Back to Journals » Infection and Drug Resistance » Volume 15
Targeted Elimination of blaNDM-5 Gene in Escherichia coli by Conjugative CRISPR-Cas9 System
Authors Li P, Wan P, Zhao R, Chen J, Li X, Li J, Xiong W, Zeng Z
Received 20 January 2022
Accepted for publication 19 March 2022
Published 8 April 2022 Volume 2022:15 Pages 1707—1716
DOI https://doi.org/10.2147/IDR.S357470
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Peisi Li,1– 3 Peng Wan,1– 3 Ruonan Zhao,1– 3 Jin Chen,1– 3 Xiaoshen Li,1– 3 Jie Li,1– 3 Wenguang Xiong,1– 3 Zhenling Zeng1– 3
1Guangdong Provincial Key Laboratory of Veterinary Pharmaceutics Development and Safety Evaluation, South China Agricultural University, Guangzhou, 510642, People’s Republic of China; 2National Laboratory of Safety Evaluation (Environmental Assessment) of Veterinary Drugs, College of Veterinary Medicine, South China Agricultural University, Guangzhou, 510642, People’s Republic of China; 3National Risk Assessment Laboratory for Antimicrobial Resistance of Animal Original Bacteria, South China Agricultural University, Guangzhou, 510642, People’s Republic of China
Correspondence: Zhenling Zeng; Wenguang Xiong, Tel +862085281204, Fax +862085284896, Email [email protected]; [email protected]
Purpose: Plasmid-borne carbapenem resistance gene blaNDM-5 accelerates the dissemination of carbapenem-resistant Enterobacteriaceae. To efficiently eliminate the blaNDM-5-harboring plasmid and sensitize the antibiotic-resistant bacteria to meropenem, we used the CRISPR-Cas9 system for combating the carbapenem-resistant Escherichia coli (E. coil).
Methods: A series of CRISPR-Cas9 plasmids was constructed, and specific guide RNAs(sgRNA) were designed to target the blaNDM-5 gene. We used chemically transformation or conjugation delivery methods, and the elimination efficiency in each recipient strains was evaluated by plate counting, PCR and quantitative real-time PCR (qPCR). Antimicrobial susceptibility test was carried out by using the broth microdilution method. In addition, we assessed the effect of the CRISPR-Cas9 system of adaptive immunity on the prevention of the exogenous resistant plasmids pNDM-5 by introducing the system into E coli J53.
Results: The results showed that pCas9, pCas9-oriT and pBAD-Cas9-oriT can effectively eliminate blaNDM-5 in E. coli with > 94.00% elimination efficiency. The blaNDM-5-harboring E. coli successfully restored their susceptibility to meropenem, with eight-fold reduction of minimum inhibitory concentration (MIC) values (from 16 μg/mL to 0.06 μg/mL). The E. coli J53 strain containing plasmid pCas9-N reduced the number of transconjugants by 26-fold.
Conclusion: The CRISPR-Cas9 system achieved plasmid clearance and simultaneous re-sensitization to meropenem in E. coli. The CRISPR-Cas9 system could block the horizontal transfer of plasmid pNDM-5. The conjugative delivery of CRISPR-Cas9 provides a new tool for the removal of resistance plasmids and sensitize the recipient to carbapenem. It provides a therapeutic approach to counteract the propagation of blaNDM-5 gene among clinical pathogens.
Keywords: antimicrobial resistance, eliminating efficiency, plasmid conjugation, re-sensitization
Introduction
Antimicrobial resistant microorganisms have become a public health concern in recent decades.1 Horizontal gene transfer (HGT) is the main contributor to the rapid dissemination of antibiotic resistance genes (ARGs).2 The emergence and dissemination of carbapenem resistance in Enterobacteriaceae mediated by plasmid-borne carbapenemase genes such as blaNDM-5 have been a concern.3 The blaNDM-5 gene is carried by conjugative plasmids that can horizontally transfer, resulting in multidrug or extensively drug-resistant phenotypes.4 Therefore, novel strategies to combat plasmid-mediated antimicrobial resistance should be developed.
