Back to Journals » Advances and Applications in Bioinformatics and Chemistry » Volume 16
Synthesis, Biological Evaluation and Molecular Docking Studies of Novel Series of Bis-1,2,4-Triazoles as Thymidine Phosphorylase Inhibitor
Authors Korol N , Holovko-Kamoshenkova OM, Slivka M, Pallah O, Onysko MY, Kryvovyaz A, Boyko NV, Yaremko OV , Mariychuk R
Received 25 April 2023
Accepted for publication 28 July 2023
Published 4 August 2023 Volume 2023:16 Pages 93—102
DOI https://doi.org/10.2147/AABC.S415961
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Nataliya Korol,1 Oksana M Holovko-Kamoshenkova,1 Mikhailo Slivka,1 Oleksandra Pallah,2 Mykhailo Yu Onysko,1 Andriy Kryvovyaz,1 Nadiya V Boyko,2 Olha V Yaremko,3 Ruslan Mariychuk4
1Organic Chemistry Department, Educational and Research Institute of Chemistry and Ecology, Uzhhorod National University, Uzhhorod, Ukraine; 2Department of Clinical and Laboratory Diagnostics and Pharmacology, Faculty of Dentistry, Uzhhorod National University, Uzhhorod, Ukraine; 3Department of Microbiology and Virology, Lviv National Stepan Gzhytsky University of Veterinary Medicine and Biotechnology, Lviv, Ukraine; 4Department of Ecology, Faculty of Humanities and Natural Science, University of Presov, Presov, Slovak Republic
Correspondence: Nataliya Korol, Organic Chemistry Department, Educational and Research Institute of Chemistry and Ecology, Uzhhorod National University, Tomchaniia Street, 13, Uzhhorod, 88001, Ukraine, Tel +380 95 704 34 56, Email [email protected]
Introduction: Heterocyclic compounds have diverse biological activities and potential in drug development. This study aims to synthesize novel compounds with two 1,2,4-triazole cores and evaluate their biological properties, particularly their inhibitory activity against thymidine phosphorylase (TP), an enzyme involved in various physiological processes.
Methods: The compounds were synthesized by reacting 5,5’-butane-bis-1,2,4-triazole derivatives with prenyl bromide. Characterization involved various techniques, including spectroscopy and elemental analysis. Antimicrobial potential was evaluated against bacteria and fungi, with comparative antibiotics as references. Inhibitory activity against TP was assessed, and molecular docking studies were conducted.
Results: Six compounds were successfully synthesized and their structures confirmed. The synthesized triazole derivatives exhibited high biological activity, with compounds 2 and 6 showing the most promising TP inhibition. Molecular docking studies revealed interactions between compound 2 and TP, involving nine amino acids.
Discussion: The synthesis of novel compounds with two 1,2,4-triazole cores contributes significantly to bis-triazole research. These compounds have potential as anti-tumor agents due to their inhibitory activity against TP, a crucial enzyme in tumor growth and metastasis. Comparative evaluation against antibiotics highlights their potency. Docking results provide insights into their interactions with TP, supporting their potential as potent TP inhibitors. Further research should focus on evaluating their efficacy in biological models, understanding their mechanisms of action, and optimizing their activities.
Conclusion: The synthesized compounds with two 1,2,4-triazole cores exhibit significant biological activity, including strong TP inhibition and broad-spectrum antimicrobial effects. These findings emphasize their potential as anti-tumor agents and the need for further exploration and optimization. Future research should focus on evaluating their efficacy in biological models, understanding their mechanisms of action, and developing more potent bis-triazole derivatives for drug discovery efforts. The combined results from assays and docking studies support the therapeutic potential of these compounds as anti-tumor agents.
Keywords: alkylations, antitumor agents, heterocycles, hydrogen bonds, molecular modelling
Introduction
The field of heterocyclic chemistry offers a wide range of valuable compounds, with biological activity being a key indicator of their significance.1,2 In fact, the incorporation of heterocyclic moieties in drug compositions has become increasingly prevalent, with approximately 90% of all drugs containing such components. The triazole ring is no exception to this trend,3–5 as it is known for its biological properties, pharmaceutical effects, and diverse synthesis pathways.6–9 Mononuclear and fused derivatives of 1,2,4-triazole have proven to be particularly valuable compounds, finding applications as antifungal, anticancer, and hypotensive agents (Figure 1).
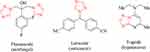 |
Figure 1 Examples of 1,2,4-triazole derivatives used in drugs. The red color serves as a visual indication of the 1,2,4-triazole cores. |
Despite the proven anticancer10 and antifungal11 activities, as well as their potential application as chemosensors,12 bis-triazoles have not been extensively explored in the pharmaceutical field.
