Volume 2020:12 Pages 3673—3689
DOI https://doi.org/10.2147/CMAR.S245233
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Chien-Feng Li
Changjiu Li,1,* Xian Fu,2,* Huadong He,1 Chao Chen,2 Yuyong Wang,2 Lugeng He2
1Department of Urology, Affiliated Hangzhou First People’s Hospital, Nanjing Medical University, Hangzhou 310006, Zhejiang Province, People’s Republic of China; 2Department of Urology, Affiliated Hangzhou First People’s Hospital, Zhejiang University, Hangzhou 310006, Zhejiang Province, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Huadong He Tel +86 18058765837
Email [email protected]
Abstract: Bladder cancer (BCa) is the 10th most prevalent malignancy worldwide and remains a crucial cause of cancer-related morbidity and mortality. Circular RNAs (circRNAs), a large class of endogenous non-coding RNAs, contain unique covalent closed structures and their biogenesis and turnover are regulated by multiple factors. Recently, multiple circRNAs have been found to serve as important factors in several biological processes such as tumorigenesis. An increasing amount of research discovered that circRNAs are dysregulated in multiple cancer tissues compared with matched normal tissues, especially in BCa, indicating that circRNAs can act as biomarkers for the diagnosis and prognosis of BCa. In this review, we focus on the biogenesis, properties, turnover, and functions of circRNAs, summarizing their potential functions and clinical implications in BCa.
Keywords: circRNA, circular RNA, biogenesis, microRNA sponge, bladder cancer, biomarker
Background
Bladder cancer (BCa) has become the 10th most prevalent malignancy worldwide. It is considered that nearly 549,000 new BCa patients and 200,000 related deaths were reported in 2018, worldwide.1 In recent years, treatment of BCa has come a long way. Apart from traditional surgery, radiotherapy, and chemotherapy, immunotherapy has also been used for the treatment of BCa patients.2 However, the morbidity and mortality of BCa patients remain high and BCa remains a crucial cause of cancer-related morbidity and mortality worldwide.3
An increasing amount of research indicates that plentiful non-coding RNAs (such as microRNAs and long non-coding RNAs) are linked to the carcinogenesis of BCa.4,5 Recently, several studies have aimed to investigate the expression profile and functions of circular RNAs (circRNAs) in BCa.6,7
CircRNAs are a large class of non-coding RNAs that have unique covalent loop structures. They were first discovered as naturally occurring in plant viroids in 1976.8 A few years later, circRNAs were also found in the cytoplasm of eukaryotic cells. Nonetheless, researchers found the expression of circRNAs to be extremely low, suggesting that they were the byproducts of aberrant splicing events9 and were short of biological functions. Recently, with the development of high-throughput RNA sequencing technologies (RNA-seq) and unique prediction techniques, tens of thousands of circRNAs have been detected in different species.10
CircRNAs are covalently closed circular molecules without 5ʹ-cap and 3ʹ-poly(A) structures11 that generally accumulate in the cytoplasm.12 As a result of the lack of a free terminus, they are inherently resistant to exonuclease degradation, which majorly recognizes the 5ʹ and 3ʹ termini. Several studies have indicated that circRNAs may instead be eliminated via RNase L upon viral infection13 or via extracellular vesicle release.14,15 Most circRNAs are encoded by known protein-coding genes16 and contain one or more exons.17 However, some circRNAs contain both intronic and exonic sequences (known as EIcircRNAs) and some of them originate from intronic sequences alone (known as ciRNAs).18,19
To this day, there is evidence to suggest that circRNAs could potentially have important roles in the occurrence and development of several human diseases, including cancer.20 Because of their stability and tissue specificity, an increasing number of circRNAs have been confirmed as diagnostic and prognostic biomarkers for a variety of diseases.21,22 In particular, circRNAs are specially significant in the process of tumorigenesis, invasion, and metastasis.23 Therefore, circRNAs could potentially act as diagnostic and therapeutic biomarkers of BCa.
In this review, we describe recent research regarding the biogenesis and function of circRNAs in BCa and their potential in the diagnosis, prognosis, and treatment of BCa.
Biogenesis of circRNAs
In all eukaryotes, removal of introns and connecting exons is a major part of the RNA splicing process, which converts the intron-containing precursor mRNAs (pre-mRNAs) into mRNAs, which lack any introns. Most eukaryotic circular RNAs are generated from pre-mRNAs. Nevertheless, the specific mechanism of circRNA biosynthesis is still unclear.
In a previous study, treatment of HeLa cells with isoginkgetin (a type of splice inhibitor) led to a significant reduction in both back-splicing and canonical splicing, indicating that circRNA biogenesis was linked with the canonical splicing machinery.24 Another research revealed that the production of circular RNAs increased following inhibition of the spliceosome, suggesting that inhibition or slowing of canonical splicing events would accelerate back-splicing events.25 Li et al found that the opportunity for back-splicing events would be increased by exon-definition complexes on long exons, indicating circRNA was an outgrowth of the spliceosome-mediated splicing event in eukaryotes.26
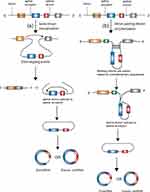
Because of the lack of 5ʹ-cap and 3ʹ-poly(A) structures, RNA circularization normally involves a back-splicing process, wherein the 3ʹ downstream splice donor (SD) is connected to the 5ʹ upstream splice acceptor (SA).27 There are two widely accepted mechanisms that can explain the back-splicing process, known as lariat-driven circularization and intron pairing-driven circularization.12 In the former model, circularization usually happens in exon-skipping events28 and needs the downstream SD to attach to the upstream SA covalently to form a lariat which also contains the skipped exons.27 If the back-splicing event takes place in the lariat before this lariat is disintegrated, a steady and covalent circRNA including the skipped exons will be produced.12 If the lariat possesses only one exon, the downstream SD of the exon attaches to its own upstream SA through a transesterification reaction, resulting in a circular RNA containing one exon without introns.29 If the lariat possesses multiple exons, the end of the last exon ligates to the start of the first exon through a transesterification reaction and thus intervening introns would be preserved, which results in an EIcircRNA. The EIciRNA can be further spliced by the spliceosome, leading to a circRNA containing several exons but no intervening introns.29 In another model, circularization is normally independent of exon-skipping events. The flanking introns are paired based on complementary sequences, which brings exons close and circularizes them (Figure 1).12
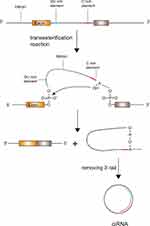
In a conventional colinear splicing reaction, intronic lariats are degraded by debranching enzymes inside the nucleus immediately. In some cases, intronic lariats can resist 2ʹ→5ʹ debranching enzymes and generate stable covalent circular structures called intronic circRNAs.19 The covalent circular structure of ciRNAs depends on a 7 nucleotides (nts) long GU-rich sequence near the splice site and another 11 nts C-rich sequence close to the branchpoint.19 During the back-splicing event, the two sequences first connect with each other to form a loop structure, the exonic and intronic segments in the connecting portion are removed by the spliceosome and the rest of introns bind together to generate a ciRNA (Figure 2).