The CRISPR-Cas9 (Clustered Regularly Interspaced Short Palindromic Repeats ‒ CRISPR-associated protein-9 nuclease) system provides an effective tool to eradicate ARGs and plasmids carrying ARGs by cleaving double-stranded DNA.5 After the sgRNA targets specific sequences and recruits Cas9 protein to form functional complex, the Cas9 protein acts as a nuclease and generates a blunt end double-strand break (DSB). Considering the lack of the error-prone non-homologous end-joining pathway in prokaryotes, Cas9-mediated DSB cannot be repaired spontaneously, resulting in DSB in plasmid sequence and thus triggering plasmid elimination.6
Existing bacterial CRISPR-Cas systems can target ARGs and observe a CRISPR-Cas mediated cytotoxicity in different bacteria.7–9 Subsequent studies have developed several CRISPR-Cas9 systems for specific elimination of plasmids in bacterial hosts that carry multiple resistant plasmids.10−13 The mobile genetic elements (MGE)-associated CRISPR-Cas9 may serve as a therapeutic approach to control the dissemination of antibiotic resistance among clinical pathogens.14,15 These works support that CRISPR-Cas9 system can solve the problem of multi-drug resistance in clinical and environmental settings. However, concerning the CRISPR-Cas9 system in clinical application, the delivery method must be developed and optimized.16 Bacterial conjugation may be a feasible way to deliver the CRISPR-Cas9 into bacteria in their natural environment.17–19 In the present study, we investigated the effect of CRISPR-Cas9 system on the elimination blaNDM-5 gene and explored the potential of CRISPR-Cas9 system deliver methods via bacterial conjugative transfer to control the dissemination of blaNDM-5 resistance.
Materials and Methods
Bacterial Strains and Plasmids
The strains and plasmids used in this study are listed in Table 1, while the primers used in this study are listed in Table 2. E. coli 01 containing blaNDM-5 and E. coli J53 were prepared for conjugation experiments.20 Donor strain E. coli 01 and recipient strain E. coli J53 grown separately in LB medium were cultured at 37°C. Equal volumes of the donor culture and the recipient culture were mixed, and then incubated at 37 °C. After 12 h, dilutions of the mixtures were plated on Luria-Bertani (LB) agar plates. Transconjugants, named as E. coli 02, were incubated at 37 °C in Luria-LB agar plates containing with chloramphenicol (50 µg/mL) and sodium azide (200 µg/mL). E. coli 02 was verified by PCR via using the primers NDM-5-F/R. E. coli strains were grown in LB broth and agar plates at 37 °C. When needed, antibiotics such as chloramphenicol (50 µg/mL), meropenem (1 µg/mL) and sodium azide (200 µg/mL) were added.
 |
Table 1 Bacterial Strains and Plasmids Used in This Study |
 |
Table 2 Primers Used in This Study |
Plasmid Construction
A 20 nt sgRNA (N-F/R) targeting the blaNDM-5 gene was designed using the CHOPCHOP tool while adding BsaI digestion sites. The primers were annealed and ligated with BsaI-digested pCas9 by using T4 DNA ligase (NEB), and the recombinant plasmids named pCas9-N.21 The recombinant plasmids were transformed into E. coli DH5α by using the heat-shock method,22 and the inserted sgRNA sequences were verified using primers DR-F/R. The plasmid pCas9 without the sgRNA cassette was used as a control plasmid.
