Back to Journals » Drug Design, Development and Therapy » Volume 16
Targets Exploration of Hydroxychloroquine for Pigmentation and Cell Protection Effect in Melanocytes: The Clue for Vitiligo Treatment
Authors Xie B , Chen Y, Hu Y, Zhao Y, Luo H, Xu J, Song X
Received 10 December 2021
Accepted for publication 19 March 2022
Published 5 April 2022 Volume 2022:16 Pages 1011—1024
DOI https://doi.org/10.2147/DDDT.S350387
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Georgios Panos
Bo Xie,1,* Yi Chen,2,* Yebei Hu,2 Yan Zhao,2 Haixin Luo,2 Jinhui Xu,1 Xiuzu Song1
1Department of Dermatology, Hangzhou Third People’s Hospital, Affiliated Hangzhou Dermatology Hospital, Zhejiang University School of Medicine, Hangzhou, 310009, People’s Republic of China; 2Department of Dermatology, Hangzhou Third Hospital Affiliated to Zhejiang Chinese Medical University, Hangzhou, 310009, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Xiuzu Song, Department of Dermatology, Hangzhou Third People’s Hospital, Affiliated Hangzhou Dermatology Hospital, Zhejiang University School of Medicine, West Lake Road 38, Hangzhou, 310009, People’s Republic of China, Tel +86-571-87823102, Email [email protected]
Objective: The treatment of vitiligo is often challenging to dermatologists. There is ample evidence to suggest that hydroxychloroquine (HCQ) is effective for vitiligo treatment; nonetheless, the underlying mechanism remains unknown. In the present study, we sought to uncover the molecular targets of HCQ by an integrated network-based pharmacologic and transcriptomic approach.
Methods: The potential targets of HCQ were retrieved from databases based on the crystal structure. Targets related to vitiligo were screened and intersected with potential targets of HCQ. A protein-protein interaction network of the intersected targets was generated. Interactions between the targets were verified by molecular docking. Moreover, human vitiligo immortalized melanocytes (PIG3V) were evaluated after treatment with HCQ (1μg/mL) for 24h. The total RNA of PIG3V was extracted and determined by RNA-seq transcriptomics for differential gene expression analysis. Network pharmacology was then used to identify the relationships between putative targets of HCQ and differentially expressed genes.
Results: Molecular docking analysis revealed four putative key targets (ACHE, PNMT, MC1R, and VDR) of HCQ played important roles in vitiligo treatment. According to the transcriptomic results, the melanosomal biogenesis-related gene BLOC1S5 was upregulated 138005.020 fold after HCQ treatment. Genes related to protein repair (MSRB3) and anti-ultraviolet (UV) effect (UVSSA) were upregulated 4.253 and 2.603 fold, respectively, after HCQ treatment.
Conclusion: The expression of the BLOC1S5 gene is significantly upregulated, indicating upregulated melanosomal biogenesis after HCQ treatment. In addition, HCQ yields a protective effect on melanocytes by upregulating genes associated with damaged protein repair (MSRB3) and anti-UV effect (UVSSA). The protective effects of HCQ are mediated by binding to putative targets ACHE, PNMT, MC1R, and VDR according to network pharmacology and docking verification.
Keywords: vitiligo, hydroxychloroquine, treatment, pigmentation, melanocyte protection
Introduction
Vitiligo is an autoimmune depigmentation disorder characterized by the loss of functional melanocytes and patchy skin pigmentation.1 It has been reported that at least 0.5% of the population suffers from vitiligo worldwide.2 Importantly, vitiligo can lead to stigma, shame and embarrassment in this patient population.3 Until now, the mechanisms that belie the pathogenesis of vitiligo remain unknown. An increasing body of evidence suggests that both the adaptive and innate immune systems are involved in the pathogenesis of vitiligo.4 Moreover, interferon (IFN) γ-inducible chemokines and CD8+ T cells can be initiated by external triggers such as ultraviolet (UV) and chemical stimuli.5 Mitochondria generate reactive oxygen species (ROS) in response to oxidative stimuli, disrupting the normal functions of organelles such as mitochondria, lysosome, endoplasmic reticulum (ER), etc.6 ER injury has been reported to lead to the generation of unfolded proteins.7 Besides, it has been established that damaged proteins and exosomes secreted by melanocytes can be recognized by antigen-presenting cells to stimulate autoreactive T cell maturation.8 The positive feedback of melanocyte-specific CD8+ T cells recruitment induced by chemokines can reportedly potentiate the autoimmune attack towards melanocytes.9 Notwithstanding that unprecedented progress has been achieved in understanding the pathophysiology, the specific mechanisms have not been clearly elucidated, accounting for the difficulty dermatologists face in treating this disease during clinical practice, hence emphasizing the need for future studies.10
According to current guidelines, topical glucocorticoids, calcineurin inhibitors, vitamin D3 derivatives, and phototherapy remain the mainstay of treatment for vitiligo. Systemic glucocorticoids are indicated with rapid progression of the lesions.1–4 Indeed, efficient vitiligo treatment is often difficult, with low repigmentation rates and high relapse rates. Currently, HCQ is recommended to treat rheumatic diseases such as systemic lupus erythematosus (SLE), rheumatoid arthritis (RA), antiphospholipid antibody syndrome, and Sjogren’s syndrome, which are autoimmune-related.11–13 In addition, HCQ is reportedly effective in treating vitiligo, chronic actinic dermatitis, chronic urticaria, dermatomyositis, vasculitis, lichen planus, etc.14 In RA and SLE patients presenting with vitiligo, it was observed that HCQ treatment also promotes pigmentation in vitiligo lesions.10,15 In addition, skin pigmentation after HCQ treatment was observed in RA and SLE patients without vitiligo.16,17 It is widely believed that HCQ exerts an anti-inflammatory effect that can disrupt T-cell receptor-related Ca2+ signaling and antigen processing.18 Importantly, Li DG et al19 showed that HCQ could protect melanocytes from autoantibody-induced damage by reducing the formation of antigen-antibody complexes.
