Back to Journals » Drug Design, Development and Therapy » Volume 18
Systematic Pharmacology and Experimental Validation to Reveal the Alleviation of Astragalus membranaceus Regulating Ferroptosis in Osteoarthritis
Authors Chen K , Yu Y, Wang Y, Zhu Y, Qin C, Xu J, Zou X, Tao T, Li Y, Jiang Y
Received 15 October 2023
Accepted for publication 22 January 2024
Published 1 February 2024 Volume 2024:18 Pages 259—275
DOI https://doi.org/10.2147/DDDT.S441350
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Manfred Ogris
Kai Chen,1 Yaohui Yu,1 Yishu Wang,1 Yi Zhu,1 Chaoren Qin,1 Jintao Xu,1 Xiangjie Zou,2 Tianqi Tao,1 Yang Li,1 Yiqiu Jiang1
1Department of Sports Medicine and Joint Surgery, Nanjing First Hospital, Nanjing Medical University, Nanjing, People’s Republic of China; 2Department of Orthopaedics, The First Affiliated Hospital of Nanjing Medical University, Nanjing, People’s Republic of China
Correspondence: Yiqiu Jiang, Nanjing First Hospital, Nanjing Medical University, 68 Changle Road, Nanjing, People’s Republic of China, Email [email protected]
Background: Astragalus membranaceus (AM) shows promise as a therapeutic agent for osteoarthritis (OA), a debilitating condition with high disability rates. OA exacerbation is linked to chondrocyte ferroptosis, yet the precise pharmacological mechanisms of AM remain unclear.
Methods: We validated AM’s protective efficacy in an anterior cruciate ligament transection (ACLT) mouse model of OA. The Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform (TCMSP) database was utilized to identify AM’s active components and their targets. FerrDb (a database for regulators and markers of ferroptosis and ferroptosis-disease associations) pinpointed ferroptosis-related targets, while GeneCards, Online Mendelian Inheritance in Man (OMIM), Pharmacogenomics Knowledgebase (PharmGKB), Therapeutic Target Database (TTD), and DrugBank sourced OA-related genes. Molecular docking analysis further validated these targets. Ultimately, the validation of the results was accomplished through in vitro experiments.
Results: AM exhibited anabolic effects and suppressed catabolism in OA chondrocytes. Network pharmacology identified 19 common genes, and molecular docking suggested quercetin, an AM constituent, interacts with key proteins like HO-1 and NRF2 to inhibit chondrocyte ferroptosis. In vitro experiments confirmed AM’s ability to modulate the NRF2/HO-1 pathway via quercetin, mitigating chondrocyte ferroptosis.
Conclusion: This study elucidates how AM regulates chondrocyte ferroptosis, impacting OA progression, providing a theoretical basis and experimental support for AM’s scientific application.
Keywords: systematic pharmacology, osteoarthritis, ferroptosis, Astragalus membranaceus, quercetin
Introduction
Osteoarthritis (OA), a prevalent musculoskeletal disorder worldwide, represents a leading cause of impaired mobility, diminished quality of life, reduced work efficiency, and escalated healthcare expenses.1,2 Pathologically, osteoarthritis encompasses various changes, such as chondrocyte senescence and hypertrophy, matrix degradation, subchondral bone remodeling, and synovial inflammation.3 Being a multifactorial condition, osteoarthritis exhibits associations with genetics, age, gender, BMI, trauma, and numerous other factors.4 Ferroptosis, an iron-dependent lipid peroxidation-mediated form of cell death discovered about a decade ago, has been extensively studied regarding its involvement in osteoarthritis progression.5 Notably, iron accumulation constitutes one of the pathological hallmarks of osteoarthritis, and the disturbance of iron homeostasis in chondrocytes due to iron overload leads to oxidative stress and subsequent ferroptosis.6 Moreover, compelling evidence suggests that the utilization of ferroptosis inhibitors can markedly mitigate matrix degradation and oxidative stress levels in chondrocytes, thus ameliorating osteoarthritis.7
Astragalus membranaceus (AM), a traditional Chinese herbal medicine with a longstanding history of therapeutic use, either as a standalone treatment or in combination with other herbs, exhibits broad efficacy in addressing diverse ailments. The principal chemical constituents of AM encompass polysaccharides, saponins, flavonoids, amino acids, trace elements, sterols, and other components. Notably, flavonoids represent the primary pharmacologically active compounds within AM, including quercetin, kaempferol, isoquercitrin, and various other derivatives.8 Among these, quercetin emerges as a prominent component of the flavonoid fraction within AM and has demonstrated its capability to modulate ferroptosis across a range of pathological conditions, such as type 2 diabetes, acute kidney injury, and epilepsy.9–11
Network pharmacology represents a burgeoning approach to drug discovery that revolves around the identification of drug components and their corresponding targets. Although traditional medicine has gained widespread usage, elucidating its precise mechanism of action remains a paramount objective. Integrating traditional medicine with network pharmacology aligns with the current paradigm shift from phenotype and symptom-based approaches to endotype and causative-based strategies.12 Considering this, leveraging network pharmacology and the traditional Chinese medicine database, this study aims to: 1) determine the therapeutic effect of Astragalus membranaceus on OA, and 2) reveal the mechanism of Astragalus membranaceus in regulating ferroptosis in chondrocytes.
Materials and Methods
Reagents
Primary antibodies targeting SOX9, MMP13, GAPDH, NRF2, HO-1, GPX4, and SLC7A11 were procured from Proteintech (Wuhan, China), while the primary antibody against COL2A1 was obtained from Affinity Biosciences (Changzhou, China). HRP-conjugated Rabbit Anti-Goat IgG(H+L) and HRP-conjugated Mouse Anti-Goat IgG(H+L) were sourced from Proteintech. Quercetin was purchased from Macklin (Shanghai, China). The Alexa Fluor 488 rabbit anti-goat IgG (H+L) secondary antibody (green) was obtained from Abcam (US). The Cell Counting Kit-8 (CCK-8) kit was supplied by Biosharp (Anhui, China). TRIzol reagent was procured from Invitrogen (CA, US). The SweScript RT II First Strand cDNA Synthesis Kit (With gDNA Remover) and 2 × Universal Blue SYBR Green qPCR Master Mix were purchased from Servicebio (Wuhan, China). The Lipid Peroxidation MDA Assay Kit was obtained from Beyotime (Shanghai, China). The Glutathione Peroxidase (GSH-PX) assay kit (Colorimetric method) and Superoxide Dismutase (SOD) assay kit (WST-1 method) were procured from Jiancheng Bioengineered Institute (Nanjing, China). Live & Dead Viability/Cytotoxicity Assay Kit for Animal Cells was purchased from KeyGEN BioTECH (Nanjing, China). Ferric ammonium citrate (FAC) and Ferrostatin-1 (Fer-1) were purchased from MedChemExpress (Shanghai, China). Mouse interleukin-1β (IL-1β), interleukin-6 (IL-6), and tumor necrosis factor alpha (TNF-α) ELISA kit were purchased from Proteintech (Wuhan, China). Ferrous Iron Colorimetric Assay Kit was purchased from Elabscience (Wuhan, China).