The biogenesis of circRNA is regulated by a number of parameters and factors such as intronic sequences, enzymes, and protein factors.30 CircRNA biogenesis is not regulated by specific exonic sequences.31 However, exons with long sequences are more likely to generate a circRNA than short exonic sequences.26 CircRNA biosynthesis relies on the flanking introns of exons, making circRNA biogenesis dynamic.17 It has been confirmed that complementary repetitive sequences in the flanking intronic sequences of exons can bring the 3ʹ SD closer to the 5ʹ SA, such as in the production of mouse Sry circRNA.32 Flanking introns of circularized exons can bring back-splice sites close to each other and further catalyze exon circularization, especially repetitive complementary sequences (such as Alu elements12) and other nonrepetitive but complementary sequences.17 About 90% of circRNAs seem to have complementary sequences in the flanking intronic sequences30 and their flanking introns are five times longer than common introns, which lead to more opportunities to catalyze exon circularization. Nevertheless, not all intronic complementary repetitive sequences can accelerate circRNA biogenesis. Some kinds of intronic base-pairing can inhibit exon circularization.33 Additionally, the efficiency of circRNA biogenesis may be affected by intronic base-pairing competition if several complementary sequences exist in one single gene.17
RNA-binding proteins (RBPs) are regulated factors which can connect with specific sequences in the flanking intronic sequences to facilitate or inhibit circRNA biosynthesis.34 Muscleblind (MBL/MBNL1) is a conserved RNA-binding protein which can participate in alternative splicing, maintain mRNA stability and regulate circRNA biosynthesis in muscles.31,35 It contains four zinc fingers that bind strongly and specifically to conserved MBL binding sites.35,36 Muscleblind has a strong link with circMbl which originates from its own second exon and the production of which increases following muscleblind overexpression.31 However, according to a recent study, contrary to previous expectations, downregulation of circRNAs was not seen in DM1 cells, thereby questioning the role of MBNLs in being significant factors in circRNA biogenesis.37
QKI is another RBP which dynamically modulates the biogenesis of circRNA and facilitates the expression of circRNAs in epithelial–mesenchymal transition (EMT).34 QKI has been found to take part in multiple biological processes such as RNA splicing and translation.38 The addition of QKI motifs is beneficial to the biogenesis of circRNA compared to conventional linearly spliced transcripts. This discovery shows that circular RNAs are both purposefully generated and modulated by specific mechanisms, indicating that they may function as significant roles during EMT.34
ADAR, an RNA-editing factor, binds double-stranded RNA (dsRNA) and protects the immune system by converting adenosines to inosines.36 Ivanov et al demonstrated that ADAR antagonized the interaction between reverse complementary sequences in the flanking introns which catalyzed circRNA generation, suggesting that ADAR might negatively regulate circRNA generation that is dependent on base pairing between reverse complementary sequences.30
Moreover, other factors (such as HNRNPL,39 FUS,40 and RBM2041) have been verified to function in the regulation of circRNA biogenesis. As research progresses more and more factors will be identified as taking part in the regulation of RNA circularization.
In conclusion, the specific mechanisms of circRNA biogenesis and regulation are still elusive. Further research is warranted to acquire a deeper insight of the circRNA biogenesis.
Turnover of circRNAs
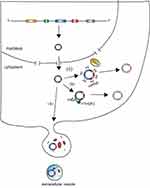
CircRNAs are covalently circular structures that are mostly resistant to RNase R. Because of the lack of 5ʹ-cap and 3ʹ-poly(A) structures, circRNAs are more stable than linear RNAs. Thus, most of the circRNAs cannot be degraded by traditional RNA degradation mechanisms (Figure 3).
Previous research revealed that ciRS-7 (also called CDR1as) can be bound by miR-671 through its miRNA binding sites and is subsequently cleaved by protein Argonaute 2 (AGO2).42 This supports the hypothesis that AGO2 might serve as an important factor by which cells cleave circRNAs. However, numerous circRNAs have been shown to not have miRNA binding sites and thus they could not be cleaved by AGO2, which suggests that AGO2-mediated cleavage is likely the turnover mechanism of only a small fraction of circRNAs. A recent study showed that GW182, a conserved factor, participates in circRNA degradation,43 but further research is needed to confirm this mechanism. Another study found that circRNAs with N6-methyladenosine (m6A) can be degraded by endoribonucleolytic cleavage via the YTHDF2-HRSP12-RNase-P/MRP pathway.44 m6A is a widespread internal modification of circRNAs,45 indicating that the YTHDF2-HRSP12-RNase-P/MRP pathway is probably a primary mechanism of circRNA turnover. Additionally, several studies have shown circRNAs to be enriched in extracellular vesicles,14,15 suggesting that extracellular vesicles release might be a mechanism of circRNA turnover. However, whether extracellular vesicles release results in a significant decrease in the levels of endogenous expression of circular RNAs remains to be answered. Interestingly, given that some circRNAs maintain their circular structures in extracellular vesicles14 and extracellular vesicles can be absorbed by other cells,15 release of circRNAs may also have a function in cell to cell communication.
Although research has begun to elucidate the mechanisms of circRNA turnover there are still many unanswered questions, including whether the release of circRNAs from extracellular vesicles is an effective mechanism of circRNA degradation. Therefore, more studies are warranted to help obtain a deeper and better understanding of circRNA turnover.
Function of circRNAs
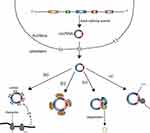
Although circRNAs were thought to be the byproducts of aberrant splicing events and thus are short of biological functions, recent research has shown that circRNAs might have some biological functions. Nevertheless, only a tiny fraction of circRNAs have verified paly significant roles in biological processes (Figure 4), with the major biological function of circRNAs being a miRNA sponge.46,47 In addition, some studies found that circRNAs can associate with proteins and subsequently directly influence protein function by acting as protein sponges or performing other functions.48–50 Moreover, a few circRNAs can act as mRNAs and guide protein synthesis via cap-independent translation.51–53 The major functions of circRNA are discussed below.
Acting as miRNA Sponges
The major biological function of circRNAs involves their role in acting as miRNA sponges. Highly abundant circRNAs with abundant miRNA response elements are more likely to bind miRNAs and compete with endogenous RNA function.24
ciRS-7, the most well-characterized circRNA, contains over 70 miR-7 binding sites and is abundant and highly conserved in many tissues, especially in neuronal tissues.46 The expression of ciRS-7 negatively regulates the miR-7 expression and subsequently accelerates the miR-7-targeted transcripts, indicating that ciRS-7 competitively binds miR-7 to its miR-7 binding sites and regulates gene expression through its miRNA sponge function.46 In the mouse brain, ciRS-7 has been found to be highly expressed, suggesting it is likely involved in neuronal function and differentiation.54 In the mouse, knock-out of ciRS-7 leads to a behavioral phenotype characterized by neuropsychiatric disorders and also leads to a reduction in miR-7 expression.54
Many other circRNAs can also serve as RNA sponges. The circular RNA cSMARCA5 may competitively bind to miR-17-3p and miR-181b-5p to facilitate TIMP3 expression, a type of tumor inhibitor.47 cSMARCA5 is found to be downregulated in HCC cells and is negatively associated with cell invasion and metastasis. Moreover, the expression level of cSMARCA5 could function as a crucial factor for evaluating the patients’ overall survival after hepatectomy. Circ-ITCH is found to inhibit BCa cell progression by sponging miR-17 and miR-224 and subsequently promoting p21 and PTEN gene expression.7
However, several studies have shown that most of the circRNAs rarely contain abundant miRNA binding sites, indicating that they may not regulate miRNA expression by functioning as miRNA sponges.16
Interaction with Proteins
CircRNAs can interact with proteins and subsequently influence protein function. CircMbl originates from the exon of the gene that encodes muscleblind and was the first circRNA to be found to act as protein sponge.31 The intronic sequences flanking circMbl have multiple putative MBL binding sites. These binding sites indicate that some MBLs might facilitate the circularization of circMbl from the second exon of MBL gene.31 Consequently, it has been suggested that there is a sophisticated autoregulatory loop between MBL and circMbl. Overexpression of MBL can reduce the production of its linear mRNA by promoting circRNA production and then this circRNA promotes expression of its own mRNA by binding to MBL.