At present, the mechanisms underlying the therapeutic effect of HCQ in vitiligo are unclear. Current evidence suggests that HCQ could be a multi-target drug given its wide range of effects. Indeed, the immunoregulatory effect of HCQ plays a key role in its pharmacological mechanisms. In recent years, the rapid development of computer technology and big data analytics has led to the advent of network pharmacology, which provides a new strategy for the research of multi-targets drugs. Network pharmacology integrates poly-pharmacology, bioinformatics and systems biology for multi-targets drug research and evaluation. Importantly, network pharmacology emphasizes the “multi-targets network/drug” pattern in contrast to the conventional “one target/ one drug” paradigm.20–23 In the present study, human vitiligo immortalized melanocytes (PIG3V) were used to conduct differential gene expression analysis by RNA-sequencing after HCQ treatment. Transcriptomic, GO annotation, and KEGG enrichment analyses were conducted to elaborate changes in gene expression and biological functions after HCQ treatment. Finally, network pharmacology and transcriptomic results were integrated to uncover the mechanisms of HCQ in treating vitiligo, providing a foothold for future studies on its potential use for vitiligo treatment.
Methods and Materials
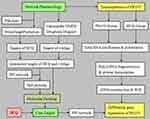
The predicted target proteins were retrieved from the database according to the crystal structure and chemical groups of HCQ. Then, vitiligo-related proteins were collected from disease databases. Data on these proteins were imported into Cytoscape software (v.3.8.2). The two groups of proteins were then intersected. A protein-protein interaction (PPI) network of the intersected proteins was constructed with the plugin Bisogenet of Cytoscape software. Subsequently, all intersected targets and hub proteins in the PPI network were chosen for molecular docking with HCQ. Then, gene expression in PIG3V was explored after treatment with HCQ. Transcriptomic, GO annotation and KEGG enrichment analyses were conducted. Finally, we integrated network pharmacology and transcriptomic results to assess the relationships between HCQ molecular docking targets and differentially expressed genes after HCQ treatment in PIG3V cells (Figure 1). According to the guidelines of Ethics Committee of Hangzhou Third People’s Hospital, the human public databases we used in this research were exempted from approval, because only studies of identifiable human specimens or data need to be approved by ethics committee.
Predicted Targets of HCQ and Identification of Vitiligo-Related Proteins and Core Targets
The chemical structure (Canonical SMILES) of HCQ was retrieved from PubChem database (https://pubchem.ncbi.nlm.nih.gov/): CCN(CCCC(C)NC1=C2C=CC(=CC2=NC=C1)Cl)CCO. The SMILES molecular formula of HCQ was imported into the SwissTargetPrediction database (http://www.swisstargetprediction.ch/index.php). Then predicted targets of HCQ were calculated and retrieved from the SwissTargetPrediction database based on the crystal structure. The species was set as “Homo sapiens”. The top 100 potential targets were selected according to the calculated parameters.24,25 Vitiligo-related proteins were searched using the keywords and species (“vitiligo” and “Homo sapiens”) across four databases, including Genecards (https://www.genecards.org/), OMIM (https://omim.org/), Drugbank (https://go.drugbank.com/) and DisGeNET (https://www.disgenet.org/). A total of 1185 vitiligo-related proteins were screened.26,27 The bisogenet plugin of Cytoscape was used for the PPI network construction. The intersected proteins between vitiligo and HCQ were entered into bisogenet. The topological features “degree”, “betweenness” and “closeness” were used to select the putative targets using the Cytoscape plugin CytoNCA.28
Computational Biology Verification
Along with the intersection targets between HCQ and vitiligo, the core targets screened in the previous step were chosen as candidates for molecular docking. The spatial interactions between target proteins and HCQ were analyzed by AutoDock (v.4.2) software. First, the 3-dimensional crystal structure of HCQ was retrieved from the Research Collaboratory for Structural Bioinformatics Protein Data Bank (RCSB PDB) database. Then, modifications such as water and ligand removal, amino acid optimization and patching, and hydrogen addition were manipulated in AutoDock. ChemBioDraw 3D (v.15.1) software was used for 3-D visualization and energy minimizing. Finally, MolegroVirtualDocker software was used to compute docking targets by comparing the conformation with the existing 3-D crystal structure of HCQ.29