Astragalus membranaceus was purchased from the pharmacy of Jiangsu Provincial Hospital of Traditional Chinese Medicine. Weigh 50 g of Astragalus each time, add 500 mL deionized water, decoct twice, 2 h each time, combine the filtrate, concentrate under reduced pressure to 5 g/mL, and store at 4 °C. Each prepared medicinal solution should be used within 2 days.
Identification of Compounds and Potential Active Targets of AM
In this study, a total of 87 compounds of AM were collected from Traditional Chinese Medicine Systems Pharmacology Database (TCMSP, https://old.tcmsp-e.com/tcmsp.php).13 Based on the screening criteria, 20 compounds with Oral Bioavailability (OB) ≥ 30%14 and drug-likeness (DL) ≥ 0.1815 were identified for pharmacological activity. The results are presented in Table S1.
Prediction of Potential Targets of AM Regulating Ferroptosis During Osteoarthritis Treatment
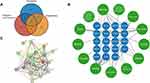
Potential targets of active ingredients in AM are collected from the TCMSP and DrugBank databases.16 927 osteoarthritis-related genes were obtained and assembled from GeneCards, Online Mendelian Inheritance in Man (OMIM), Pharmacogenomics Knowledgebase (PharmGKB), Therapeutic Target Database (TTD), and DrugBank using the keyword “Osteoarthritis”. 267 ferroptosis-related genes were collected through FerrDb17 and those reported in previous studies. After obtaining ferroptosis, osteoarthritis-related genes, and potential targets of AM, 19 intersecting genes were obtained, as shown in Figure 1A.
Bioactive Component-Target Network and Protein-Protein Interaction Network (PPI) Construction
A total of 13 bioactive components and 19 co-target genes were used to construct the component-target network and the network was visualized through Cytoscape 3.9.1.18
To construct a protein-protein interaction network (PPI) for the hub genes, we used the Search Tool for the Retrieval of Interacting Genes (STRING) database. The gene symbols for the upregulated and downregulated genes were uploaded into the database, and the PPI network was constructed based on experimentally validated interactions and high-confidence interactions predicted by computational meth. We used a confidence score of 0.4 (medium confidence) as the cutoff for the interactions. The resulting PPI network was visualized using Cytoscape software, and the network topology and properties were analyzed using NetworkAnalyzer, a plugin for Cytoscape.
Functional and Pathway Enrichment of Gene Modules
Biological functions and signaling pathways were enriched for nineteen genes using gene ontology (GO) analysis and the Kyoto encyclopedia of genes genomes (KEGG) pathway.19,20 p value < 0.05 was considered to be significant. The results of the functional and pathway enrichment analyses were used to generate hypotheses about the biological mechanisms underlying the observed gene expression changes. These hypotheses were further assessed through experimental validation.
Molecule Docking
The SDF files of the compounds were acquired from PubChem (https://pubchem.ncbi.nlm.nih.gov/) based on their CAS numbers, while the protein PDB files were retrieved from RCSB PDB (https://www.rcsb.org/). For protein structures not available in the RCSB database, the AlphaFold Protein Structure Database was utilized to obtain the structure file. To conduct molecular docking between the identified components of AM and target proteins, the online tool CB-DOCK2 was employed.21 A lower Score value indicated a higher likelihood of interaction.
Cell Culture and Treatment
Articular cartilage was obtained from the knee joint of 4-week-old mice, rinsed three times with 1 × PBS, and subjected to digestion in a 37°C incubator using 0.2% type II collagenase for a duration of 8 to 12 hours. After collagenase digestion of cartilage tissue, the resulting cell suspension was filtered through a 48μm cell strainer and centrifuged at 400g for 10 minutes at room temperature. Subsequently, the pellet was washed with 5 mL PBS, centrifuged again, and resuspended in 15 mL complete culture medium. Subsequent experiments were conducted utilizing chondrocytes derived from the second (P2) or third (P3) generation of mice. The CCK-8 assay was performed to assess cell viability and determine the optimal concentration of drug intervention. Experimental protocols adhered to the manufacturer’s instructions. The control group remained untreated, while the FAC group received an intervention of 100 μM FAC for 24 hours. In the drug group, cells were pretreated with 2.5, 5 μM Quercetin or 1 μM Fer-1 for 24 hours, followed by exposure to 100 μM FAC for an additional 24 hours.
Immunofluorescence
To detect the expression and localization of NRF2 in cells, immunofluorescence was performed. Cells were fixed with 4% paraformaldehyde for 20 minutes, permeabilized with 0.1% Triton X-100 for 5 minutes and blocked with 5% BSA for 30 minutes at room temperature. The cells were then incubated with a rabbit polyclonal antibody against protein NRF2 (1:500 dilution, Proteintech, Wuhan, China) overnight at 4°C. After washing with PBS, the cells were incubated with an Alexa Fluor 488-conjugated goat anti-rabbit secondary antibody (1:1000 dilution) for 1 hour at room temperature. Nuclei were counterstained with DAPI. Fluorescence images were acquired using a fluorescence microscope (Zeiss) and analyzed using ImageJ software. Quantitative analysis of nuclear translocation was achieved by using the plug-in Coloc 2 for ImageJ. Manders’ Colocalization Coefficients (MCC) was used to quantify the co-localization of NRF2 and nuclei of chondrocytes.22
Animal Models and Experimental Design
All animal experiments in this experiment have been approved by the Experimental Animal Ethics Committee of Nanjing First Hospital (DWSY-2101052). Thirty 8-week-old C57BL/6 male mice were purchased from the Animal Experiment Center of Nanjing Medical University. All mice were acclimatized for one week in a constant humidity room at a temperature of (22 ± 2) °C and 50 ± 5% relative humidity under an alternate 12-h light/dark cycle. The osteoarthritis model in 20 mice was induced with the Anterior Cruciate Ligament Transection (ACLT),23 the remaining 10 mice were operated on the sham. Each group of mice is set up with one redundancy to prevent unexpected situations caused by feeding or surgery. Starting from the third day after surgery, the mice in the AM treatment group were intragastrically administered with AM at a dose of 80 mg/kg every week, while the mice in the control group and ACLT group were intragastrically administered with an equal volume of normal saline. After four weeks, the mice were anesthetized with pentobarbital and executed, and the intact knee joint was taken for subsequent experiments. All laboratory mice were disposed in accordance with the American Veterinary Medical Association (AVMA) Guidelines for the Euthanasia of Laboratory Animals (2020).