In our previous studies, we successfully synthesized fused 1,2,4-triazolium salts13–16 using a low-cost, efficient, and easy workup electrophilic cyclization methodology.17 Building upon this work, our current aim is to obtain new derivatives of 5,5’-butane-bis-1,2,4-triazole, considering the potential enhancement of their biological properties due to the presence of both the butane bridge and the triazole moiety.18 Furthermore, we seek to investigate their inhibitory activity against thymidine phosphorylase (TP).
Thymidine phosphorylase is an angiogenic enzyme that plays a role in pyrimidine salvage to repair RNA and DNA degradation.19 It is known to be associated with various conditions, such as bone loss, sepsis,20 disseminated intravascular coagulation,21 and even as a diagnostic marker for COVID-19.22 Recent investigations23–28 have revealed that many different heterocyclic compounds exhibit effective inhibition of thymidine phosphorylase and demonstrate potent anti-tumor activity.25,29–33
In line with these findings, our research focuses on the synthesis of novel compounds containing two 1,2,4-triazole cores. We aim to evaluate their biological properties, specifically their inhibitory activity against thymidine phosphorylase, through a comprehensive analysis. Additionally, molecular docking studies will be conducted to gain insights into the interactions between the synthesized compounds and the thymidine phosphorylase enzyme.
These combined approaches and techniques will allow us to assess the significance of the obtained compounds and facilitate the design of further transformations aimed at increasing their biological activity. Ultimately, our goal is to explore the potential of these compounds as anti-tumor agents.
Materials and Methods
Chemistry
All reagents were obtained from commercial suppliers and used without any further purification, only hexyl-, heptyl- and octyl-isothiocyanates were obtained according to the protocol described in.34 The melting points were determined on Stuart SMP30 instrument. 1H NMR (400 MHz) and 13C NMR (100 MHz) spectra were recorded in (CD3)2SO as a solvent and TMS as an internal standard on Varian VXR 400. Elemental analyses were performed on Elementar Vario MICRO cube analyzer.
General Procedure for the Synthesis of Compounds 1–6
To a solution of 5,5’-Butane-1,4-diyl(4-substituted-4H-1,2,4-triazole-3-thioles (10.0 mmol) in ethanol (20 mL) was added 12.0 mmol of potassium hydroxide. The mixture was heated. Prenyl bromides (12.0 mmol) in ethanol (5 mL) were added to the cooled solution of triazole. The mixture was boiled for 1 h. After cooling, the precipitated product was filtered, washed with deionized water, and purified by crystallization from ethanol.
5,5’-Butane-1,4-Diylbis(4-Methyl-3-[(3-Methylbut-2-en-1-yl)sulfanyl])-4H-1,2,4-Triazole (1)
White powder (recrystallized from EtOH), Yield: 87%; MW 420.64 g/mol; m.p. 106–107 °C; 1H NMR (400 MHz, DMSO) δ [ppm] 1.44 (6H, s, N–CH3), 1.62 (6H, s, =C–CH3), 1.71 (6H, s, =C–CH3), 2.73 (4H, s, - CH2-CH2), 3.44 (4H, s, - CH2-CH2), 3.61 (4H, d, J = 7.8, - CH2-CH=), 5.26 (2H, t, J = 7.5, - CH2-CH=). 13C NMR (75MHz, DMSO) δ [ppm] 17.66, 24.50, 25.79, 26.11, 30.42, 32.26, 119.40, 137.10, 148.89, 156.16; Anal. found for C20H32N6S2: C, 56.95%, N, 19.74%, H, 7.52%. Calculated: C, 57.11%, H, 7.67%, N, 19.98%, S, 15.25%.
5,5’-Butane-1,4-Diylbis(4-Hexyl-3-[(3-Methylbut-2-en-1-yl)sulfanyl])-4H-1,2,4-Triazole (2)
White powder (recrystallized from EtOH), Yield: 92%; MW 560.90 g/mol; m.p. 114–115°C; 1H NMR (400 MHz, DMSO) δ [ppm] 0.96 (6H, s), 1.25 (6H, s), 1.52 (14H, m), 1.64 (6H, s), 1.78 (6H, s), 2.73 (4H, s), 3.70 (4H, d, J = 7.8, - CH2-CH=), 3.83 (4H, t, J = 7.2), 5.29 (2H, t, J = 7.5, -CH2-CH=). 13C NMR (75MHz, DMSO) δ [ppm] 14.24, 17.76, 22.40, 24.40, 25.76, 26.03, 26.49, 29.96, 31.13, 32.21, 43.54, 119.35, 137.26, 148.78, 155.51; Anal. found for C30H52N6S2: C, 64.08%, N, 15.04, H, 9.27%. Calculated: C, 64.24%, H, 9.34%, N, 14.98%, S, 11.43%.