Circ-Foxo3 is another well-characterized circRNA molecule that can serve as a protein scaffold. Levels of circ-Foxo3 have been shown to be increased in the heart tissue of aged patients and decreased in several cancer tissues,48–50 suggesting circ-Foxo3 might play a significant role in apoptosis. The murine double minute 2 protein (MDM2) is an important protein in the inhibition of apoptosis. MDM2 can bind both p53 and Foxo3, facilitate ubiquitination of p53 and Foxo3, and promote their degradation.55 Circ-Foxo3 can act as a protein scaffold to bind p53 and MDM2, promote the combination of p53 and MDM2, and accelerate p53 degradation. However, Foxo3, another MDM2 target protein, can avoid this degradation, which can lead to the induction of Foxo3’s downstream molecule Puma which facilitates apoptosis.55 Furthermore, circ-Foxo3 might associate with CDK2 and p21, and upregulate the formation of circ-Foxo3-p21-CDK2 ternary complex which blocks the cell cycle transition from G1 to S phase.55 In mammalian heart tissue, increased circ-Foxo3 facilitates cardiac senescence by associating with ID1 and E2F1 which participate in cell senescence, and FAK and HIF1ɑ which participate in the stress response.48
Circ-Dnmt1, circ-Ccnb1, and circACC1 are all thought to play important roles in tumor progression.51,56,57 Circ-Dnmt1 can upregulate the nuclear translocation of p53 and Auf1 by associating with them, and this induces cellular autophagy, restrains cellular senescence, and promotes cell proliferation.56 Circ-Ccnb1 can associate with both Cyclin B1 (Ccnb1) and Cyclin-dependent kinase 1 (Cdk1) and form a circ-Ccnb1-Ccnb1-Cdk1 complex. This complex inhibits the function of Ccnb1 in cell proliferation, migration, and invasion, and subsequently inhibits tumor growth.57 Finally, research has shown that circACC1 can directly bind to the β1 and ƴ1 subunits of AMPK, promoting stabilization of AMPK holoenzyme and facilitating glycolysis and β-oxidation. Moreover, a positive relationship has been found between circACC1 and tumor progression, both in vitro and in vivo,51 indicating circACC1 might have an important role in tumor progression. In addition, ci-ankrd52, a type of ciRNA that is abundant in the nucleus, can promote the efficiency of transcription by associating with RNA Pol II.19
Translation
Because of the lack of 5ʹ-cap and 3ʹ-poly(A) structures, circRNAs were initially thought to be untranslatable. Recently, researchers found that circRNAs can act as mRNAs to guide protein synthesis if they contain internal ribosome entry sites (IRESs), which they showed both in vitro and in vivo by using artificial circRNAs containing IRESs.52,53 Although over 7000 endogenous circRNAs are predicted to contain both IRES and open reading frames (ORFs),58 which means they might have protein-coding abilities, very few circRNAs were actually proved to guide protein synthesis.
Circ-ZNF609, a type of circRNA that specifically regulates myoblast proliferation, has been found to contain a 753-nt long ORF.58 A small proportion of circ-ZNF609 was confirmed to be able to load onto heavy polysomes, indicating that circ-ZNF609 might have coding ability. Translation of tagged circular transcripts in vitro and in vivo confirmed this hypothesis.59 However, whether these proteins encoded by circ-ZNF609 have a specific molecular activity has not been verified. In addition, unlike the zinc-finger protein 609, the counterpart encoded by linear ZNF609 mRNA, which has two zinc-finger domains, the protein produced from circZNF609 does not contain these zinc-finger domains, indicating that it might have completely different functions compared to its linear counterpart.
In addition to circRNAs containing IRESs, circRNAs with m6A could also have the potential to guide protein synthesis.60,61 Researchers have shown that circRNAs translation through m6A is directly initiated by initiation factor eIF4G2 (a non-canonical eIF4G protein) and is significantly increased by methyltransferase METTL3/METTL14.61 Interestingly, heat shock stress enhances circRNAs translation, particularly m6A-driven circRNAs translation, suggesting that circRNAs translation driven by m6A might have a significant role during cellular stress responses.
Although thousands of circRNAs are predicted to have both IRESs and ORFs or m6A sites,58,61 which indicates that they probably have translation potential, few endogenous circRNAs have been confirmed to have this ability. The function of circRNA-encoded proteins remains unclear. Some evidence indicates that circRNA-encoded proteins could possibly play a role in stress responses.61 However, the efficiency of cap-independent translation is very low and translation of circRNAs might therefore be limited.
CircRNA Immunogenicity
Exogenous circRNAs could have potential immunogenicity in mammalian cells and elicit immune protection against viral infection.62 The immunogenicity of exogenous circRNAs depends on the circular structure of the circRNA instead of the base sequence or ribosome binding. Exogenous circRNAs are sensed by retinoic-acid-inducible gene-I (RIG-I) which can specifically recognize viruses and function in the process of immune protection against them.62 However, Wesselhoeft et al carried out additional purifications of exogenous circRNAs and showed that exogenous circRNAs do not stimulate cellular RNA sensors such as TLRs or RIG-I.63 Chen et al, however, demonstrated that exogenous circRNAs with further purification do stimulate RIG-I, although not with the same effectiveness as dsRNA with 5ʹ-triphosphate.64 To date, whether circRNAs have definitive immunogenicity remains unclear. Research indicates that possibly only some exogenous circRNAs are immunogenic in environment-, cell-, and time-specific cases.65 Exogenous circRNAs may, therefore, become effective tools for therapeutic interventions. Further studies are warranted to confirm more details regarding circRNAs immunogenicity such as which circRNAs are immunogenic and how do endogenous circRNAs achieve self-tolerance.
circRNA in Extracellular Vesicles
Extracellular vesicles (EVs) have been proved involved in transferring functional proteins and RNAs between cells and modulating recipient cell behaviors.66 Some studies have proven that circRNAs can be enriched in EVs and can act as biomarkers for the diagnosis of human diseases.
In serum samples, thousands of circRNAs have been found enriched in EVs to distinguish between patients and healthy people.67 Xu et al found 209 upregulated EV-carried circRNAs and 66 downregulated EV-carried circRNAs in patients with endometrial cancer compared with that in healthy people.68 Among 209 upregulated EV-carried circRNAs, has_circ_0109046 and has_circ_0002577 contained multiple miRNA binding sites, indicating that these EV-carried circRNAs may function as competing endogenous RNAs in receipt cells after uptaking EVs from cancer cells. Moreover, these circRNAs may play important roles in many processes such as proliferation, invasion, and drug resistance of cancer cells. has_circ_0109046 exhibited tumor-suppressive properties that made colorectal cancer cells sensitive to chemotherapy.67 Interestingly, by transferring circ_0000338 via extracellular vesicles into HCT116-P cells, HCT116-P cells showed higher chemoresistance than normal control, indicating that chemoresistance can be transferred via circRNAs in extracellular vesicles.
However, there are also several limitations that can not be ignored. As thousands of EV-carried circRNAs in serum samples and tumor heterogeneity, it is hard to identify the tissue of origin. Also, considering low concentrations of EV-carried circRNAs in serum samples, many difficulties need to be solved in clinical testing.
CircRNAs and BCa
Although the biogenesis and functions of circRNAs are largely unclear, an increasing number of studies have verified that circRNAs are dysregulated in multiple cancer tissues compared with matched normal tissues, especially in BCa. With more detailed research being carried out, several circRNAs known to participate in the progression of BCa have been identified.