Immunoblotting
For cell samples, post-treatment, each culture dish underwent three washes with cold PBS. Subsequently, the cells were lysed using RIPA buffer (ThermoFisher Scientific, Illinois, US) supplemented with protease inhibitor (Beyotime, Shanghai, China) and phosphatase inhibitor (MCE, Shanghai, China). The resulting supernatant was collected after centrifugation at 12,000g for 10 minutes. To facilitate subsequent immunoprecipitation, the supernatant was heated at 95°C for 5 minutes upon the addition of loading buffer and reducing buffer (ThermoFisher Scientific, Illinois, US).
For tissue samples, RIPA Lysis Buffer (Beyotime, Shanghai, China) was utilized. Following sacrifice, the skin, and patellar ligament were dissected to expose the knee joint cavity completely. The anterior and posterior cruciate ligaments were transected using a surgical scalpel to separate the tibia from the femur. Cartilage tissue was scraped from the femoral condyle and tibial plateau surfaces with a surgical scalpel and briefly stored in normal saline. The cartilage was sufficiently frozen, ground in liquid nitrogen, and lysed by adding lysate after adequate grinding. The subsequent steps mirrored those used for cellular protein extraction.
Electrophoresis and membrane transfer steps were carried out according to the recommended protocols by Bio-Rad. The membrane was blocked using 5% skim milk for 1 hour. The primary antibody was incubated overnight at the recommended dilution ratio and 4°C. Following three washes with TBST, the membrane was incubated with the secondary antibody at room temperature for 1 hour, followed by three additional washes with TBST. The final exposure was performed using an ECL luminescent solution.
Malondialdehyde (MDA), Glutathione (GSH), Superoxide Dismutase (SOD), Cellular Ferrous Iron (Fe2+) Concentration, and Live and Dead Viability Assay
The MDA assay was employed to assess lipid peroxidation levels as a measure of oxidative stress. It quantitatively measured lipid peroxidation levels, reflecting the extent of oxidative stress in the samples. GSH is the most important antioxidant sulfhydryl substance in cells and plays an important role in antioxidant, protein sulfhydryl protection, and amino acid transmembrane transport. Superoxide dismutase (SOD) is an antioxidant metalloenzyme that exists in organisms. It can catalyze the dismutation of superoxide anion radicals to generate oxygen and hydrogen peroxide and plays a vital role in the balance of oxidation and antioxidants in the body. Ferrous ions (Fe2+) in cells are one of the key factors in the occurrence of ferroptosis. Excessive Fe2+ can promote the Fenton reaction and generate highly active oxygen free radicals. These free radicals can cause lipid peroxidation, damage the cell membrane structure, and ultimately leading to cell death.24 All experimental procedures were performed according to the manufacturer’s instructions.
Isolation of Chondrocyte RNA and Real-Time qPCR
The total RNA of chondrocytes was extracted using TRIzol reagent (Invitrogen, US) according to the manufacturer’s instructions. The quality of isolated RNA was assessed using the NanoDrop 2000 spectrophotometer (ThermoFisher Scientific, US). Samples with A260/A280 ratios ranging between 1.8 and 2.0 were deemed as qualified specimens. Reverse transcription and complementary DNA (cDNA) amplification were performed using the SweScript All-in-One RT SuperMix for qPCR purchased from ServiceBio (Wuhan, China). Following the manufacturer’s instructions, cDNA amplification was conducted using the 2× Universal Blue SYBR Green qPCR Master Mix. The reactions were performed on the Applied Biosystems QuantStudio 5 system (ThermoFisher Scientific, Illinois, US). In the qPCR experiment, the reaction program was set as an initial denaturation at 95°C for 30 seconds, followed by 40 cycles consisting of denaturation at 95°C for 15 seconds, annealing at 60°C for 10 seconds, extension at 72°C for 30 seconds. The steps for the melting curve analysis were conducted according to the default settings of the instrument’s program. The primer sequences for genes assessed with chondrocytes were shown in Table S2, and GAPDH was used for reference. The comparative threshold cycle (Ct) technique (2−∆∆Ct method) was used to calculate the relative fold gene expression of samples.
Histopathological Examination
Intact mouse knee joints were fixed in 4% paraformaldehyde for 24 hours, followed by decalcification in 10% EDTA, pH 7.4 decalcifying solution for 2 weeks, with the decalcifying solution, changed 3 to 4 times a week. After decalcification, the knee joints were sent to Wuhan ServiceBio for embedding and sectioning. Immunohistochemistry was performed using Elabscience 2-step plus Poly-HRP Anti Rabbit/Mouse IgG Detection System (with DAB Solution) according to the manufacturer’s instructions.
HE and safranin staining first followed by deparaffinization, the slides were placed in a slide straining rack and immersed in xylene for 5 minutes to remove the paraffin. This step was repeated twice to ensure complete deparaffinization. After deparaffinization, the slides were submerged in a series of descending concentrations of alcohol (100%, 95%, 80%, and 70%) for 5 minutes each to rehydrate the joint sections. To stain the nuclei, the sections were immersed in a hematoxylin solution for approximately 3–5 minutes and then rinsed with running tap water to remove excess stain. Differentiation was achieved by placing the slides in an acid alcohol solution for 2–5 seconds, followed by rinsing with tap water. 0.5% Ammonia Water Solution was used for bluing hematoxylin, followed by rinsing with tap water. Next, the slides were immersed in eosin solution for 1–2 minutes to stain the cytoplasm and extracellular components. After staining, the slides were rinsed gently with distilled water. All sections were dehydrated through a series of ascending alcohol concentrations (70%, 80%, 95%, and 100%) for a few minutes each to remove excess water. The sections were then immersed in Fast Green solution for 5 minutes to counterstain the non-cartilage components, such as muscle and bone, providing a green coloration. After that, the sections were immersed in Safranin O solution for 15–30 seconds and then quickly dehydrated with four tanks of anhydrous ethanol. Safranin O selectively binds to cartilage proteoglycans, staining them red. This step provides visual contrast and allows for the evaluation of cartilage integrity and proteoglycan content. A mounting medium, such as a synthetic resin, was applied to the tissue sections, and glass coverslips were placed over them.