Bacterial conjugation via plasmid RP4 requires the origin of transfer (oriTRP4), while the other proteins necessary for conjugation were obtained from E. coli S17-1, which contains chromosomally integrated genes from the natural conjugative plasmid RP4. The OriTRP4 fragment was amplified from pBBR1MCS-2 by using primers oriT-XbaI-F/R and inserted into XbaI-digested pCas9 and pBAD-Cas9, resulting in conjugative plasmids pCas9-oriT and pBAD-Cas9-oriT.23 Subsequently, 20 nt sgRNA ligated with BsaI-digested pCas9-oriT and pBAD-Cas9-oriT were prepared and named as pCas9-oriT-N and pBAD-Cas9-oriT-N by using T4 DNA ligase (NEB).21 The recombinant plasmids were transformed into E. coli S17-1 by using the heat-shock method.17,22 The plasmid pCas9-oriT and pBAD-Cas9-oriT, which lack the sgRNA cassette, were used as a control plasmid.
Plasmid Transformation and pNDM-5 Eliminative Efficiency
Chemically competent E. coli 02 samples were prepared using 0.1 M MgCl2–CaCl2 and competent cells were transformed with plasmids pCas9-N by using the heat-shock method at 42 °C for 90s.22 In addition, different plasmid types have different replicons and backbone structures, and the effects of different plasmid types (IncFII and lncX3) were analyzed.4,24 In brief, chemically competent E. coli 03 was transformed with plasmids pCas9-N. The culture was diluted and plated on LB agar plates with chloramphenicol (50 µg/mL) only or the combination of chloramphenicol (50 µg/mL) and meropenem (1 µg/mL) to determine the number of colony-forming units (CFU). Elimination efficiency was calculated using the equation (1 – colonies grown on chloramphenicol and meropenem plates/ colonies grown on chloramphenicol plates) × 100%. The empty plasmid pCas9 was used as the negative control. Each experiment was performed in triplicate.
To further analyze the efficiency of pCas9 on eliminating blaNDM-5 gene, we detected the change of pNDM-5 copy number at each time point via qPCR. Genomic DNA was extracted using TIANamp bacteria DNA kit (Tiangen, Beijing, China). qPCR was performed using SYBRP remix ExTaqII (Takara, Dalian, China) with primers 16S-F/-R and qPCR-NDM-F/R.25 The 2−ΔΔCTmethod was used to calculate the change of pNDM-5 copy number in the experimental group compared with the control group. All reactions were run in triplicate.
Conjugation Assays Plasmids and Eliminating Experiment
Donor strain E. coli S17-1 containing plasmid pCas9-oriT-N and recipient strain E. coli 02 grown separately in LB medium were cultured overnight and grew at an OD600 value of 0.4 at 37 °C.17,26 Equal volumes of the donor culture and the recipient culture were mixed, and then incubated at 37 °C. After 12 h, dilutions of the mixtures were plated on LB agar plates with chloramphenicol (50 µg/mL) only or the combination of chloramphenicol (50 µg/mL) and sodium azide (200 µg/mL) to select donor and transconjugant clones. The plates were incubated overnight at 37 °C. Colonies grown on the plates were enumerated, and the CFU were calculated. Conjugation efficiency was calculated by dividing the number of transconjugant strains by the number of donor strains. A total of 24 transconjugant strains were randomly picked for evaluating the eliminating efficiency via PCR. The plasmids pCas9-oriT and pNDM-5 were detected with primers pCas9-F/R and NDM-5-F/R, respectively. The empty plasmid pCas9-oriT was used as negative control. Each experiment was performed in triplicate.
Eliminating Condition – Incubation Time and Arabinose Concentrations
We verified the feasibility of CRISPR-Cas9 system via conjunctive delivery method. Furthermore, we determined whether efficient clearance can be achieved in other conjunctive plasmids. Notably, we then evaluated the target eliminating efficiency under different arabinose induction concentrations and incubation time.27 After introducing pBAD-Cas9-oriT-N into E. coli 02 via conjugation, transconjugant strains were selected using LB agar plates supplemented with chloramphenicol (50 µg/mL), and PCR confirmation was carried out using primers pCas9-F/R and NDM-5-F/R. Overnight culture of pBAD-Cas9-oriT-N positive E. coli 02 was diluted 100-fold in 2 mL of LB broth containing 0.01%, 0.10%, 1.00% or 2.00% arabinose in combination with chloramphenicol (50 µg/mL). The cultures were incubated for 6 h at 37 °C with shaking at 180–200 rpm. After 6 h, the cultures were diluted and simultaneously plated on LB agar plates containing chloramphenicol (50 µg/mL) only or combination of chloramphenicol (50 µg/mL) and meropenem (1 µg/mL) to determine the number of CFU. Elimination efficiency was calculated using the equation (1–colonies grown on chloramphenicol and meropenem plates/ colonies grown on chloramphenicol plates) × 100%. The experiments were conducted using three randomly picked pBAD-Cas9-oriT-N positive E. coli 02 colonies.