Expression of circRNAs in BCa
With the development of high-throughput sequencing and biochip technology, thousands of circular RNAs exceptionally expressed in BCa tissues and cell lines have been identified.
Wu et al analyzed BCa tissues from four different patients and matched adjacent non-cancerous bladder tissues by microarray analysis. They identified 433 circRNAs as being significantly differentially expressed, of which 169 were downregulated and 264 were upregulated.69 Zhong et al carried out a high-throughput microarray assay on four pairs of BCa tissues and adjacent non-tumor tissues and detected that 285 circRNAs (60.8%) were upregulated while 184 circRNAs (39.2%) were downregulated in BCa tissues.70 Liu et al carried out a microarray analysis to investigate circRNA expression profile and identified 734 circRNAs as being differentially expressed between BCa tissues and matched normal bladder tissues, and of which 256 (34.9%) were downregulated, and 478 (65.1%) were upregulated.71
Moreover, Li et al performed a high-throughput RNA sequencing on three BCa tissues and matched adjacent normal bladder tissues and identified 571 dysregulated circRNAs.72 Of them, 524 (91.8%) were downregulated and 47 (8.2%) were upregulated. Li’s group revealed that, while 316 circRNAs were differentially expressed between high-grade BCa tissues and normal controls, 244 circRNAs were altered between the high-grade BCa group and the low-grade BCa group.73 Among them, 42 circRNAs overlapped between the groups, suggesting that they might be useful for investigating the physiopathological mechanisms of BCa progression.
Although many studies have aimed to investigate the expression of circRNAs in BCa, they often have some limitations. For example, the sample size of these studies is small and they often lack expression profiles in different histological grades and pathological stages. Larger scale sequencing is warranted to obtain more details regarding the landscape of circRNAs expression in BCa.
CircRNAs as Biomarkers
Early diagnosis and treatment of BCa can effectively improve patient survival. Clinical manifestation of early stage BCa is not typical; hematuria is the most common symptom, but this can easily lead to confusion with other urinary diseases. Urinary cytology and cystoscopy are reliable BCa diagnostic methods; however, cystoscopy is an invasive examination, while traditional urinary cytology has a low sensitivity and is also prone to false-positive results. With the development of high-throughput sequencing and biochip technology, an increasing number of circRNAs have been shown to have potential as diagnostic biomarkers (Table 1).
 |
Table 1 ircRNAs as Biomarkers in BCa |
CircRNAs in Serum/Urine of BCa Patients
Serum and urine are more readily available than tissue samples in clinical practice. Therefore, it would be more convenient to detect circRNAs from the serum or urine.
Chi et al showed that the expression levels of circRNA_000285 were decreased in serum samples from BCa patients’ compared to those in healthy serum samples.74 Xu et al found that circPTK2 was frequently upregulated in BCa tissues.75 By comparing pre- and post-operative blood samples of 40 patients, they found that expression levels of circPTK2 were decreased in postoperative blood samples, which indicates that circPTK2 could have potential in clinical practice. In addition, Chen et al confirmed that circPRMT5 was enriched in the exosomes of BCa patients’ serum and urine samples, through comparison of 71 BCa patients’ serum samples and 18 BCa patients’ urine samples with those of 36 and 14 healthy donors, respectively.76
CircRNAs as Biomarkers in BCa Metastasis
CircACVR2A (hsa_circ_0001073) has been found to have reduced expression in BCa tissues and cell lines compared with that in normal tissues.77 Low expression of circACVR2A is linked with positive lymphatic metastasis of BCa, indicating that circACVR2A could be a potential biomarker for BCa metastasis. Yan et al found that circPICALM was downregulated in BCa tissues and cell lines.78 It was further confirmed that low circPICALM expression contributed to a higher pathological stage, histological grade, and lymphatic metastasis of BCa.
In addition, circ-VANGL1 has also been shown to be overexpressed in BCa tissues and cell lines.79 Circ-VANGL1 expression was related to some clinicopathological features of BCa, such as a high T stage, high histological grade, and lymphatic metastasis. Liang et al confirmed that circRNA_0058063 was augmented in BCa tissues compared with normal tissues and high expression of circRNA_0058063 was significantly correlated with higher pathological stage and increased likelihood of lymphatic metastasis.80
CircRNAs as Biomarkers in Prediction of BCa Response to Systemic Chemotherapy
Several studies have found that numerous circRNAs are correlated with chemotherapy sensitivity of BCa. Chi et al demonstrated that circ_000285 expression was markedly suppressed in BCa tissues and serum of BCa patients compared to that in normal controls.74 Expression of circ_000285 was also linked with tumor size, lymphatic metastasis, and distant metastasis. Interestingly, the expression level of circRNA_000285 was reduced in cisplatin-resistant patients compared to that in cisplatin-sensitive patients. Chen et al found that circFNTA was upregulated in BCa tissues and cell lines.81 It was further confirmed that high expression of circFNTA was significantly correlated with cisplatin chemoresistance. Su et al demonstrated that hypoxia-elevated circELP3 was associated with progression and cisplatin resistance of BCa.82 Depletion of circELP3 by siRNA treatment can facilitate cisplatin sensitivity in BCa cells. Expression of circHIPK3 was proven to be significantly decreased in BCa tissues.83 Low circHIPK3 expression is associated with gemcitabine resistance in BCa, suggesting that circHIPK3 might be a useful biomarker for the evaluation of BCa chemotherapeutic efficacy. Gemcitabine and cisplatin treatment remains the primary chemotherapy treatment for advanced BCa patients and resistance to these chemotherapeutics leads to a higher likelihood of recurrence and worse overall survival. Therefore, it is necessary to find some effective biomarkers to predict an individual patient’s response to systemic chemotherapy for BCa.
CircRNAs as Biomarkers in Prognosis of BCa
It is crucial to identify patients with poor prognosis and to thereby implement effective treatments. There is therefore an urgent need to uncover new biomarkers to evaluate the prognosis of BCa. CircRNAs are expressed differently in tumors of different histological grades and pathological stages, and in lymphatic metastasis or distant metastasis, which means circRNAs have potential as biomarkers in the prognosis of BCa.
For instance, Lin et al showed that levels of expression of circLPAR1 (hsa_circ_0087960) were downregulated in MIBC tissues compared with those in adjacent normal tissues.84 Patients with low circLPAR1 expression had a reduced overall survival. The median disease-specific survival time was 52.4 months for the low circLPAR1 expression group and 56.0 months for the high circLPAR1 expression group. Circ-BPTF, a circRNA originating from the BPTF gene, was remarkably upregulated in BCa tissues compared to adjacent normal tissues.85 The high expression of circ-BPTF was significantly correlated with worse overall survival and a higher likelihood of recurrence, which means that circ-BPTF might act as a biomarker for evaluating prognosis. CircRIP2, which originates from the RIP2 gene, was found as a conserved and downregulated circRNA in BCa tissues and cell lines.86 Higher expression of circRIP2 was significantly negatively correlated with a higher pathological stage and metastasis. According to analysis by Kaplan–Meier curves, patients with high circRIP2 expression had a better overall survival than those with low circRIP2 expression. CircFUT8 has also been found to have reduced expression in BCa tissues and cell lines compared with that in normal tissues.87 Overall survival analysis showed that patients with low circFUT8 expression had poor prognosis.