Enzyme-Linked Immunosorbent Assays (ELISAs) for Mouse IL-1β, IL-6 and TNF-α
The circulating levels of IL-1β, IL-6, and TNF-α in the serum collected from each group of mice in the OA model were determined using the enzyme-linked immunosorbent assay (ELISA). In brief, inflammation factor levels were assessed using commercial assay kits in accordance with the manufacturer’s instructions. All samples were assayed in triplicate within each group of six mice, and each experiment was conducted a minimum of three times. In vitro experiments measuring inflammation factor levels were also conducted following the manufacturer’s protocols.
Statistical Analysis
All quantitative data were collected by repeating the experiment three times independently unless otherwise stated and statistically analyzed using SPSS 20® (SPSS Science Inc™, US). One-way analysis of variance (ANOVA) was required to compare the means of three or more groups, while independent tests (t-test, unpaired two-tailed) were used to check for differences between the two groups. Statistical significance was considered if p < 0.05. All statistical plots were designed using GraphPad Prism 9 (GraphPad Software Inc, San Diego, CA, USA), and data are shown as mean ± SD.
Results
AM Ameliorates Osteoarthritis Induced by ACLT in 6-Week C57BL/6N Mice
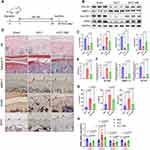
This study utilized ACLT surgery in mice to establish a model of osteoarthritis, aiming to investigate the protective effects of AM intervention. A total of 27 mice were assigned to three groups: sham-operated, ACLT, and ACLT with intraperitoneal AM administration (Figure 2A). Western blot analysis revealed a significant increase in catabolic proteins, MMP13, and a decrease in anabolic and chondrogenic proteins, COL2A1 in the ACLT group compared to the sham-operated group at 4 weeks post-surgery. However, AM treatment demonstrated remarkable protective effects against ACLT-induced osteoarthritis, as evidenced by the restoration of protein expression levels (Figure 2B and 1C). The PCR results of articular cartilage samples were consistent with the Western blot findings (Figure 2H).
Histological evaluations using HE staining, Safranin O and fast green staining were performed to assess proteoglycan content and tissue structure in the mouse joint cartilage. Representative images of these staining methods are shown in Figure 2D. The sham-operated group exhibited intense red staining of Safranin O, indicative of preserved proteoglycans, along with smooth articular surfaces and no signs of cartilage erosion or bone damage. Conversely, the ACLT group displayed characteristic features of osteoarthritis, including articular surface fibrillation, erosion, loss of proteoglycan content, and subchondral bone sclerosis. Notably, AM treatment significantly ameliorated proteoglycan loss and surface erosion in the articular cartilage, as supported by the ORASI score quantification (Figure 2E).
Immunohistochemistry was conducted to assess the expression levels of MMP13 and ACAN in the articular cartilage of the sham-operated, ACLT, and AM+ACLT groups. The results demonstrated that AM treatment mitigated the increase in MMP13 expression and the decrease in Acan expression induced by ACLT (Figure 2F). The expression of the important anti-ferroptosis protein GPX4 in knee articular cartilage can also be restored by AM treatment (Figure 2F), which is consistent with the results of Western Blot. Through in vivo experiments in mice, we substantiated that ACLT induced knee inflammation, whereas AM treatment exerted a protective effect against these detrimental effects. The decreased expression of anti-ferroptosis key proteins SLC7A11and GPX4caused by osteoarthritis was also reversed with the intervention of AM. ELISA was utilized to analyze inflammatory factors in the cartilage of mouse knee joints. The outcomes revealed a noteworthy reduction in the levels of IL-1β, IL-6, and TNF-α within the articular cartilage following treatment with AM in mice subjected to ACLT (Figure 2G).
Overall, our findings from this mouse model confirmed the protective effects of AM intervention in the context of ACLT-induced osteoarthritis, highlighting its potential as a therapeutic approach for this condition.
Screening for Bioactive Components and Targets Related to Osteoarthritis, AM, and Ferroptosis
Through a rigorous screening process utilizing the Traditional Chinese Medicine Systems Pharmacology (TCMSP) database, a total of 20 active ingredients from AM were identified based on the criteria of oral bioavailability (OB) ≥ 30% and drug-likeness (DL) ≥ 0.18 (Table S1). The TCMSP further provided 396 predicted targets associated with these twenty active ingredients. To expand the scope of potential targets, we searched GeneCards, OMIM, PharmGKB, TTD, and DrugBank databases, retrieving 927 disease-associated genes after removing duplicate entries. By integrating the osteoarthritis-related genes, potential targets of AM, and ferroptosis-related genes, a Venn diagram analysis highlighted 19 intersecting genes of particular interest (Figure 1A). This intersection of gene sets provides valuable insights into the molecular associations and potential mechanisms underlying the effects of AM in the context of osteoarthritis and ferroptosis.
Bioactive Component Targets and PPI Network
Utilizing the Cytoscape software, we constructed a comprehensive drug-target network to explore the interactions between AM active ingredients and their corresponding target genes (Figure 1B). The network encompassed 53 edges and 32 nodes, with the yellow nodes representing the active ingredients of AM and the blue nodes representing the target genes. To further investigate the functional relationships and potential pathways associated with these target genes, we uploaded the 19 target genes to the STRING.db database and employed mean confidence intervals to construct a protein-protein interaction (PPI) network (Figure 1C), The number of connected nodes and the connection score of each protein node are shown in Table S2. This network comprised 121 edges and nineteen nodes, and its construction involved screening criteria such as “Betweenness > 5.26, Closeness > 0.79, Degree > 12, Eigenvector > 0.22, LAC > 9.49, Network > 11” (averages for all criteria). Notably, the Cytoscape plugin, CytoNCA, was employed to identify core genes based on various topological measures. As a result, HO-1, TP53, PTGS2, and VEGFA were identified as crucial core proteins, warranting further investigation and research focus. These findings provide a basis for subsequent studies aimed at elucidating the potential mechanisms and key pathways associated with the therapeutic effects of AM in the context of osteoarthritis.