Thereafter, the target eliminating efficiency under different arabinose incubation times was evaluated. Similarly, the overnight cultures of pBAD-Cas9-oriT-N positive E. coli 02 colonies were diluted 100-fold in 2 mL of LB broth containing 1.00% arabinose and chloramphenicol (50 µg/mL), followed by incubation at 37 °C with shaking for 16 h. At 0, 2, 4, 6, 8 and 16 h, 100 µL of culture were removed, diluted, and plated out for colony counting. The curing efficiency was calculated as above.
Antimicrobial Susceptibility Tests
The MIC of meropenem was determined using the broth microdilution method in accordance with the guide lines of Clinical and Laboratory Standards Institute.28 The strains contained original E. coli 02 and the corresponding plasmid-eliminated strains eliminating blaNDM-5 gene. E. coli ATCC 25922 was routinely used as quality control strain. Each experiment was performed in triplicate.
The CRISPR-Cas9 System Preventing Plasmid Acquisition
We used E. coli J53 containing plasmid pCas9 as the recipient to assess whether the integration of CRISPR-Cas9 can impede the acquisition of pNDM-5 via conjugation, and the donor was E. coli J53 contained plasmid pNDM-5. The conjugation assay was performed as described above. The blocking of blaNDM-5 transfer was evaluated based on the number of transconjugants. Each experiment was performed in triplicate.
Statistical Analysis
Statistical analyses were performed using the GraphPad Prism software (version 6, GraphPad Software, CA, United States).
Results
Construction of CRISPR-Cas9 Plasmids Targeting blaNDM-5
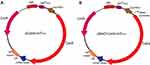
Plasmid pCas9-N harbors essential elements, including Cas9, tracrRNA and crRNA, in combination with the 20 nt sgRNA. To test the utility of the CRISPR-Cas9 system via conjugation, we used pCas9 and pBAD-Cas9 as the backbone to construct the pCas9-oriT and pBAD-Cas9-oriT, which ligate the oriTRP4, and plasmid mapping as shown in Figure 1.
Elimination of Plasmid by Using the pCas9-sgRNA
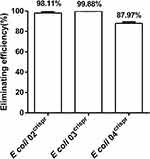
We firstly examined whether plasmid elimination can be achieved by targeting blaNDM-5. The pCas9-N were transformed into E. coli 02 bearing pNDM-5 plasmid with IncFII-type. The result showed that 98.11% bacterial population lost their corresponding target genes (Figure 2). Then, the elimination efficiency under high copy number pressure was verified by transforming the pCas9-N into E. coli 03 containing the high copy numbers plasmid pUC19-blaNDM-5. The result showed that 99.88% bacterial lost their corresponding resistant genes. In addition, pCas9-N targeted the different plasmid type, and the pCas9-N was transformed into E. coli 04 with IncX3-type. pCas9-N targeted IncFII and IncX3 could also remove pNDM-5 plasmids, and the elimination efficiency reached 87.00%. Plasmid removal could be achieved through CRISPR-Cas9 mediated cleavage with different pNDM-5 plasmid.
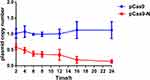
To accurately evaluate the elimination efficiency, qPCR-based detection of the blaNDM-5 gene was conducted at each time point in pCas9-N and pCas9 control groups, and the results showed that the elimination efficiency of the experimental group was greater than 80.00% at the 16th hour and continued to increase until the 24th hour, and a substantial plasmid reduction had occurred in E. coli 02 (Figure 3).