CircRNAs Suppress Tumor Progression
Tumorigenesis is the result of a complex interaction between multiple genes, factors, and signaling pathways. Significant research is still required to elucidate the specific mechanisms of tumorigenesis as well as the function of different factors and genes in cancer progression. Multiple circRNAs have been confirmed to play important roles in the progression of BCa, and thus could help provide a better understanding of the mechanisms underlying cancer progression (Table 2).
 |
Table 2 Dysregulated circRNAs in BCa |
CircHIPK3 (hsa_circ_0000284), which originates from exon 2 in the gene HIPK3, is widely and abundantly expressed in human cells. CircHIPK3 regulates cell growth by sponging several miRNAs.88 CircHIPK3 has been found to promote cancer cell proliferation in multiple types of cancer, such as gastric cancer, but, conversely, to suppress cancer growth in ovarian cancer.89,90 Mechanistically, circHIPK3 acts as a cancer inhibitor in BCa and was found to have reduced expression in BCa tissues. CircHIPK3 serves as a sponge for miR-558, which directly suppresses heparanase (HPSE) expression in BCa tissues. Low expression of circHIPK3 promotes proliferation, migration, and angiogenesis in cancer cells by interacting with the miR-558/HPSE axis.72 CircACVR2A (hsa_circ_0001073) has also been found to have reduced expression in BCa tissues and cell lines compared with that in normal tissues.77 CircACVR2A can bind to miR-626, rescuing the suppressing effect of miR-626 on the expression of EYA4, subsequently repressing proliferation, invasion, and metastasis of BCa in vitro and in vivo. Circ-ITCH is also a well-researched circRNA that functions as a miRNA sponge, suppressing tumor growth in several cancers such as ovarian cancer and BCa.7,91 Yang et al demonstrated that circ-ITCH has low levels of expression in BCa cell lines and tissues. Circ-ITCH functions as a miRNA sponge for miR-17 and miR-224 and thus, to some extent, reduces the inhibitory effects of miR-17/miR-224 on p21 and PTEN. Overexpression of circ-ITCH suppressed proliferation and metastasis of BCa cells and inhibited the cell cycle by interacting with the miR-17/miR-224/p21/PTEN axis and upregulating the expression of p21 and PTEN.37 CircMTO1 (hsa_circRNA_0007874/hsa_circRNA_104135) can directly interact with miR-221 to negatively regulate its expression.92 Overexpression of circMTO1 decreased cancer aggressiveness and inhibited EMT in cancer cells by sponging miR-221.
Liu et al found that circFNDC3B (hsa_circ_0006156) is downregulated in BCa cell lines and tissues93 and that circFNDC3B can directly sponge miR-1178-3p, thus decreasing the expression of G3BP2. They also found that circFNDC3B can downregulate the phosphorylation of SRC and FAK and subsequently influence the SRC/FAK signaling pathway. Overexpression of circFNDC3B restrained tumor cell proliferation and lymphatic metastasis in BCa. Another circRNA, circSLC8A1 (hsa_circ_0000994), has also been shown to be frequently downregulated in BCa cell lines and human BCa tissues, but to be highly abundant in the early stages of BCa. CircSLC8A1 has a key function as a miRNA sponge for miR-130b/miR-494, regulating the expression of PTEN, and subsequently negatively regulating cell proliferation and invasion of cancer cells.94 Overexpression of circSLC8A1 activates the signaling of PTEN, which has been commonly identified as an anti-oncogene, as PTEN mutations or deletions are often observed in multiple cancers.95 Finally, a study using high-throughput RNA sequencing found circUBXN7 (hsa_circ_0001380) to also be downregulated in BCa tissues and cell lines.88 Liu et al observed that decreased expression of circUBXN7 in BCa was correlated with a high T stage and a high histological grade.96 Further research showed that circUBXN7 binds to miR-1247-3p and functions as a miRNA sponge, subsequently modulating the miR-1247-3p/B4GALT3 axis and inhibiting growth and metastasis in BCa cells.
CircRNAs Promote Tumor Progression
Zhong et al observed that circRNA-MYLK acts as a crucial factor in the growth and metastasis of BCa cells.97 CircRNA-MYLK inhibited the downstream activity of miR-29 by acting as a competing endogenous RNA (ceRNA), and thus eliminating the miR-29 induced inhibition of the target gene VEGFA, which facilitates EMT and activates the downstream Ras/ERK signaling pathway in BCa progression. Further research then showed that overexpression of circRNA-MYLK accelerates growth, angiogenesis, invasion, and metastasis of BCa cells, while downregulation of circRNA-MYLK has the opposite effect. CircCEP128 (hsa_circ_0102722) has been found to be significantly upregulated in BCa tissues compared to adjacent normal tissues. CircCEP128 functions as a molecular sponge of miR-145-5p and inhibits the suppressor effect of miR-145-5 on the expression of SOX11, which inhibits apoptosis and facilitates the proliferation of BCa cells.69 Furthermore, Sun’s group observed that circCEP128 facilitates the expression of MYD88 and downstream proteins in the MAPK pathway by modulating miR-145-5p, further accelerating the cell cycle and repressing apoptosis in BCa cells.98
Chong et al indicated that circTCF25 (hsa_circ_0041103) can associate with miR-103a-3p and miR-107 to promote the expression of CDK6.70 CDK6 is hyperactive in multiple cancers, continuously facilitating cell cycle entry of cancer cells by triggering the transition from G1 to S phase.99 High expression levels of circTCF25 and CDK6 in BCa cells facilitate tumorigenesis, tumor progression, and invasion of BCa tissues. CircPTK2 (hsa_circ_0003221) is also expressed at higher levels in BCa tissues compared with adjacent normal tissues. Experiments on circPTK2 overexpression showed that upregulation of circPTK2 facilitates cell proliferation and migration.75 However, this particular study did not include any in vivo experiments, nor did it look into molecular mechanisms, which reduces its credibility.
Hsa_circ_0068871, a circRNA originating from the FGFR3 gene, was remarkably upregulated in BCa tissues and cell lines compared to adjacent normal tissues and normal bladder epithelial cell lines.100 Mao et al demonstrated that hsa_circ_0068871 was beneficial to the development of BCa by competitively interacting with miR-181a-5p which affects the levels of expression of FGFR3 and activates STAT3 signaling. Circular RNA TFRC (cTFRC, has-circ-0001445) serves as a tumor promoter in BCa tissues and cell lines.101 cTFRC interacts with miR-107, promoting the expression of the miR107 target gene TFRC and eventually accelerating the proliferation and invasion in BCa. Liu et al verified that the expression of circDOCK1 (hsa_circ_0020394) was increased in BCa tissues. Further investigation confirmed that circDOCK1 has a miRNA sponge role for miR-132-3p, thereby promoting the progression of BCa via modulation of the circDOCK1/miR-132-3p/Sox5 axis.71
Discussion and Prospects
In this review, we described the recent research on the biogenesis, turnover, and function of circRNAs and their potential in the diagnosis, prognosis, and treatment of BCa. As we summarized in this review, circRNA function as important factors in the progression of BCa, playing roles in proliferation, hypoxic adaptation, and angiogenesis, as well as in migration and drug resistance in BCa cells. As a result, circRNAs provide a new perspective on the diagnosis, prognosis, and treatment of BCa.
It is worth noting that circRNAs have great potential for use in tumor therapy. Given the covalent circular structure and high stability of circRNAs, exogenous circRNAs could be ideal at delivering therapeutic interventions. Additionally, since multiple circRNAs act as key factors in several biological processes, they could also potentially be used as drug targets.