PIG3V Cells Cultivation and HCQ Treatment
The Human vitiligo melanocyte cell line PIG3V was a gift from Professor Chunying Li (Xijing Hospital of Air Force Medical University, Xi’an, China) and cultured (2×105 cells/mL) in a 6-well plate (2mL/well) supplemented with DMEM/F12, 10% FBS, basic fibroblast growth factor (10ng/mL), phorbol myristate acetate (10ng/mL), and penicillin/streptomycin (10000U/mL, 10000μg/mL) at 37°C in 5% CO2 for 24 hours.30 The cultured PIG3V cells were randomly divided into an HCQ group and a control group. Cells in the HCQ group were treated with 1μg/mL HCQ (Sigma-Aldrich Corp., St. Louis, MO, USA) for 24 hours. The HCQ powder was dissolved in phosphate-buffered saline (PBS). In contrast, cells in the control group were treated with PBS for 24 hours. Finally, the total RNA of PIG3V cells was purified and extracted with TRIzol reagent (Invitrogen, Carlsbad, CA, USA).
mRNA Library Construction and Sequencing and Analysis
RNA samples extracted from PIG3V cells were quantified by NanoDrop (Wilmington, DE, USA) and Bioanalyzer (Agilent, CA, USA).31,32 Poly (A) RNA was extracted from total RNA by 2 rounds of purification with Dynabeads Oligo (Thermo Fisher, CA, USA). Small pieces of Poly A RNA were returned through Magnesium RNA Fragmentation Module (NEB, USA) under the condition of 94°C for 5 minutes. Subsequently, SuperScript Reverse Transcriptase (Invitrogen, USA) was used to reverse-transcribe the cleaved RNA pieces into cDNA. Then, U-labeled second-stranded DNAs were synthesized. After addition of A-base to each strand, index read preparation, and heat-labile UDG enzyme treatment, PCR amplification was conducted. PCR consisted of preheating at 95°C for 3 minutes; 8 cycles of 98°C for 15 seconds, annealing at 60°C for 15 seconds and extension at 72°C for 5 minutes. Ultimately, RNA sequencing was performed according to the protocol of Illumina Novaseq™ 6000 (LC-Bio Technology CO., Ltd., Hangzhou, China).33,34 Genes differential expression analysis was performed by DESeq2 software between two different groups. After calculation, differentially expressed genes were screened by fold change (FC)>2 or FC<0.5 and p value<0.05. These genes underwent GO and KEGG functional enrichment analyses. Hypergeometric test was used to calculate p value in GO and KEGG enrichment.35,36
The Relationships Between HCQ Targets and Differentially Expressed Genes of PIG3V After HCQ Treatment
According to the value of | log2(FC) |, the top 30 upregulated and downregulated genes in PIG3V cells were selected. To screen genes related to synthesis, transport, metabolism of melanin, melanocyte protection and other mechanisms associated with vitiligo, the functions of these genes/proteins and previous studies were retrieved from the databases of UniProt (https://www.uniprot.org/) and NCBI PubMed (https://www.ncbi.nlm.nih.gov/). Then these differentially expressed genes along with docking HCQ targets were input into Cytoscape software. A PPI network was constructed to explore the interactions between HCQ targets and vitiligo-related differentially expressed genes.37,38
Results
The Putative Targets of HCQ
The predicted targets of HCQ were computed and retrieved from databases by analyzing the 2 and 3-dimensional chemical structure (Figure 2A). The top 100 potential targets (Figure 2B) were retrieved according to the index of possibility. These predicted targets were mainly classified as G protein-coupled receptors (29%), kinases (26%) and surface antigens (13%) (Figure 2C).
 |
Figure 2 Predicted targets of HCQ. (A) 3 dimensional and 2 dimensional structure of HCQ; (B) top 100 predicted targets of HCQ from databases; (C) categories of the predicted targets. |
Vitiligo-Related Proteins and Topological Network Analysis
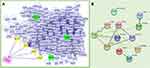
The intersection between vitiligo-related proteins (n=1185) and top 100 potential HCQ targets yielded 15 proteins (Table 1). A PPI network of the intersected targets was generated by Cytoscape using the plugin Bisogenet. Then, 1686 additional proteins closely linked to the 15 intersected proteins were retrieved from Bisogenet. Finally, a total of 42,371 interactions (edges) between 1686 targets (nodes) were identified (Figure 3A). A topological degree greater than 62 was used to screen vital nodes in these 1686 targets (Figure 3B). Then, 459 nodes and 18,251 edges were screened according to a betweenness value greater than 734.84 and closeness value greater than 0.50. The 78 hub proteins and their 1452 interactions which may play important roles in the therapeutic effects of HCQ in treating vitiligo, were retrieved (Figure 3C). The top 10 hub proteins are listed in Table 2.