Elimination of Plasmid Using the pCas9-oriT-sgRNA via Conjugation
E. coli S17-1 samples containing pCas9-oriT-N and pCas9-oriT were used as donor cells. E. coli 02 samples were used as recipient cells. The conjugation efficiency was estimated at (2.29±0.77) *10−2, while the elimination efficiency was (1–1/24) ×100%=95.83% (Figure 4). Therefore, the CRISPR-Cas9 system via conjunctive delivery method could be achieved with high elimination efficiency.
Elimination of Plasmid by Using the pBAD-Cas9-oriT-sgRNA via Conjugation
We initially investigated pBAD-Cas9-oriT-N elimination efficiency in E. coli 02 under different arabinose induction concentrations (0.01%, 0.10%, 1.00% and 2.00%). The results showed that after 6 h of incubation, the elimination efficiencies reached 97.34%, 99.84%, 99.86% and 99.74%. (Figure 5A). In addition, we examined elimination efficiencies at different incubation times. The results showed that under 1.00% arabinose treatment, the elimination efficiencies reached 76.80%, 99.85%, 99.87%, 99.69% and 68.37% at 2, 4, 6, 8 and 16 h, respectively (Figure 5B). Based on both arabinose concentration and incubation time, 1.00% arabinose and induction time of 6 h were the best eliminating conditions with 99.00% pNDM-5 removal, which achieved efficient clearance with conjunctive plasmids by CRISPR-Cas9.
Antimicrobial Susceptibility Tests
We examined the MIC of meropenem for E coli 02 and corresponding plasmid-eliminated strain. The results showed that the MIC of meropenem in the eliminated isolates were reduced by eight-fold from 16 µg/mL to 0.06 µg/mL. The elimination of blaNDM-5 gene effectively restored the carbapenem susceptibility.
Plasmid Conjugation Blocking
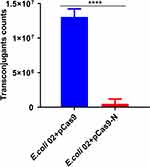
E. coli 02, which carries the conjugation plasmid pNDM-5, was used as a donor. The E. coli J53, which was transformed with a control plasmid pCas9 or a recombinant plasmid pCas9-N, was used as recipient. As shown in Figure 6, the E. coli J53 containing plasmid pCas9-N reduced the number of transconjugants by 26 times compared with the control plasmid pCas9 (1.31E+07 vs 5.00E+05 CFU/mL; p<0.0001). The results demonstrated that the CRISPR-Cas9 system in the recipient cell could block the horizontal transfer of the conjugative plasmid pNDM-5.
Discussion
The HGT of ARGs plays an essential role in disseminating antimicrobial resistance.29 The occurrence and dissemination of carbapenem resistance gene blaNDM-5 in bacteria makes the treatment of clinical infections challenging.30 The re-sensitization of the sensitivity to traditional antibiotics might be more effective than searching for new antimicrobials.31 Therefore, new strategies to prevent the dissemination of blaNDM-5 should be developed.