Although many advances have been made in the field of circRNA research, many limitations remain to be improved. Firstly, the biogenesis, turnover, mechanisms of action, and functions of circRNAs are still unclear. Compared with plentiful circRNAs whose functions are unknown, the function of only a few circRNAs has been verified. Also, studies investigating the circRNA expression profiles of BCa cancer tissues compared to normal tissues in the same diseased bladder have presented inconclusive results. The sample size of these studies is small and they often lack expression profiles of different histological grades and pathological stages. In addition, few studies have compared circRNA expression in serum and urine samples of BCa patients in comparison to healthy people. Serum and urine are more readily available than tissue samples in clinical practice. It would be much more convenient if we could diagnose BCa by circRNAs in serum and urine. In some studies, sensitivity and specificity of these biomarkers are not addressed. The use of circRNAs as diagnostic and prognostic biomarkers is still a long way from clinical application. Finally, because of the complexity of tumor pathogenesis, the exact function of circRNAs in BCa remains unclear. Studies in the field of circular RNA remain in their infancy and much work still needs to be done in order to obtain the answers to these questions.
Acknowledgments
This work was supported by the Science and Technology Planning Project of Zhejiang Province, China (Grant No. 2017C37145).
Author Contributions
Changjiu Li contributed to the literature research, drafting, interpretation and writing of manuscript. Xian Fu and Huadong He contributed to the supervision and writing of manuscript. Lugeng He, Chao Chen and Yuyong Wang contributed to the literature research of manuscript. All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Disclosure
The authors declare no conflict of interest.
References
1. Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
2. Bellmunt J, Powles T, Vogelzang NJ. A review on the evolution of PD-1/PD-L1 immunotherapy for bladder cancer: the future is now. Cancer Treat Rev. 2017;54:58–67. doi:10.1016/j.ctrv.2017.01.007
3. Antoni S, Ferlay J, Soerjomataram I, et al. Bladder cancer incidence and mortality: a global overview and recent trends. Eur Urol. 2017;71(1):96–108. doi:10.1016/j.eururo.2016.06.010
4. Zhang J, Wang L, Mao S, et al. miR-1-3p contributes to cell proliferation and invasion by targeting glutaminase in bladder cancer cells. Cell Physiol Biochem. 2018;51(2):513–527. doi:10.1159/000495273
5. Zhan Y, Chen Z, Li Y, et al. Long non-coding RNA DANCR promotes malignant phenotypes of bladder cancer cells by modulating the miR-149/MSI2 axis as a ceRNA. J Exp Clin Cancer Res. 2018;37(1):273. doi:10.1186/s13046-018-0921-1
6. Yuan W, Zhou R, Wang J, et al. Circular RNA Cdr1as sensitizes bladder cancer to cisplatin by upregulating APAF1 expression through miR-1270 inhibition. Mol Oncol. 2019;13(7):1559–1576. doi:10.1002/1878-0261.12523
7. Yang C, Yuan W, Yang X, et al. Circular RNA circ-ITCH inhibits bladder cancer progression by sponging miR-17/miR-224 and regulating p21, PTEN expression. Mol Cancer. 2018;17:18.
8. Sanger HL, Klotz G, Riesner D, et al. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci U S A. 1976;73(11):3852–3856. doi:10.1073/pnas.73.11.3852
9. Cocquerelle C, Mascrez B, Hetuin D, et al. Mis-splicing yields circular RNA molecules. FASEB J. 1993;7(1):155–160. doi:10.1096/fasebj.7.1.7678559
10. Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi:10.1038/nature11928
11. Vicens Q, Westhof E. Biogenesis of circular RNAs. Cell. 2014;159(1):13–14. doi:10.1016/j.cell.2014.09.005
12. Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. Rna. 2013;19(2):141–157. doi:10.1261/rna.035667.112
13. Liu CX, Li X, Nan F, et al. Structure and degradation of circular RNAs regulate PKR activation in innate immunity. Cell. 2019;177(4):865–880.e821. doi:10.1016/j.cell.2019.03.046
14. Preusser C, Hung LH, Schneider T, et al. Selective release of circRNAs in platelet-derived extracellular vesicles. J Extracell Vesicles. 2018;7(1):1424473. doi:10.1080/20013078.2018.1424473
15. Lasda E, Parker R. Circular RNAs co-precipitate with extracellular vesicles: a possible mechanism for circRNA clearance. PLoS One. 2016;11(2):e0148407. doi:10.1371/journal.pone.0148407
16. Guo JU, Agarwal V, Guo H, Bartel DP. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15(7):409. doi:10.1186/s13059-014-0409-z
17. Zhang XO, Wang HB, Zhang Y, et al. Complementary sequence-mediated exon circularization. Cell. 2014;159(1):134–147. doi:10.1016/j.cell.2014.09.001
18. Li Z, Huang C, Bao C, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22(3):256–264. doi:10.1038/nsmb.2959
19. Zhang Y, Zhang XO, Chen T, et al. Circular intronic long noncoding RNAs. Mol Cell. 2013;51(6):792–806. doi:10.1016/j.molcel.2013.08.017
20. Han D, Li J, Wang H, et al. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology (Baltimore, Md). 2017;66(4):1151–1164. doi:10.1002/hep.29270
21. Zuo L, Zhang L, Zu J, et al. Circulating circular RNAs as biomarkers for the diagnosis and prediction of outcomes in acute ischemic stroke. Stroke. 2019;Strokeaha119027348.
22. Zhao Z, Li X, Gao C, et al. Peripheral blood circular RNA hsa_circ_0124644 can be used as a diagnostic biomarker of coronary artery disease. Sci Rep. 2017;7:39918. doi:10.1038/srep39918
23. Vo JN, Cieslik M, Zhang Y, et al. The Landscape of Circular RNA in Cancer. Cell. 2019;176(4):869–881.e813. doi:10.1016/j.cell.2018.12.021
24. Kristensen LS, Andersen MS, Stagsted LVW, et al. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20(11):675–691.
25. Liang D, Tatomer DC, Luo Z, et al. The output of protein-coding genes shifts to circular RNAs when the Pre-mRNA processing machinery is limiting. Mol Cell. 2017;68(5):940–954.e943. doi:10.1016/j.molcel.2017.10.034
26. Li X, Liu S, Zhang L, et al. A unified mechanism for intron and exon definition and back-splicing. Nature. 2019;573(7774):375–380. doi:10.1038/s41586-019-1523-6
27. Czubak K, Sedehizadeh S, Kozlowski P, et al. An overview of circular RNAs and their implications in myotonic dystrophy. Int J Mol Sci. 2019;20(18):4385. doi:10.3390/ijms20184385
28. Chen I, Chen CY, Chuang TJ. Biogenesis, identification, and function of exonic circular RNAs. Wiley Interdiscip Rev RNA. 2015;6(5):563–579. doi:10.1002/wrna.1294
29. Holdt LM, Kohlmaier A, Teupser D. Molecular roles and function of circular RNAs in eukaryotic cells. Cell Mol Life Sci. 2018;75(6):1071–1098. doi:10.1007/s00018-017-2688-5
30. Ivanov A, Memczak S, Wyler E, et al. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. 2015;10(2):170–177. doi:10.1016/j.celrep.2014.12.019
31. Ashwal-Fluss R, Meyer M, Pamudurti NR, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56(1):55–66. doi:10.1016/j.molcel.2014.08.019
32. Capel B, Swain A, Nicolis S, et al. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell. 1993;73(5):1019–1030. doi:10.1016/0092-8674(93)90279-Y
33. Liang D, Wilusz JE. Short intronic repeat sequences facilitate circular RNA production. Genes Dev. 2014;28(20):2233–2247. doi:10.1101/gad.251926.114
34. Conn SJ, Pillman KA, Toubia J, et al. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160(6):1125–1134. doi:10.1016/j.cell.2015.02.014
35. Wang ET, Treacy D, Eichinger K, et al. Transcriptome alterations in myotonic dystrophy skeletal muscle and heart. Hum Mol Genet. 2019;28(8):1312–1321. doi:10.1093/hmg/ddy432
36. Eisenberg E, Levanon EY. A-to-I RNA editing - immune protector and transcriptome diversifier. Nat Rev Genet. 2018;19(8):473–490.