 |
Table 1 The Intersection of HCQ Predicted Targets and Vitiligo-Related Proteins |
 |
Table 2 The Hub Proteins Screened in PPI Network |
Computational Biological Verification by Molecular Docking
The intersected 15 proteins and the top 10 hub proteins of the PPI network were chosen as target candidates for molecular docking verification (Tables 1 and 2), providing them visual explanations of spatial interactions with HCQ. A docking score below −20 demonstrated that HCQ could effectively combine with target proteins. Acetylcholinesterase (ACHE), Phenylethanolamine N-methyltransferase (PNMT), Melanocortin receptor 1 (MC1R), and Vitamin D3 receptor (VDR) were identified by molecular docking analysis as the putative targets of HCQ during vitiligo treatment (Table 3). The 4 key targets exhibited the most solid chemical binding forces and spatial conjunctions with HCQ. Importantly, the docking results showed that ionic bonds, hydrogen bonds and π-π stacking interactions were the predominant chemical forces. For instance, the hydroxyl, carbonyl and amino groups within HCQ formed hydrogen bonds with target proteins. The benzene and aromatic rings of HCQ formed π-π stacking interactions with target proteins (Figure 4).
 |
Table 3 The Docking Results of Target Candidates |
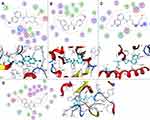 |
Figure 4 Molecular docking verification. Target candidates (A) ACHE, (B) VDR, (C) MC1R, and (D) PNMT were shown interacting with HCQ molecule (represented by a green ball-and-stick model). |
RNA Sequencing Analysis of PIG3V After HCQ Treatment Suggests a Protective Effect
PIG3V is an immortalized cell line derived from human vitiligo melanocytes. To better understand the state of PIG3V cells after HCQ (1μg/L) treatment for 24 hours, we analyzed the transcriptome of PIG3V. We found 108 and 97 DEGs were upregulated and downregulated by 2 fold or more in PIG3V cells, respectively, after HCQ treatment compared to the PIG3V control (p value<0.05). The top 30 upregulated and down-regulated genes in PIG3V cells were selected according to the value of | log2(FC) | and displayed in Table 4. As for the melanin synthesis pathway, BLOC1S5 was highly upregulated by 138005.020 fold after HCQ treatment compared to the control group, suggesting that melanin synthesis was significantly enhanced. In addition, two upregulated DEGs, MSRB3 and UVSSA, were enriched in melanocyte protection, which indicated an antioxidant effect in PIG3V cells after HCQ treatment. In addition, many upregulated genes were involved in modulating the activity of the immune system during acute-phase reactions such as ORM1, SAA1, SAA2, HP, FGA, FGB, FGG, CRP, AMBP, PTPRC, SERPINA1, and SERPINA3. These “acute-phase genes” may be related to the antibiotic effect of HCQ, acting as guards against the invasion of harmful microorganisms such as plasmodium.39 In contrast to “acute-phase genes”, some “chronic-phase genes” were downregulated, including COL1A1, DCN, and ELN. The downregulated genes were associated with decreased collagen synthesis and fibrosis, which are features of chronic inflammation. This finding may be a potential mechanism of HCQ in treating RA and other rheumatic diseases. Moreover, current evidence suggests that COL1A1 and ELN downregulation are associated with skin aging.40,41 KEGG pathway analysis showed that platelet activation, protein digestion, absorption, and herpes simplex virus 1 infection were significantly enriched pathways. GO enrichment analysis showed that GO terms, including acute-phase response, collagen-containing extracellular matrix, and extracellular matrix organization, were significantly enriched for molecular functions (Figure 5). However, it remains unclear whether these “acute-phase genes” are linked to vitiligo.
 |
Table 4 Differential Gene Expression of PIG3V After HCQ Treatment |
PPI Network Between HCQ Targets and Differentially Expressed Genes
To validate that ACHE, PNMT, MC1R, and VDR were targets of HCQ during vitiligo treatment, the differentially expressed genes in PIG3V cells were detected by RNA sequencing. Except for acute phase genes and other genes detected by RNA sequencing, genes potentially related to vitiligo or melanocyte (including BLOC1S5, MSRB3 and UVSSA) underwent PPI network analysis with docked HCQ targets, yielding a total of 163 nodes (proteins) along with 1019 edges (interactions) (Figure 6).