Gene Ontology and KEGG Pathway Enrichment Analysis
The GO enrichment analysis performed on the 19 genes yielded a comprehensive set of 1619 functional categories. Among these categories, Figure 3A illustrates the top 12 most significant biological processes, which predominantly encompass functions associated with “response to reactive oxygen species”, “response to oxygen levels”, “cellular response to metal ion”, and others. This analysis provides valuable insights into the biological mechanisms underlying the effects of these target genes in the context of osteoarthritis.
To further investigate the signaling pathways relevant to osteoarthritis and their association with the identified 19 target genes, we conducted pathway mapping using the KEGG database. This analysis revealed a total of 135 signaling pathways. Figure 3B showcases the top 10 significant pathways, including “Fluid shear stress and atherosclerosis”, “Kaposi sarcoma-associated herpesvirus infection”, “Chemical carcinogenesis reactive oxygen species”, “HIF-1 signaling pathway”, “Relaxin signaling pathway”, and others. These findings shed light on potential signaling cascades and molecular pathways that may contribute to the pathogenesis and progression of osteoarthritis.
Molecular Docking of Bioactive Components and Proteins Encoding by Hub Genes
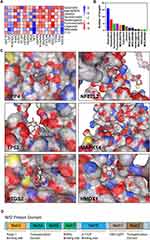
To gain insights into the molecular interactions between components in AM and the target genes, we conducted molecular docking experiments utilizing the top 9 components based on the number of targets and the set of 19 target genes. For each ligand-protein docking result, we selected the model with the smallest vina score to represent the binding between the compound and the target protein. Figure 4A illustrates the comprehensive results of all docking experiments. Notably, DPP4, HO-1 (HMOX1), p38 (MAPK14), NRF2 (NFE2L2), PTGS2, and TP53 exhibited significant binding potential (Vina Score < −7) with multiple AM components.
Among the components, Quercetin emerged as the most enriched in terms of target interactions (Figure 4B), demonstrating remarkable binding capabilities with a wide range of potential targets. The binding model of quercetin with these target proteins is visually depicted in Figure 4C, shedding light on the structural details of the quercetin-target interactions, in addition, schematic diagrams of the interaction structures from more angles are described in the Supplementary Material.
In our docking simulation investigating the interaction between quercetin and NRF2, we observed that quercetin forms hydrogen bonds, ionic interactions, and cation-pi interactions with specific amino acids, namely PHE39, ARG42, ARG43, TYR46, GLU47, LYS50, VAL523, GLU526, GLN527, ASP528, ASP530, HIS531, and ASP534 of the NRF2 protein. The Neh2 domain of NRF2 spans amino acids 28 to 82, and it is intriguing to note that the DGR/Kelch domain of KEAP1 interacts with the Neh2 domain of NRF2, resulting in the ubiquitination of NRF2. This spatial proximity suggests that quercetin may modulate NRF2 ubiquitination by potentially influencing the binding between the KEAP1 DGR/Kelch domain and the NRF2 Neh2 domain (Figure 4D).
The observed interactions between quercetin and specific amino acids within NRF2 provide valuable insights into the potential molecular mechanisms underlying the effects of quercetin on NRF2 activity and ubiquitination. By potentially interfering with the Keap1-NRF2 interaction, quercetin may contribute to the regulation of NRF2 activity and its downstream signaling pathways. These findings highlight the potential therapeutic implications of quercetin in modulating NRF2-mediated cellular processes and warrant further investigation into its precise mode of action in the context of osteoarthritis.
Quercetin Relieved FAC-Induced Oxidative Stress and Ferroptosis of Chondrocytes in vitro
Based on molecular docking results and PPI results, we hypothesized that AM ameliorated osteoarthritis through the regulation of the p38/NRF2/HO-1 axis by quercetin to relieve chondrocyte oxidative stress and ferroptosis. Ferric ammonium citrate (FAC) is a physiological form of iron that does not bind to transferrin. It induces intracellular iron overload, leading to cellular iron death, making it a common inducer of iron-dependent cell death.7 After 100 μM FAC treatment for 24 h, chondrocytes in the FAC group showed a significant trend of ferroptosis, an increase in catabolism, and a decrease in anabolism in chondrocytes. 2.5μM Quercetin pretreatment significantly increased the expression of anti-ferroptosis protein GPX4 (p = 0.0225) and increased the expression of SLC7A11 (p = 0.0884) in chondrocytes after FAC treatment. As the concentration of quercetin increased, the expression of SLC7A11 also increased significantly at 5 μM (p < 0.0001). Furthermore, this recovery of anti-ferroptosis proteins was dose-dependent (p = 0.0021; p = 0.0435). Quercetin also shows concentration-dependent chondroprotective effects on the main proteins of chondrocyte differentiation, extracellular matrix synthesis and catabolism (Figure 5A–F). Moreover, compared with the addition of the ferroptosis inhibitor Fer-1, the protection of quercetin had the same trend.
To investigate the mitochondrial function of chondrocytes, MitoTracker Red staining was performed. MitoTracker Red is a fluorescent dye that selectively stains active mitochondria, providing a visual representation of the mitochondrial network and allowing for the evaluation of mitochondrial activity. The results showed that in the control group and Fer-1 treated group, chondrocytes displayed well-distributed, elongated, and interconnected mitochondrial networks with intense MitoTracker Red staining. However, in the experimental group, chondrocytes exhibited fragmented and condensed mitochondrial networks, with weak MitoTracker Red staining (Figure 5G and H). This mitochondrial damage could be ameliorated by Quercetin in a dose-dependent manner.
To assess the ferroptosis status of chondrocytes, we measured the levels of MDA, GSH, SOD, and Live and Dead Viability assay in both the control and experimental groups. The MDA levels were significantly higher in the experimental group compared to the control group. Similar to Fer-1 pretreatment, pretreatment with Quercetin significantly reduced MDA and increased the content of SOD; In the reduction of MDA, Quercetin showed a significant dose dependence (p = 0.0004), while in the improvement of SOD, although higher concentrations of quercetin increased the content of SOD, it did not show significant statistical significance (p = 0.1455) (Figure 5I and J). Conversely, the GSH levels were significantly lower in the FAC group compared to the control group, suggesting a decrease in antioxidant defense capacity. Quercetin can increase GSH content in a dose-dependent manner (Figure 5K). Using a fluorescence microscope to observe the results of Calcein AM and PI fluorescence staining, it was found that quercetin can effectively improve the cell death caused by FAC similar to the ferroptosis inhibitor group. This protective effect can be increased with the increase of Quercetin concentration (Figure 5L).