In the present study, we investigated the potential of the CRISPR-Cas9 system to counteract plasmid-borne blaNDM-5 gene in wild-type plasmids (pNDM-5) and the high-copy numbers of plasmids (pUC19-NDM-5). The results showed that our pCas9 system could efficiently remove several carbapenem resistance plasmids by targeting blaNDM-5 genes and re-sensitize them to meropenem after the removal of plasmids pNDM-5. Comparing engineered high copy number plasmid (pUC19-NDM-5) to wild-type plasmid (pNDM-5) in elimination efficiency, The elimination efficiency of the high-copy plasmid is slightly higher than that of the wild-type plasmid. We guess that the high-copy plasmid is an engineered plasmid with a clear plasmid background, while the wild-type plasmid is clinically isolated and the background is more complicated. The pCas9 system can also eliminate other plasmid types such as lncX3. Hence, the content of plasmid backbone may have an influence on elimination efficiency. Our results were consistent with the previous findings that showing CRISPR-Cas9 can eliminate the resistance plasmid.15,32–34 Designing multiple sgRNAs that target different ARG subtypes and even different ARG types can prevent antimicrobial resistance.35,36 Multiple sgRNA for potentially targeting different gene, such as replication, partition, conjugation and plasmid stability, is a new strategy.12
However, an optimal deliver strategy to introduce the CRISPR-Cas9 system into the target bacterial populations remains a great challenge for plasmid or resistance gene elimination in the clinical application.37 Most of the previous studies used physical or chemical methods, such as electroporation and chemical transformation, but these approaches are restricted to the Laboratory research and are not be applied in the clinical application. Considering that bacterial conjugation is critical for the dissemination of antimicrobial resistance, we aimed to explore conjugative plasmids to eliminate antibiotic resistance plasmids in bacteria.17 Our results demonstrated that the blaNDM-5-harbouring plasmids in the recipient can be eliminated and sensitized to antibiotics via conjugative delivery of CRISPR-Cas9. For two recombinant conjugative plasmids, the elimination efficiencies reached 95.00%. Moreover, the conjugative delivery of CRISPR-Cas9 antimicrobials may be adaptable for the exact targeting of defined multidrug-resistant bacteria.15,17 Although our conjugative system has achieved compelling results, some limitations, such as the low conjugation efficiency (usually <10−1), limits the utility of this conjugative system to neutralize resistance at a population level. Conjugation efficiency may be influenced by time and biofilm structure.38 Research has shown that transfer efficiency can be artificial increased using glass beads in vitro.39 Similarly, F plasmid as a helper plasmid mediates efficient transfer.18 Further research should focus on screening broad-host-range conjugative plasmids and enhance conjugation efficiency.17 Considering that bacterial conjugation is a naturally occurring process, this strategy could be optimized for clinical application. Furthermore, phages and nanoparticles can be effectively used for the delivery of the CRISPR-Cas9 system into the bacterial pathogens.13,40 In addition to plasmid elimination, the CRISPR-Cas9 could further provide immunity in E. coli against the acquisition of plasmid-mediated ARGs.14,34
Conclusion
We provide a proof of principle that a novel CRISPR-Cas9 system eliminate carbapenem genes and plasmids to interrupt the HGT of ARGs. This method aims to achieve proactive prevention of the dissemination of carbapenem resistance in clinical pathogens.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grantno.31872524) and Local innovative and Research Teams Project of Guangdong Pearl River Talents Program (grant number 2019BT02N054).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Hernando-Amado S, Coque TM, Baquero F, Martinez JL. Defining and combating antibiotic resistance from one health and global health perspectives. Nat Microbiol. 2019;4(9):1432–1442. doi:10.1038/s41564-019-0503-9
2. Lerminiaux NA, Cameron ADS. Horizontal transfer of antibiotic resistance genes in clinical environments. Can J Microbiol. 2019;65(1):34–44. doi:10.1139/cjm-2018-0275
3. Xie S, Fu S, Li M, et al. Microbiological characteristics of carbapenem-resistant Enterobacteriaceae clinical isolates collected from county hospitals. Infect Drug Resist. 2020;13:1163–1169. doi:10.2147/IDR.S248147
4. Yin D, Lin Y, Li Z, et al. Horizontal transfer of antibiotic resistance genes in clinical environments. Infect Drug Resist. 2020;65:3929–3935. doi:10.2147/IDR.S277997
5. Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9 Science. 2014;346(6213):1258096.
6. Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable Dual-RNA–Guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–821. doi:10.1126/science.1225829
7. Bikard D, Euler CW, Jiang W, et al. Exploiting CRISPR-Cas nucleases to produce sequence-specific antimicrobials. Nat Biotechnol. 2014;32(11):1146–1150. doi:10.1038/nbt.3043
8. Citorik RJ, Mimee M, Lu TK. Sequence-specific antimicrobials using efficiently delivered RNA-guided nucleases. Nat Biotechnol. 2014;32(11):1141–1145. doi:10.1038/nbt.3011
9. Yosef I, Manor M, Kiro R, Qimron U. Temperate and lytic bacteriophages programmed to sensitize and kill antibiotic-resistant bacteria. Proc Natl Acad Sci USA. 2015;112(23):7267–7272. doi:10.1073/pnas.1500107112
10. Buckner MMC, Ciusa ML, Piddock LJV. Strategies to combat antimicrobial resistance: anti-plasmid and plasmid curing. FEMS Microbiol Rev. 2018;42(6):781–804. doi:10.1093/femsre/fuy031
11. Lauritsen I, Porse A, Sommer MOA, Norholm MHH. A versatile one-step CRISPR-Cas9 based approach to plasmid-curing. Microb Cell Fact. 2017;16(1):135. doi:10.1186/s12934-017-0748-z
12. Wang PX, He DM, Li BY, et al. Eliminating mcr-1-harbouring plasmids in clinical isolates using the CRISPR/Cas9 system. J Antimicrob Chemother. 2019;74(9):2559–2565. doi:10.1093/jac/dkz246
13. Liu H, Li H, Liang Y, et al. Phage-delivered sensitisation with subsequent antibiotic treatment reveals sustained effect against antimicrobial resistant bacteria. Theranostics. 2020;10(14):6310–6321. doi:10.7150/thno.42573
14. He YZ, Yan JR, He B, et al. A transposon-associated CRISPR/Cas9 system specifically eliminates both chromosomal and plasmid-borne mcr-1 in Escherichia coli. Antimicrob Agents Ch. 2021;65(10). doi:10.1128/AAC.01054-21
15. He YZ, Kuang X, Long TF, et al. Re-engineering a mobile-CRISPR/Cas9 system for antimicrobial resistance gene curing and immunization in Escherichia coli. J Antimicrob Chemother. 2021;77:74–82. doi:10.1093/jac/dkab368
16. Lino CA, Harper JC, Carney JP, Timlin JA. Delivering CRISPR: a review of the challenges and approaches. Drug Deliv. 2018;25(1):1234–1257. doi:10.1080/10717544.2018.1474964
17. Dong H, Xiang H, Mu D, Wang D, Wang T. Exploiting a conjugative CRISPR/Cas9 system to eliminate plasmid harbouring the mcr-1 gene from Escherichia coli. Int J Antimicrob Agents. 2019;53(1):1–8. doi:10.1016/j.ijantimicag.2018.09.017
18. Reuter A, Hilpert C, Dedieu-Berne A, et al. Targeted-antibacterial-plasmids (TAPs) combining conjugation and CRISPR/Cas systems achieve strain-specific antibacterial activity. Nucleic Acids Res. 2021;49(6):3584–3598. doi:10.1093/nar/gkab126
19. Rodrigues M, McBride SW, Hullahalli K, Palmer KL, Duerkop BA. Conjugative delivery of CRISPR-Cas9 for the selective depletion of antibiotic-resistant enterococci. Antimicrob Agents Chemother. 2019;63(11). doi:10.1128/AAC.01454-19
20. Yi H, Cho YJ, Yong D, Chun J. Genome sequence of Escherichia coli J53, a reference strain for genetic studies. J Bacteriol. 2012;194(14):3742–3743. doi:10.1128/JB.00641-12
21. Su T, Liu F, Gu P, et al. A CRISPR-Cas9 assisted non-homologous end-joining strategy for one-step engineering of bacterial genome. Sci Rep. 2016;6:37895. doi:10.1038/srep37895
22. Chan WT, Verma CS, Lane DP, Gan SK. A comparison and optimization of methods and factors affecting the transformation of Escherichia coli. Biosci Rep. 2013;33:6. doi:10.1042/BSR20130098
23. Kovach ME, Elzer PH, Hill DS, et al. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene. 1995;166(1):175–176. doi:10.1016/0378-1119(95)00584-1
24. Zhao Q, Berglund B, Zou HY, et al. Dissemination of bla(NDM-5) via IncX3 plasmids in carbapenem-resistant Enterobacteriaceae among humans and in the environment in an intensive vegetable cultivation area in eastern China. Environ Pollut. 2021;273:116370. doi:10.1016/j.envpol.2020.116370
25. Vetrovsky T, Baldrian P, Neufeld J. The variability of the 16S rRNA gene in bacterial genomes and its consequences for bacterial community analyses. PLoS One. 2013;8(2):e57923. doi:10.1371/journal.pone.0057923
26. Strand TA, Lale R, Degnes KF, et al. Improved host-independent plasmid system for RK2-based conjugal transfer. PLoS One. 2014;9(3):e90372. doi:10.1371/journal.pone.0090372
27. Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177(14):4121–4130. doi:10.1128/jb.177.14.4121-4130.1995