37. Czubak K, Taylor K, Piasecka A, et al. Global increase in circular RNA levels in myotonic dystrophy. Front Genet. 2019;10:649. doi:10.3389/fgene.2019.00649
38. Hall MP, Nagel RJ, Fagg WS, et al. Quaking and PTB control overlapping splicing regulatory networks during muscle cell differentiation. Rna. 2013;19(5):627–638. doi:10.1261/rna.038422.113
39. Fei T, Chen Y, Xiao T, et al. Genome-wide CRISPR screen identifies HNRNPL as a prostate cancer dependency regulating RNA splicing. Proc Natl Acad Sci. 2017;114(26):E5207–E5215.
40. Errichelli L, Dini Modigliani S, Laneve P, et al. FUS affects circular RNA expression in murine embryonic stem cell-derived motor neurons. Nat Commun. 2017;8:14741.
41. Khan MA, Reckman YJ, Aufiero S, et al. RBM20 Regulates Circular RNA Production From the Titin Gene. Circ Res. 2016;119(9):996–1003. doi:10.1161/CIRCRESAHA.116.309568
42. Hansen TB, Wiklund ED, Bramsen JB, et al. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J. 2011;30(21):4414–4422. doi:10.1038/emboj.2011.359
43. Jia R, Xiao MS, Li Z, et al. Defining an evolutionarily conserved role of GW182 in circular RNA degradation. Cell Discov. 2019;5:45.
44. Park OH, Ha H, Lee Y, et al. Endoribonucleolytic cleavage of m(6)A-Containing RNAs by RNase P/MRP Complex. Mol Cell. 2019;74(3):494–507.e498. doi:10.1016/j.molcel.2019.02.034
45. Zhou C, Molinie B, Daneshvar K, et al. Genome-wide maps of m6A circRNAs identify widespread and cell-type-specific methylation patterns that are distinct from mRNAs. Cell Rep. 2017;20(9):2262–2276. doi:10.1016/j.celrep.2017.08.027
46. Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi:10.1038/nature11993
47. Yu J, Xu QG, Wang ZG, et al. Circular RNA cSMARCA5 inhibits growth and metastasis in hepatocellular carcinoma. J Hepatol. 2018;68(6):1214–1227. doi:10.1016/j.jhep.2018.01.012
48. Du WW, Yang W, Chen Y, et al. Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur Heart J. 2017;38(18):1402–1412. doi:10.1093/eurheartj/ehw001
49. Shen Z, Zhou L, Zhang C, et al. Reduction of circular RNA Foxo3 promotes prostate cancer progression and chemoresistance to docetaxel. Cancer Lett. 2020;468:88–101. doi:10.1016/j.canlet.2019.10.006
50. Xing Y, Zha WJ, Li XM, et al. Circular RNA circ-Foxo3 inhibits esophageal squamous cell cancer progression via the miR-23a/PTEN axis. J Cell Biochem. 2019.
51. Li Q, Wang Y, Wu S, et al. CircACC1 regulates assembly and activation of AMPK complex under metabolic stress. Cell Metab. 2019;30(1):157–173.e157. doi:10.1016/j.cmet.2019.05.009
52. Chen CY, Sarnow P. Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science (New York, N Y). 1995;268(5209):415–417. doi:10.1126/science.7536344
53. Wang Y, Wang Z. Efficient backsplicing produces translatable circular mRNAs. Rna. 2015;21(2):172–179. doi:10.1261/rna.048272.114
54. Piwecka M, Glazar P. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science. 2017;357:6357.
55. Du WW, Fang L, Yang W, et al. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ. 2017;24(2):357–370. doi:10.1038/cdd.2016.133
56. Du WW, Yang W, Li X, et al. A circular RNA circ-DNMT1 enhances breast cancer progression by activating autophagy. Oncogene. 2018;37(44):5829–5842. doi:10.1038/s41388-018-0369-y
57. Fang L, Du WW, Awan FM, et al. The circular RNA circ-Ccnb1 dissociates Ccnb1/Cdk1 complex suppressing cell invasion and tumorigenesis. Cancer Lett. 2019;459:216–226. doi:10.1016/j.canlet.2019.05.036
58. Chen X, Han P, Zhou T, et al. circRNADb: a comprehensive database for human circular RNAs with protein-coding annotations. Sci Rep. 2016;6:34985. doi:10.1038/srep34985
59. Legnini I, Di Timoteo G, Rossi F, et al. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol Cell. 2017;66(1):22–37.e29. doi:10.1016/j.molcel.2017.02.017
60. Meyer KD, Patil DP, Zhou J, et al. 5ʹ UTR m(6)A promotes cap-independent translation. Cell. 2015;163(4):999–1010. doi:10.1016/j.cell.2015.10.012
61. Yang Y, Fan X, Mao M, et al. Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res. 2017;27(5):626–641. doi:10.1038/cr.2017.31
62. Chen YG, Kim MV, Chen X, et al. Sensing self and foreign circular RNAs by intron identity. Mol Cell. 2017;67(2):228–238.e225. doi:10.1016/j.molcel.2017.05.022
63. Wesselhoeft RA, Kowalski PS, Parker-Hale FC, et al. RNA circularization diminishes immunogenicity and can extend translation duration in vivo. Mol Cell. 2019;74(3):508–520.e504. doi:10.1016/j.molcel.2019.02.015
64. Chen YG, Chen R, Ahmad S, et al. N6-Methyladenosine modification controls circular RNA immunity. Mol Cell. 2019;76(1):96–109.e109. doi:10.1016/j.molcel.2019.07.016
65. Basavappa MG, Cherry S. Going in circles: the Black Box of circular RNA immunogenicity. Mol Cell. 2019;76(1):3–5. doi:10.1016/j.molcel.2019.08.027
66. Shah R, Patel T, Freedman JE. Circulating extracellular vesicles in human disease. N Engl J Med. 2018;379(10):958–966. doi:10.1056/NEJMra1704286
67. Li Y, Zheng Q, Bao C, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25(8):981–984. doi:10.1038/cr.2015.82
68. Xu H, Gong Z, Shen Y, et al. Circular RNA expression in extracellular vesicles isolated from serum of patients with endometrial cancer. Epigenomics. 2018;10(2):187–197. doi:10.2217/epi-2017-0109
69. Wu Z, Huang W, Wang X, et al. Circular RNA CEP128 acts as a sponge of miR-145-5p in promoting the bladder cancer progression via regulating SOX11. Mol Med. 2018;24(1):40.
70. Zhong Z, Lv M, Chen J. Screening differential circular RNA expression profiles reveals the regulatory role of circTCF25-miR-103a-3p/miR-107-CDK6 pathway in bladder carcinoma. Sci Rep. 2016;6:30919. doi:10.1038/srep30919
71. Liu P, Li X, Guo X, et al. Circular RNA DOCK1 promotes bladder carcinoma progression via modulating circDOCK1/hsa-miR-132-3p/Sox5 signalling pathway. Cell Prolif. 2019;52(4):e12614. doi:10.1111/cpr.12614
72. Li Y, Zheng F, Xiao X, et al. CircHIPK3 sponges miR-558 to suppress heparanase expression in bladder cancer cells. EMBO Rep. 2017;18(9):1646–1659.