Quercetin Alleviates Ferroptosis in Mice Chondrocytes via the NRF2/HO-1 Pathway
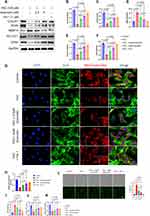
The transcription factor NRF2 plays a pivotal role in regulating cellular antioxidant defense mechanisms. Upon activation, NRF2 translocated to the nucleus and binds to antioxidant response elements, thereby orchestrating the expression of various antioxidant genes. In this study, Western blot analysis was employed to evaluate the protein expression levels of p38, NRF2, and HO-1. The results indicated that exposure to 100 μM FAC increased NRF2 expression, while the expression of HO-1 was not significantly affected. However, pretreatment with quercetin at concentrations of 2.5 μM and 5 μM increased the expression of both NRF2 and HO-1 (Figure 6A), and the increase in HO-1 expression is concentration-dependent (p = 0.0030). Notably, the phosphorylation of p38 did not exhibit significant alterations upon quercetin pretreatment. Immunofluorescence staining further corroborated these findings by demonstrating a prominent enhancement in NRF2 nucleation following quercetin pretreatment, in line with our initial hypothesis (Figure 6B). To quantify the nuclear translocation of NRF2, Manders’ Colocalization Coefficients were employed (Figure 6C). Additionally, the results suggested that quercetin pretreatment facilitated the translocation of NRF2 in a dose-dependent manner. Consistent with Western blot findings, immunofluorescence results reveal an increase in the average fluorescence intensity of NRF2 with Quercetin treatment (Figure 6D). Furthermore, Figure 6E illustrated that pretreatment with Quercetin ameliorated the decrease in cell viability induced by FAC.
To confirm our hypothesis based on the docking results, co-immunoprecipitation (co-IP) was used to investigate the effect of quercetin on the NRF2-KEAP1 interaction. As shown in the results, the binding of KEAP1 to NRF2 was dose-dependently reduced with quercetin intervention (Figure 6F).
These observations provide valuable insights into the molecular mechanisms underlying the protective effects of quercetin on chondrocytes. The upregulation of NRF2 and HO-1 expression following quercetin pretreatment suggests its potential role in enhancing the cellular antioxidant defense system. The nuclear translocation of NRF2 further supports the hypothesis that quercetin exerts its protective effects through the activation of NRF2-mediated antioxidant pathways. The demonstrated improvement in cell viability further emphasizes the potential therapeutic implications of quercetin in mitigating the detrimental effects induced by FAC exposure.
Discussion
Osteoarthritis remains a significant challenge for individuals and society at large, highlighting the pressing need for affordable, efficient, and minimally invasive therapeutic interventions to impede or reverse disease progression. This study aimed to elucidate the potential molecular mechanisms underlying the therapeutic effects of AM, a traditional Chinese herbal medicine, in osteoarthritis treatment. To achieve this, we utilized an ACLT-induced mouse model of osteoarthritis and demonstrated the ability of AM to enhance anabolism while inhibiting catabolism in osteoarthritic cartilage. Subsequently, we conducted an extensive investigation of the major drug components of AM, identifying a total of 396 potential drug targets through the TCMSP database. Additionally, we curated 928 genes associated with osteoarthritis targets using the GeneCards, PharmGkb, TTD, and DrugBank databases. Furthermore, we identified 267 genes related to ferroptosis from the FerrDb. Through comprehensive analysis, we identified 19 intersecting genes as the most promising potential targets for further investigation. Among these genes, iNOS has been implicated in promoting cartilage destruction and cellular damage by catalyzing excess NO production.25 Selective inhibitors of COX2 have been widely employed for pain relief in osteoarthritis.26 NRF2 and HO-1 play critical roles in enhancing chondrocyte resistance against oxidative stress by serving as major mediators of antioxidant responses.27 In a study by HY Sun et al, inhibition of the p38-MAPK pathway in chondrocytes exhibited significant improvements in chondrocyte apoptosis and a reduction in the expression of inflammatory factors.28
To elucidate the underlying mechanism by which AM exerts its therapeutic effects in osteoarthritis, we performed comprehensive GO and KEGG enrichment analyses. Our findings revealed that the 19 target genes were prominently enriched in biological processes related to response to reactive oxygen species, response to oxygen levels, cellular response to metal ions, and other key functions. These enrichments strongly suggest that the mitigation of ferroptosis may represent a significant mechanism through which AM ameliorates osteoarthritis.
To further elucidate the molecular-level mechanism underlying the therapeutic action of AM in osteoarthritis, we employed two complementary approaches for cross-validation. Firstly, we utilized CytoNCA to identify four core targets, namely HO-1, TP53, PTGS2, and VEGFA, within a protein-protein interaction (PPI) network constructed from the 19 potential targets. Additionally, we employed CB-Dock, a highly effective molecular docking tool, to perform docking simulations between AM’s nine major drug components and the 19 potential targets, including Quercetin, Kaempferol, and Jaranol. The docking results provided robust confirmation of our hypotheses. Notably, Quercetin, as one of the key active ingredients, demonstrated significant binding affinity (<-7 kcal/mol) with nearly all the 19 targets. Among the 19 potential targets, HO-1, p38, MAPK8, NRF2, NOS2, and PTGS2 displayed strong binding interactions with the majority of the active ingredients. Similarly, GO enrichment analysis of these 19 targets found that these targets were mainly enriched in the biological process of cells responding to oxidative stress, including p38/NRF2/HO-1. In the KEGG pathway enrichment, p38/NRF2/HO-1 is also included in the “Chemical carcinogenesis - reactive oxygen species” pathway. Notably, the p38/NRF2/HO-1 axis has emerged as a crucial pathway in the pathogenesis of various diseases in recent years.29 For instance, in KRAS mutant colorectal cancer, cetuximab has been shown to enhance RSL3-induced ferroptosis in tumor cells by modulating the p38/NRF2/HO-1 pathway.30 Wang et al discovered that Madecassoside can prevent LPS-induced acute liver failure in mice by inhibiting the p38/NF-κB pathway and activating the NRF2/HO-1 pathway to augment intracellular antioxidant levels.31 Dai et al found that quercetin could improve quinolone-induced cytotoxicity and apoptosis by activating the p38/NRF2/HO-1 pathway and inhibiting the ROS/mitochondrial apoptosis pathway.32 In addition, p38/NRF2/HO-1 has also been confirmed to inhibit obesity-related metabolic syndrome by alleviating adipose tissue inflammation and oxidative stress.33 Considering the accumulated knowledge and recent advancements in the field, we postulate that p38, NRF2, and HO-1 may represent the core targets involved in the therapeutic effects of AM.