Discussions
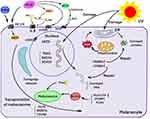
In the present study, the transcriptomic results showed that BLOC1S5 gene expression exhibited the highest fold change (138005.020 fold) after PIG3V cells were treated with HCQ. It has been established that BLOC1S5 encodes a subunit of the biogenesis of the lysosome-related organelles complex (BLOC-1), which is involved in melanosomal biogenesis.42 Oculocutaneous depigmentation is widely acknowledged as one of the characteristics of Hermansky-Pudlak Syndrome (HPS). Recent studies have demonstrated the presence of pathogenic BLOC1S5 variants in HPS.43,44 Moreover, it has been shown that knockdown of BLOC1S5 in zebrafish led to retinal depigmentation.45 In addition, a significant association between single nucleotide polymorphism (SNP) of 3 genes, including BLOC1S5, involved in skin pigmentation and 25-(OH)D serum concentration has been found.46 In the present study, network pharmacology analysis demonstrated that Vitamin D3 receptor (VDR) was a predicted target of HCQ, which may play a role similar to 25-(OH)D. In addition, another docking target of HCQ, MC1R, is a documented G-protein-coupled receptor that plays a vital role in skin pigmentation. It has been reported that α melanocyte-stimulating hormone (α-MSH) could stimulate cAMP signaling and melanin production after combination with MC1R, enhancing the repair of damaged DNA and protein after UV exposure.45 The MC1R/cAMP/MITF signaling pathway is well-established to regulate UV-induced pigmentation.46 Although no direct relationship between MC1R/VDR and BLOC1S5 was found in our PPI network, the highly upregulated BLOC1S5 indicated modulation of this pathway (Figure 7). Further studies are warranted in the future to validate this finding.
Current evidence suggests that in vitiligo patients, organelle functions of melanocytes are damaged in response to external triggers such as UV and chemical stimuli, leading to the attack of the body’s immune system.7 Our transcriptomic analysis showed that DEGs, including MSRB3 and UVSSA, were upregulated 2 fold or more after treatment with HCQ. These two genes are widely acknowledged for protein and DNA repair and maintaining organelle homeostasis. It has been shown that MSRB3 encodes methionine-R-sulfoxide reductase (MSR). In response to environmental stress, many reactive oxygen species (ROS) are produced. ROS can oxidize methionine (Met) residues in protein peptides to form methionine sulfoxide [Met(O)], which leads to protein function impairment. Importantly, MSR could catalyze the reduction of Met(O) to restore the function of proteins in response to oxidative damage. Accordingly, MSRB3 plays a pivotal role in cell protection under environmental stress.47 Moreover, it has been shown that UVSSA encodes for the UV-stimulated scaffold protein A, a transcription-coupled nucleotide excision repair (TCNER) factor, in response to UV damage. TCNER stabilizes gene ERCC6 by recruiting the enzyme USP7 into the TCNER complex, preventing UV-induced ERCC6 degradation by proteasomes.48 TCNER induces the removal of RNA polymerase II (RNA pol II) from the active genes for transcription. Subsequently, ubiquitination at UV-damaged sites can accelerate RNA pol II recalling to nucleotide excision repair machinery.49 According to the molecular docking and PPI network analysis results (Figure 6), the MC1R-MSRB3/UVSSA axis could be an essential mechanism of HCQ in the process of melanocyte protection against oxidative stress, and the MC1R agonist could serve as a cutaneous pigmentation promoter. An increasing body of evidence suggests that activation of MC1R alleviates oxidative stress and neuronal apoptosis through protein kinase R-like endoplasmic reticulum kinase (PERK)-nuclear factor erythroid 2-related factor 2 (NRF2) pathway, exhibiting an antioxidant role in the unfolded protein response (UPR) system. As the UPR system is initiated, impaired (unfolded, misfolded, etc.) proteins are restored by enzymes through a series of reactions.7,50
As molecular docking targets of HCQ, ACHE and PNMT potentially play important roles in vitiligo treatment. Non-neuronal acetylcholine (ACh) is well-recognized to regulate human keratinocyte (KCs) functions, including cell differentiation, cell-cell interaction, secretion, and mitosis. Besides, ACh plays an important role in immune regulation. It has been established that the Ach receptor is expressed in both KCs and melanocytes.51 A study reported that the average levels of ACh were higher in areas of skin depigmentation compared to controls. Interestingly, the level of ACh was significantly decreased after treatment, which showed a significant positive correlation with the severity of vitiligo.52 A study by Taieb et al53 substantiated the toxic effect of ACh on melanocytes. In addition to ACHE, PNMT, which catalyzes the synthesis of adrenaline (AD) from noradrenaline (NA), plays an important role in vitiligo. It has been shown that KCs could synthesize NA and AD. Current evidence suggests that KCs in the lesion area of vitiligo patients synthesized 4 times more NA than normal skin area, with low PNMT activity.54 Therefore, increasing the activity of ACHE and PNMT to reduce skin ACh and NA may be one of the mechanisms underlying the efficacy of HCQ in vitiligo treatment. Nonetheless, further studies are required to increase the robustness of our findings.