73. Li W, Li Y, Sun Z, et al. Comprehensive circular RNA profiling reveals the regulatory role of the hsa_circ_0137606/miR1231 pathway in bladder cancer progression. Int J Mol Med. 2019;44(5):1719–1728. doi:10.3892/ijmm.2019.4340
74. Chi BJ, Zhao DM, Liu L, et al. Downregulation of hsa_circ_0000285 serves as a prognostic biomarker for bladder cancer and is involved in cisplatin resistance. Neoplasma. 2019;66(2):197–202. doi:10.4149/neo_2018_180318N185
75. Xu ZQ, Yang MG, Liu HJ, et al. Circular RNA hsa_circ_0003221 (circPTK2) promotes the proliferation and migration of bladder cancer cells. J Cell Biochem. 2018;119(4):3317–3325. doi:10.1002/jcb.26492
76. Chen X, Chen RX, Wei WS, et al. PRMT5 circular RNA promotes metastasis of urothelial carcinoma of the bladder through sponging miR-30c to induce epithelial-mesenchymal transition. Clin Cancer Res. 2018;24(24):6319–6330. doi:10.1158/1078-0432.CCR-18-1270
77. Dong W, Bi J, Liu H, et al. Circular RNA ACVR2A suppresses bladder cancer cells proliferation and metastasis through miR-626/EYA4 axis. Mol Cancer. 2019;18(1):95. doi:10.1186/s12943-019-1025-z
78. Yan D, Dong W, He Q, et al. Circular RNA circPICALM sponges miR-1265 to inhibit bladder cancer metastasis and influence FAK phosphorylation. EBioMedicine. 2019;48:316–331. doi:10.1016/j.ebiom.2019.08.074
79. Zeng Z, Zhou W, Duan L, et al. Circular RNA circ-VANGL1 as a competing endogenous RNA contributes to bladder cancer progression by regulating miR-605-3p/VANGL1 pathway. J Cell Physiol. 2019;234(4):3887–3896.
80. Liang H, Huang H, Li Y, et al. CircRNA_0058063 functions as a ceRNA in bladder cancer progression via targeting miR-486-3p/FOXP4 axis. Biosci Rep. 2020;40(3). doi:10.1042/BSR20193484.
81. Chen J, Sun Y, Ou Z, et al. Androgen receptor-regulated circFNTA activates KRAS signaling to promote bladder cancer invasion. EMBO Rep. 2020;21(4):e48467.
82. Su Y, Yang W, Jiang N, et al. Hypoxia-elevated circELP3 contributes to bladder cancer progression and cisplatin resistance. Int J Biol Sci. 2019;15(2):441–452. doi:10.7150/ijbs.26826
83. Xie F, Zhao N, Zhang H, et al. Circular RNA CircHIPK3 promotes gemcitabine sensitivity in bladder cancer. J Cancer. 2020;11(7):1907–1912. doi:10.7150/jca.39722
84. Lin G, Sheng H, Xie H, et al. circLPAR1 is a novel biomarker of prognosis for muscle-invasive bladder cancer with invasion and metastasis by miR-762. Oncol Lett. 2019;17(3):3537–3547. doi:10.3892/ol.2019.9970
85. Bi J, Liu H, Cai Z, et al. Circ-BPTF promotes bladder cancer progression and recurrence through the miR-31-5p/RAB27A axis. Aging. 2018;10(8):1964–1976. doi:10.18632/aging.101520
86. Su Y, Feng W, Shi J, et al. circRIP2 accelerates bladder cancer progression via miR-1305/Tgf-β2/smad3 pathway. Mol Cancer. 2020;19(1):23.
87. He Q, Yan D, Dong W, et al. circRNA circFUT8 upregulates krüpple-like factor 10 to inhibit the metastasis of bladder cancer via sponging miR-570-3p. Mol Ther Oncol. 2020;16:172–187. doi:10.1016/j.omto.2019.12.014
88. Zheng Q, Bao C, Guo W, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7:11215. doi:10.1038/ncomms11215
89. Liu WG, Xu Q. Upregulation of circHIPK3 promotes the progression of gastric cancer via Wnt/beta-catenin pathway and indicates a poor prognosis. Eur Rev Med Pharmacol Sci. 2019;23(18):7905–7912. doi:10.26355/eurrev_201909_19004
90. Teng F, Xu J, Zhang M, et al. Comprehensive circular RNA expression profiles and the tumor-suppressive function of circHIPK3 in ovarian cancer. Int J Biochem Cell Biol. 2019;112:8–17. doi:10.1016/j.biocel.2019.04.011
91. Luo L, Gao YQ, Sun XF. Circular RNA ITCH suppresses proliferation and promotes apoptosis in human epithelial ovarian cancer cells by sponging miR-10a-alpha. Eur Rev Med Pharmacol Sci. 2018;22(23):8119–8126. doi:10.26355/eurrev_201812_16503
92. Li Y, Wan B, Liu L, et al. Circular RNA circMTO1 suppresses bladder cancer metastasis by sponging miR-221 and inhibiting epithelial-to-mesenchymal transition. Biochem Biophys Res Commun. 2019;508(4):991–996. doi:10.1016/j.bbrc.2018.12.046
93. Liu H, Bi J, Dong W, et al. Invasion-related circular RNA circFNDC3B inhibits bladder cancer progression through the miR-1178-3p/G3BP2/SRC/FAK axis. Mol Cancer. 2018;17(1):161. doi:10.1186/s12943-018-0908-8
94. Lu Q, Liu T, Feng H, et al. Circular RNA circSLC8A1 acts as a sponge of miR-130b/miR-494 in suppressing bladder cancer progression via regulating PTEN. Mol Cancer. 2019;18(1):111.
95. Tamguney T, Stokoe D. New insights into PTEN. J Cell Sci. 2007;120(Pt 23):4071–4079. doi:10.1242/jcs.015230
96. Liu H, Chen D, Bi J, et al. Circular RNA circUBXN7 represses cell growth and invasion by sponging miR-1247-3p to enhance B4GALT3 expression in bladder cancer. Aging. 2018;10(10):2606–2623. doi:10.18632/aging.101573
97. Zhong Z, Huang M, Lv M, et al. Circular RNA MYLK as a competing endogenous RNA promotes bladder cancer progression through modulating VEGFA/VEGFR2 signaling pathway. Cancer Lett. 2017;403:305–317. doi:10.1016/j.canlet.2017.06.027
98. Sun M, Zhao W, Chen Z, et al. Circular RNA CEP128 promotes bladder cancer progression by regulating Mir-145-5p/Myd88 via MAPK signaling pathway. Int J Cancer. 2019;145(8):2170–2181.
99. Choi YJ, Anders L. Signaling through cyclin D-dependent kinases. Oncogene. 2014;33(15):1890–1903. doi:10.1038/onc.2013.137
100. Mao W, Huang X, Wang L, et al. Circular RNA hsa_circ_0068871 regulates FGFR3 expression and activates STAT3 by targeting miR-181a-5p to promote bladder cancer progression. J Exp Clin Cancer Res. 2019;38(1):169. doi:10.1186/s13046-019-1136-9
101. Su H, Tao T, Yang Z, et al. Circular RNA cTFRC acts as the sponge of MicroRNA-107 to promote bladder carcinoma progression. Mol Cancer. 2019;18(1):27. doi:10.1186/s12943-019-0951-0
102. Liu F, Zhang H, Xie F, et al. Hsa_circ_0001361 promotes bladder cancer invasion and metastasis through miR-491-5p/MMP9 axis. Oncogene. 2020;39(8):1696–1709. doi:10.1038/s41388-019-1092-z
103. Liu T, Lu Q, Liu J, et al. Circular RNA FAM114A2 suppresses progression of bladder cancer via regulating ∆NP63 by sponging miR-762. Cell Death Dis. 2020;11(1):47. doi:10.1038/s41419-020-2226-5
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.