Quercetin, one of the main active ingredients of Astragalus membranaceus, has attracted widespread attention due to its various biological activities such as hypoglycemic, anti-inflammatory, and anti-tumor. Additionally, emerging evidence has highlighted the involvement of ferroptosis, a recently recognized form of programmed cell death characterized by iron accumulation, reactive oxygen species (ROS) generation, and lipid peroxidation, in the pathogenesis of osteoarthritis, particularly in chondrocytes.7,34,35 Consistent with these recent studies, our findings support the notion that oxidative stress and lipid peroxidation represent prominent features of osteoarthritic chondrocytes.
In vitro experiments, we demonstrated that Quercetin improved the catabolic indexes induced by FAC, decreased cartilage phenotypic molecules such as SOX9 and COL2A1, and decreased GPX4 and SLC7A11. Under the use of ferroptosis inhibitors, the improvement in these indicators is similar to the effects produced by quercetin. Mechanical overloading, a recognized causal factor in osteoarthritis, has been shown to promote ferroptosis by reducing the antioxidant capacity of chondrocytes through PIZEO1/GPX4.36 The MitoTracker Red showed that quercetin ameliorated the mitochondrial damage induced by FAC. The results of MDA, GSH, and SOD assays also confirmed that Quercetin ameliorated lipid peroxidation and enhanced the antioxidant capacity of chondrocytes. Furthermore, although our results did not confirm that quercetin could regulate p38 in FAC-treated chondrocytes, the immunofluorescence results showed that quercetin significantly increased the nuclear translocation of NRF2. The increased nuclear translocation of NRF2, a major transcription factor regulating cellular anti-oxidative stress, suggests that this may be one of the main pathways by which Quercetin attenuates ferroptosis in chondrocytes. Co-IP results showed that quercetin intervention can reduce the interaction between KEAP1 and NRF2, which helps NRF2 enter the nucleus and exert its anti-ferroptosis effect. However, although our results confirmed the relationship between NRF2 and Quercetin, the biological processes underlying the effect of Quercetin on NRF2 remain unclear.
The present study’s major limitation is that we have only identified Quercetin as a major active component of AM, which inhibits chondrocyte ferroptosis through modulation of the NRF2/HO-1 pathway, thereby improving osteoarthritis. While Quercetin has been confirmed as one of the main bioactive components responsible for the biological effects of Astragalus membranaceus, systems pharmacology methods may not provide the necessary evidence for its role in mediating the pharmacological actions of Astragalus membranaceus.37–39 Therefore, it is still essential to include objective methods, such as GC-MS, to further substantiate the hypothesis that Quercetin is the primary component of Astragalus membranaceus responsible for inhibiting chondrocyte ferroptosis. Herbal medicines usually have an extraordinarily complex chemical composition and not all compounds are pharmacologically active; in fact, many importantly act through synergistic effects between pharmacologically active compounds. Therefore, further experiments are needed to confirm the interactions between the different compounds in AM and their effects on osteoarthritis.40
In summary, our research pioneers the revelation that Astragalus Membranaceus enhances chondrocyte resilience against ferroptosis, consequently mitigating osteoarthritis. This protective effect is attributed to quercetin, a key medicinal component of AM, substantiated by comprehensive in vitro and in vivo experiments. Our findings introduce innovative perspectives for exploring the therapeutic potential of AM in osteoarthritis management, laying the groundwork for further investigations into its application.
Data Sharing Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
The animal study was reviewed and approved by the Animal Ethics Committee of Nanjing Medical University (DWSY-2101052). All the experiments were conducted under the guidance of the “Guideline for the Care and Use of Laboratory Animals” set by the National Institute for Health of China.
Acknowledgments
We appreciated the laboratory and equipment provided by Central Laboratory, Nanjing First Hospital.
Funding
This research is funded by the National Natural Science Foundation, 81601954 to Yiqiu Jiang.
Disclosure
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Mandl LA. Osteoarthritis year in review 2018: clinical. Osteoarthritis Cartilage. 2019;27(3):359–364. doi:10.1016/j.joca.2018.11.001
2. Hunter DJ, Bierma-Zeinstra S. Osteoarthritis. Lancet. 2019;393(10182):1745–1759. doi:10.1016/S0140-6736(19)30417-9
3. Miao Y, Chen Y, Xue F, et al. Contribution of ferroptosis and GPX4’s dual functions to osteoarthritis progression. EBioMedicine. 2022;76:103847. doi:10.1016/j.ebiom.2022.103847
4. Ansari MY, Ahmad N, Haqqi TM. Oxidative stress and inflammation in osteoarthritis pathogenesis: role of polyphenols. Biomed Pharmacother. 2020;129:110452. doi:10.1016/j.biopha.2020.110452
5. Stockwell BR. Ferroptosis turns 10: emerging mechanisms, physiological functions, and therapeutic applications. Cell. 2022;185(14):2401–2421. doi:10.1016/j.cell.2022.06.003
6. Zhang S, Xu J, Si H, Wu Y, Zhou S, Shen B. The role played by ferroptosis in osteoarthritis: evidence based on iron dyshomeostasis and lipid peroxidation. Antioxidants. 2022;11(9). doi:10.3390/antiox11091668
7. Yao X, Sun K, Yu S, et al. Chondrocyte ferroptosis contribute to the progression of osteoarthritis. J Orthop Translat. 2021;27:33–43. doi:10.1016/j.jot.2020.09.006