28. CLSI. Performance Standards for Antimicrobial Susceptibility Testing.
29. Maiden MCJ. Horizontal genetic exchange, evolution, and spread of antibiotic resistance in bacteria. Clin Infect Dis. 1998;27(Suppl s1):S12–20. doi:10.1086/514917
30. Sun P, Xia W, Liu G, et al. Characterization of blaNDM-5-Positive Escherichia coli prevalent in a university hospital in Eastern China. Infect Drug Resist. 2019;12:3029–3038. doi:10.2147/IDR.S225546
31. Van Norman GA. Drugs, devices, and the FDA: part 1: an overview of approval processes for drugs. JACC Basic Transl Sci. 2016;1(3):170–179. doi:10.1016/j.jacbts.2016.03.002
32. Hao MJ, He YZ, Zhang HF, et al. CRISPR-Cas9-mediated carbapenemase gene and plasmid curing in carbapenem-resistant Enterobacteriaceae. Antimicrob Agents Ch. 2020;64(9). doi:10.1128/AAC.00843-20
33. Tagliaferri TL, Guimaraes NR, Pereira MDPM, et al. Exploring the potential of CRISPR-Cas9 under challenging conditions: facing high-copy plasmids and counteracting beta-lactam resistance in clinical strains of Enterobacteriaceae. Front Microbiol. 2020;11:578. doi:10.3389/fmicb.2020.00578
34. Wan P, Cui S, Ma Z, et al. Reversal of mcr-1-mediated colistin resistance in Escherichia coli by CRISPR-Cas9 system. Infect Drug Resist. 2020;13:1171–1178. doi:10.2147/IDR.S244885
35. Cao J, Wu L, Zhang S-M, et al. An easy and efficient inducible CRISPR/Cas9 platform with improved specificity for multiple gene targeting. Nucleic Acids Res. 2016;44(19):e149. doi:10.1093/nar/gkw660
36. Yuan TT, Zhong Y, Wang YG, et al. Generation of hyperlipidemic rabbit models using multiple sgRNAs targeted CRISPR/Cas9 gene editing system. Lipids Health Dis. 2019;18:45
37. Cheng H, Zhang F, Ding Y. CRISPR/Cas9 delivery system engineering for genome editing in therapeutic applications. Pharmaceutics. 2021;13(10):1649. doi:10.3390/pharmaceutics13101649
38. Hausner M, Wuertz S. High rates of conjugation in bacterial biofilms as determined by quantitative in situ analysis. Appl Environ Microbiol. 1999;65(8):3710–3713. doi:10.1128/AEM.65.8.3710-3713.1999
39. Hamilton TA, Pellegrino GM, Therrien JA, et al. Efficient inter-species conjugative transfer of a CRISPR nuclease for targeted bacterial killing. Nat Commun. 2019;10(1):4544. doi:10.1038/s41467-019-12448-3
40. Wan F, Draz MS, Gu MJ, Yu W, Ruan Z, Luo QX. Novel Strategy to Combat Antibiotic Resistance: A Sight into the Combination of CRISPR/Cas9 and Nanoparticles. Pharmaceutics. 2021;13(3):352.
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.