Conclusions
This present study uncovered HCQ targets (ACHE, PNMT, MC1R, and VDR) during vitiligo treatment after screening by network pharmacology and molecular docking. PIG3V cells were used to explore the mechanisms of HCQ treatment through transcriptomic analysis. The results of transcriptomic study further showed that through the above targets, HCQ significantly promoted the expression of genes related to melanin synthesis. In addition, the expression of the genes which are related to DNA and protein damage repair was also significantly up-regulated, indicating the protective effect of HCQ on vitiligo melanocytes. In silico methods were used to identify the relationships between HCQ targets and differentially expressed genes in PIG3V cells. These findings provided the theoretical basis for the mechanisms of HCQ in treating vitiligo (Figure 7). Nevertheless, there were some limitations in our study. The specific mechanisms and signal pathways of the MC1R/VDR-BLOC1S5 axis and MC1R-MSRB3/UVSSA axis involved in the pigmentation process and melanocytes protection effect were not explored. In addition, the effect of HCQ on ACHE and PNMT and further pathways were not assessed, warranting further studies.
Funding
This work was supported by [National Natural Science Foundation of China] under Grant [number 81872517].
Disclosure
The authors declare that they have no conflicts of interest.
References
1. Ezzedine K, Eleftheriadou V, Whitton M, et al. Vitiligo. Lancet. 2015;386:74–84. doi:10.1016/S0140-6736(14)60763-7
2. Bergqvist C, Ezzedine K. Vitiligo: a review. Dermatology. 2020;236:571–592. doi:10.1159/000506103
3. Alikhan A, Felsten LM, Daly M, et al. Vitiligo: a comprehensive overview part I. Introduction, epidemiology, quality of life, diagnosis, differential diagnosis, associations, histopathology, etiology, and work-up. J Am Acad Dermatol. 2011;65:473–491. doi:10.1016/j.jaad.2010.11.061
4. Picardo M, Dell’Anna ML, Ezzedine K, et al. Vitiligo. Nat Rev Dis Primers. 2015;1:15011. doi:10.1038/nrdp.2015.11
5. Strassner JP, Harris JE. Understanding mechanisms of autoimmunity through translational research in vitiligo. Curr Opin Immunol. 2016;43:81–88. doi:10.1016/j.coi.2016.09.008
6. Laddha NC, Dwivedi M, Mansuri MS, et al. Role of oxidative stress and autoimmunity in onset and progression of vitiligo. Exp Dermatol. 2014;23:352–353. doi:10.1111/exd.12372
7. Xie B, Song XZ. The impaired unfolded protein-premelanosome protein and transient receptor potential channels-autophagy axes in apoptotic melanocytes in vitiligo. Pigment Cell Melanoma Res. 2022;35:6–17. doi:10.1111/pcmr.13006
8. Al-Shobaili HA, Rasheed Z. Mitochondrial DNA acquires immunogenicity on exposure to nitrosative stress in patients with vitiligo. Hum Immunol. 2014;75:1053–1061. doi:10.1016/j.humimm.2014.09.003
9. Xie H, Zhou F, Liu L, et al. Vitiligo: how do oxidative stress-induced autoantigens trigger autoimmunity? J Dermatol Sci. 2016;81:3–9. doi:10.1016/j.jdermsci.2015.09.003
10. Joo K, Park W, Kwon SR, et al. Improvement of vitiligo in a patient with rheumatoid arthritis after hydroxychloroquine treatment. Int J Rheum Dis. 2015;18:679–680. doi:10.1111/1756-185X.12442
11. Fanouriakis A, Kostopoulou M, Alunno A, et al. 2019 update of the EULAR recommendations for the management of systemic lupus erythematosus. Ann Rheum Dis. 2019;78(6):736–745. doi:10.1136/annrheumdis-2019-215089
12. Smolen JS, Landewé RBM, Bijlsma JWJ, et al. EULAR recommendations for the management of rheumatoid arthritis with synthetic and biological disease-modifying antirheumatic drugs: 2019 update. Ann Rheum Dis. 2020;79(6):685–699. doi:10.1136/annrheumdis-2019-216655
13. Ramos-Casals M, Brito-Zerón P, Bombardieri S, et al. EULAR recommendations for the management of Sjögren’s syndrome with topical and systemic therapies. Ann Rheum Dis. 2020;79:3–18. doi:10.1136/annrheumdis-2019-216114
14. Chew CY, Mar A, Nikpour M, et al. Hydroxychloroquine in dermatology: new perspectives on an old drug. Australas J Dermatol. 2020;61:e150–e157. doi:10.1111/ajd.13168
15. Niebel D, Braegelmann C, Bieber T, et al. Vitiligo-like depigmentation subsequent to subacute cutaneous lupus erythematosus and hydroxychloroquine treatment. J Dtsch Dermatol Ges. 2020;18:1470–1473.