8. Commission CP. Pharmacopoeia of the People’s Republic of China. China Medical Science Press; 2020.
9. Li D, Jiang C, Mei G, et al. Quercetin alleviates ferroptosis of pancreatic beta cells in type 2 diabetes. Nutrients. 2020;12(10):2954. doi:10.3390/nu12102954
10. Wang Y, Quan F, Cao Q, et al. Quercetin alleviates acute kidney injury by inhibiting ferroptosis. J Adv Res. 2021;28:231–243. doi:10.1016/j.jare.2020.07.007
11. Xie R, Zhao W, Lowe S, et al. Quercetin alleviates kainic acid-induced seizure by inhibiting the Nrf2-mediated ferroptosis pathway. Free Radic Biol Med. 2022;191:212–226. doi:10.1016/j.freeradbiomed.2022.09.001
12. Nogales C, Mamdouh ZM, List M, Kiel C, Casas AI, Schmidt H. Network pharmacology: curing causal mechanisms instead of treating symptoms. Trends Pharmacol Sci. 2022;43(2):136–150. doi:10.1016/j.tips.2021.11.004
13. Ru J, Li P, Wang J, et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J Cheminform. 2014;6:13. doi:10.1186/1758-2946-6-13
14. Xu X, Zhang W, Huang C, et al. A novel chemometric method for the prediction of human oral bioavailability. Int J Mol Sci. 2012;13(6):6964–6982. doi:10.3390/ijms13066964
15. Jia CY, Li JY, Hao GF, Yang GF. A drug-likeness toolbox facilitates ADMET study in drug discovery. Drug Discov Today. 2020;25(1):248–258. doi:10.1016/j.drudis.2019.10.014
16. Wishart DS, Feunang YD, Guo AC, et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46(D1):D1074–D1082. doi:10.1093/nar/gkx1037
17. Zhou N, Bao J. FerrDb: a manually curated resource for regulators and markers of ferroptosis and ferroptosis-disease associations. Database. 2020;2020. doi:10.1093/database/baaa021
18. Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi:10.1101/gr.1239303
19. Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The gene ontology consortium. Nat Genet. 2000;25(1):25–29. doi:10.1038/75556
20. Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. doi:10.1093/nar/28.1.27
21. Liu Y, Yang X, Gan J, et al. CB-Dock2: improved protein–ligand blind docking by integrating cavity detection, docking and homologous template fitting. Nucleic Acids Res. 2022;50(W1):W159–W164. doi:10.1093/nar/gkac394
22. Dunn KW, Kamocka MM, McDonald JH. A practical guide to evaluating colocalization in biological microscopy. Am J Physiol Cell Physiol. 2011;300(4):C723–C742. doi:10.1152/ajpcell.00462.2010
23. Kawaguchi H. Endochondral ossification signals in cartilage degradation during osteoarthritis progression in experimental mouse models. Mol Cells. 2008;25(1):1.
24. Jiang X, Stockwell BR, Conrad M. Ferroptosis: mechanisms, biology and role in disease. Nat Rev Mol Cell Biol. 2021;22(4):266–282. doi:10.1038/s41580-020-00324-8
25. Ahmad N, Ansari MY, Haqqi TM. Role of iNOS in osteoarthritis: pathological and therapeutic aspects. J Cell Physiol. 2020;235(10):6366–6376. doi:10.1002/jcp.29607
26. Lane NE. Pain management in osteoarthritis: the role of COX-2 inhibitors. J Rheumatol Suppl. 1997;49:20–24.
27. Chen Z, Zhong H, Wei J, et al. Inhibition of Nrf2/HO-1 signaling leads to increased activation of the NLRP3 inflammasome in osteoarthritis. Arthritis Res Ther. 2019;21(1):1–13. doi:10.1186/s13075-019-2085-6
28. Sun H-Y, Hu K-Z, Yin Z-S. Inhibition of the p38-MAPK signaling pathway suppresses the apoptosis and expression of proinflammatory cytokines in human osteoarthritis chondrocytes. Cytokine. 2016;90:135–143. doi:10.1016/j.cyto.2016.11.002
29. Yang R, Gao W, Wang Z, et al. Polyphyllin I induced ferroptosis to suppress the progression of hepatocellular carcinoma through activation of the mitochondrial dysfunction via Nrf2/HO-1/GPX4 axis. Phytomedicine. 2024;122:155135. doi:10.1016/j.phymed.2023.155135
30. Yang J, Mo J, Dai J, et al. Cetuximab promotes RSL3-induced ferroptosis by suppressing the Nrf2/HO-1 signalling pathway in KRAS mutant colorectal cancer. Cell Death Dis. 2021;12(11):1–11. doi:10.1038/s41419-021-04367-3
31. Wang W, Wu L, Li Q, et al. Madecassoside prevents acute liver failure in LPS/D-GalN-induced mice by inhibiting p38/NF-κB and activating Nrf2/HO-1 signaling. Biomed. Pharmacother. 2018;103:1137–1145. doi:10.1016/j.biopha.2018.04.162
32. Dai C, Zhang Q, Shen L, et al. Quercetin attenuates quinocetone-induced cell apoptosis in vitro by activating the P38/Nrf2/HO-1 pathway and inhibiting the ROS/Mitochondrial apoptotic pathway. Antioxidants. 2022;11(8):1498. doi:10.3390/antiox11081498
33. Wang Z, Ka S-O, Lee Y, Park B-H, Bae EJ. Butein induction of HO-1 by p38 MAPK/Nrf2 pathway in adipocytes attenuates high-fat diet induced adipose hypertrophy in mice. Eur J Pharmacol. 2017;799:201–210. doi:10.1016/j.ejphar.2017.02.021
34. Lv Z, Han J, Li J, et al. Single cell RNA-seq analysis identifies ferroptotic chondrocyte cluster and reveals TRPV1 as an anti-ferroptotic target in osteoarthritis. EBioMedicine. 2022;84:104258. doi:10.1016/j.ebiom.2022.104258
35. Sun K, Hou L, Guo Z, et al. JNK-JUN-NCOA4 axis contributes to chondrocyte ferroptosis and aggravates osteoarthritis via ferritinophagy. Free Radic Biol Med. 2023;200:87–101. doi:10.1016/j.freeradbiomed.2023.03.008
36. Wang S, Li W, Zhang P, et al. Mechanical overloading induces GPX4-regulated chondrocyte ferroptosis in osteoarthritis via Piezo1 channel facilitated calcium influx. J Adv Res. 2022;41:63–75. doi:10.1016/j.jare.2022.01.004
37. Liu R, Xu S, Li J, Hu Y, Lin Z. Expression profile of a PAL gene from Astragalus membranaceus var. Mongholicus and its crucial role in flux into flavonoid biosynthesis. Plant Cell Rep. 2006;25(7):705–710. doi:10.1007/s00299-005-0072-7
38. Fu J, Wang Z, Huang L, et al. Review of the botanical characteristics, phytochemistry, and pharmacology of Astragalus membranaceus (Huangqi). Phytother Res. 2014;28(9):1275–1283. doi:10.1002/ptr.5188
39. Sheng Z, Jiang Y, Liu J, Yang B. UHPLC–MS/MS analysis on flavonoids composition in Astragalus membranaceus and their antioxidant activity. Antioxidants. 2021;10(11):1852. doi:10.3390/antiox10111852
40. Li C. Multi-compound pharmacokinetic research on Chinese herbal medicines: approach and methodology. Zhongguo Zhong Yao Za Zhi. 2017;42(4):607–617. doi:10.19540/j.cnki.cjcmm.2017.0016
 © 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.