16. Walsh DS, Farley MF, Beard JS, et al. Systemic lupus erythematosus: nephritis, dilated cardiomyopathy, and extensive cutaneous depigmentation responsive to hydroxychloroquine. J Am Acad Dermatol. 1995;33:828–830. doi:10.1016/0190-9622(95)91842-6
17. Kwak D, Grimes Pearl E. A case of hyperpigmentation induced by hydroxychloroquine and quinacrine in a patient with systemic lupus erythematosus and review of the literature. Int J Womens Dermatol. 2020;6:268–271. doi:10.1016/j.ijwd.2020.06.009
18. Khan DA. Alternative agents in refractory chronic urticaria: evidence and considerations on their selection and use. J Allergy Clin Immunol Pract. 2013;1(5):433–440. doi:10.1016/j.jaip.2013.06.003
19. Li DG, Hu WZ, Ma HJ, et al. Hydroxychloroquine protects melanocytes from autoantibody-induced injury by reducing the binding of antigen-antibody complexes. Mol Med Rep. 2016;14:1275–1282. doi:10.3892/mmr.2016.5354
20. Ma C, Xu T, Sun X, et al. Network pharmacology and bioinformatics approach reveals the therapeutic mechanism of action of baicalein in hepatocellular carcinoma. Evid Based Complement Alternat Med. 2019:7518374. doi:10.1155/2019/7518374
21. Luo TT, Lu Y, Yan SK, et al. Network pharmacology in research of Chinese medicine formula: methodology, application and prospective. Chin J Integr Med. 2020;26(1):1–9. doi:10.1007/s11655-019-3064-0
22. Ding M, Ma W, Wang X, et al. A network pharmacology integrated pharmacokinetics strategy for uncovering pharmacological mechanism of compounds absorbed into the blood of Dan-Lou tablet on coronary heart disease. J Ethnopharmacol. 2019;242:112055. doi:10.1016/j.jep.2019.112055
23. Zhang H, Yan ZY, Wang YX, et al. Network pharmacology-based screening of the active ingredients and potential targets of the genus of Pithecellobium marthae (Britton & Killip) Niezgoda & Nevl for application to Alzheimer’s disease. Nat Prod Res. 2019;33(16):2368–2371. doi:10.1080/14786419.2018.1440222
24. Otasek D, Morris JH, Bouças J, et al. Cytoscape automation: empowering workflow-based network analysis. Genome Biol. 2019;20(1):185–200. doi:10.1186/s13059-019-1758-4
25. Xie B, Geng Q, Xu J, et al. The multi-targets mechanism of hydroxychloroquine in the treatment of systemic lupus erythematosus based on network pharmacology. Lupus. 2020;29:1704–1711. doi:10.1177/0961203320952541
26. Stelzer G, Rosen R, Plaschkes I, et al. The genecards suite: from gene data mining to disease genome sequence analysis. Curr Protoc Bioinform. 2016;54. doi:10.1002/cpbi.5
27. Xie B, Lu H, Jinhui X, et al. Targets of hydroxychloroquine in the treatment of rheumatoid arthritis. A network pharmacology study. Joint Bone Spine. 2021;88:105099. doi:10.1016/j.jbspin.2020.105099
28. Martin A, Ochagavia ME, Rabasa LC, et al. BisoGenet: a new tool for gene network building, visualization and analysis. BMC Bioinform. 2010;11(1):91–99. doi:10.1186/1471-2105-11-91
29. Rizvi SM, Shakil S, Haneef M. A simple click by click protocol to perform docking: autoDock 4.2 made easy for non-bioinformaticians. EXCLI J. 2013;12:831–857.
30. Tian J, Wang Y, Ding M, et al. The formation of melanocyte apoptotic bodies in vitiligo and the relocation of vitiligo autoantigens under oxidative stress. Oxid Med Cell Longev. 2021;2021:7617839. doi:10.1155/2021/7617839
31. Beekman R, Chapaprieta V, Russiñol N, et al. The reference epigenome and regulatory chromatin landscape of chronic lymphocytic leukemia. Nat Med. 2018;24(6):868–880. doi:10.1038/s41591-018-0028-4
32. Wang LG, Wang SQ, Li W. RSeQC: quality control of RNA-seq experiments. Bioinformatics. 2012;28:2184–2185. doi:10.1093/bioinformatics/bts356
33. Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from next-generation sequencing data. Nucleic Acids Res. 2010;38:e164. doi:10.1093/nar/gkq603
34. Florea L, Song L, Salzberg SL. Thousands of exon skipping events differentiate among splicing patterns in sixteen human tissues. F1000Res. 2013;2:188. doi:10.12688/f1000research.2-188.v1
35. Robinson MD, McCarthy DJ, Smyth GK. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi:10.1093/bioinformatics/btp616
36. Pertea M, Pertea GM, Antonescu CM, et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 2015;33:290–295. doi:10.1038/nbt.3122
37. Li AP, Liu Y, Cui T, et al. Uncovering the mechanism of Astragali Radix against nephrotic syndrome by intergrating lipidomics and network pharmacology. Phytomedicine. 2020;77:153274. doi:10.1016/j.phymed.2020.153274
38. Li T, Zhang W, Hu E, et al. Integrated metabolomics and network pharmacology to reveal the mechanisms of hydroxysafflor yellow A against acute traumatic brain injury. Comput Struct Biotechnol J. 2021;19:1002–1013. doi:10.1016/j.csbj.2021.01.033
39. Raju MVJ, Kamaraju RS, Sritharan V, et al. Continuous evaluation of changes in the serum proteome from early to late stages of sepsis caused by Klebsiella pneumoniae. Mol Med Rep. 2016;13:4835–4844. doi:10.3892/mmr.2016.5112
40. Bigot N, Beauchef G, Hervieu M, et al. NF-κB accumulation associated with COL1A1 transactivators defects during chronological aging represses type I collagen expression through a −112/-61-bp region of the COL1A1 promoter in human skin fibroblasts. J Invest Dermatol. 2012;132:2360–2367. doi:10.1038/jid.2012.164
41. Osborne R, Carver RS, Mullins LA, et al. Practical application of cellular bioenergetics to the care of aged skin. Br J Dermatol. 2013;169:32–38. doi:10.1111/bjd.12439
42. Rossberg W, Saternus R, Wagenpfeil S, et al. Human pigmentation, cutaneous vitamin D synthesis and evolution: variants of genes (SNPs) involved in skin pigmentation are associated with 25(OH)D serum concentration. Anticancer Res. 2016;36:1429–1437.
43. Pennamen P, Le L, Tingaud-Sequeira A, et al. BLOC1S5 pathogenic variants cause a new type of Hermansky-Pudlak syndrome. Genet Med. 2020;22:1613–1622. doi:10.1038/s41436-020-0867-5
44. Zhong Z, Wu Z, Zhang J, et al. A novel BLOC1S5-related HPS-11 patient and zebrafish with bloc1s5 disruption. Pigment Cell Melanoma Res. 2021;34:1112–1119. doi:10.1111/pcmr.12995
45. Rachmin I, Ostrowski SM, Weng QY, et al. Topical treatment strategies to manipulate human skin pigmentation. Adv Drug Deliv Rev. 2020;153:65–71. doi:10.1016/j.addr.2020.02.002
46. Fu S, Luo X, Wu X, et al. Activation of the melanocortin-1 receptor by NDP-MSH attenuates oxidative stress and neuronal apoptosis through PI3K/Akt/Nrf2 pathway after intracerebral hemorrhage in mice. Oxid Med Cell Longev. 2020;2020:8864100. doi:10.1155/2020/8864100
47. Zhang XH, Weissbach H. Origin and evolution of the protein-repairing enzymes methionine sulphoxide reductases. Biol Rev Camb Philos Soc. 2008;83(3):249–257. doi:10.1111/j.1469-185X.2008.00042.x
48. Wienholz F, Zhou D, Turkyilmaz Y, et al. FACT subunit Spt16 controls UVSSA recruitment to lesion-stalled RNA Pol II and stimulates TC-NER. Nucleic Acids Res. 2019;47:4011–4025. doi:10.1093/nar/gkz055
49. Higa M, Zhang X, Tanaka K, et al. Stabilization of Ultraviolet (UV)-stimulated scaffold protein A by interaction with ubiquitin-specific peptidase 7 is essential for transcription-coupled nucleotide excision repair. J Biol Chem. 2016;291:13771–13779. doi:10.1074/jbc.M116.724658
50. Jackson E, Heidl M, Imfeld D, et al. Discovery of a highly selective MC1R agonists pentapeptide to be used as a skin pigmentation enhancer and with potential anti-aging properties. Int J Mol Sci. 2019;20:6143. doi:10.3390/ijms20246143
51. Wessler I, Kilbinger H, Bittinger F, et al. The non-neuronal cholinergic system in humans: expression, function and pathophysiology. Life Sci. 2003;72:2055–2061. doi:10.1016/S0024-3205(03)00083-3
52. Said ER, Nagui NA, Rashed LA, et al. Oxidative stress and the cholinergic system in non-segmental vitiligo: effect of narrow band ultraviolet b. Photodermatol Photoimmunol Photomed. 2021;37:306–312. doi:10.1111/phpp.12653
53. Taieb A. Intrinsic and extrinsic pathomechanisms in vitiligo. Pigment Cell Res. 2000;13(suppl 8):41–47. doi:10.1034/j.1600-0749.13.s8.9.x
54. Schallreuter KU, Wood JM, Pittelkow MR, et al. Increased monoamine oxidase A activity in the epidermis of patients with vitiligo. Arch Dermatol Res. 1996;288:14–18. doi:10.1007/BF02505037
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.