5,5’-Butane-1,4-Diylbis(4-Heptyl-3-[(3-Methylbut-2-en-1-yl)sulfanyl])-4H-1,2,4-Triazole (3)
White powder (recrystallized from EtOH), Yield: 94%; MW 588.96 g/mol; m.p. 119–120 °C; 1H NMR (400 MHz, DMSO) δ [ppm] 0.85 (6H, s), 1.24 (20H, s), 1.58 (6H, m), 1.71 (6H, m), 2.33 (4H, s), 2.70 (4H, m), 3.69 (4H, d, J = 7.9), 3.89 (4H, s), 5.29 (2H, t, J = 7.5). 13C NMR (75MHz, DMSO) δ [ppm] 14.24, 17.76, 22.40, 24.40, 25.76, 26.03, 26.49, 27.17, 27.30, 29.96, 31.13, 32.21, 43.54, 119.35, 137.26, 148.78, 155.51; Anal. found for C32H56N6S2: C, 65.18%, N, 14.33%, H, 9.61%. Calculated: C, 65.26%, H, 9.58%, N, 14.27%, S, 10.89%.
5,5’-Butane-1,4-Diylbis(4-Octyl-3-[(3-Methylbut-2-en-1-yl)sulfanyl])-4H-1,2,4-Triazole (4)
White powder (recrystallized from EtOH), Yield: 90%; MW 617.01 g/mol; m.p. 120–122 °C; 1H NMR (400 MHz, DMSO) δ [ppm] 0.86 (6H, s), 1.24–1.32 (20H, m), 1.65–1.75 (20H, m), 2.85 (4H, t, J = 7.3), 3.51 (4H, d, J = 6.95), 3.94 (4H, t, J = 7.3), 5.61 (2H, t, J = 6.95). 13C NMR (75MHz, DMSO) δ [ppm] 14.0, 17.9, 22.6, 25.8, 25.9, 26.2,27.1, 27.2, 29.3, 29.4, 31.8, 35.2, 50.1, 119.2, 138.0, 143.2, 150.9; Anal. found for C34H60N6S2: C, 66.21%, N, 13.68%, H, 9.77%. Calculated: C, 66.18%, H, 9.80%, N, 13.62%, S, 10.39%.
5,5’-Butane-1,4-Diylbis(4-Phenyl-3-[(3-Methylbut-2-en-1-yl)sulfanyl])-4H-1,2,4-Triazole (5)
White powder (recrystallized from EtOH), Yield: 97%; MW 544.78g/mol; m.p. 136–137 °C; 1H NMR (400 MHz, DMSO) δ [ppm] 1.45 (4H, s), 1.53 (6H, s), 1.63 (6H, s), 2.43 (4H, s), 3.65 (4H, d, J = 7.6), 5.22 (2H, t, J = 7.0), 7.32 (4H, s), 7.54 (6H, s). 13C NMR (75MHz, DMSO) δ [ppm] 17.93, 24.61, 25.80, 31.14, 119.13, 127.72, 130.25, 133.69, 137.42, 149.97, 155.55; Anal. found for C30H36N6S2: C, 66.09%, N, 15.46%, H, 6.63%. Calculated: C, 66.14%, H, 6.66%, N, 15.43%, S, 11.77%.
5,5’-Butane-1,4-Diylbis(4-(4-Nitrophenyl)-3-[(3-Methylbut-2-en-1-yl)sulfanyl])-4H-1,2,4-Triazole (6)
Yellow powder (recrystallized from EtOH), Yield: 93%; MW 634.77g/mol; m.p. 141–142 °C; 1H NMR (400 MHz, DMSO) δ [ppm] 1.49 (4H, s), 1.55 (6H, s), 1.62 (6H, s), 3.65 (4H, d, J = 8.0), 5.18 (2H, t, J = 7.6), 5.57 (1H, s), 6.62 (2H, m), 6.84 (2H, m), 7.72 (2H, d, J = 8.8), 7.92 (1H, d, J = 9.1), 8.39 (4H, d, J = 8.8). 13C NMR (75MHz, DMSO) δ [ppm] 17.86, 24.58, 25.58, 25.73, 31.63, 112.82, 118.87, 125.46, 126.69, 128.00, 129.38, 137.67, 138.94, 148.22, 149.59, 155.39; Anal. found for C30H34N8O4S2: C, 56.73%, N, 17.59%, H, 5.42%. Calculated: C, 56.76%, H, 5.40%, N, 17.65%, O, 10.08%, S, 10.10%.
Biological Section
Comparative antibiotics were commercially obtained from India («HiMedia», India). Dimethyl Sulfoxide (DMSO) was used to prepare stock solutions of test compounds. Bacterial strains (Staphylococcus aureus Ks-02, Pseudomonas aeruginosa Kp-02, Escherichia coli Ke-06, Proteus mirabili Km-02, Enterococcus faecalis Ke-05, Salmonella enterica Ksa-32, Enterobacter cloacae Kc-12, Klebsiella pneumonia Kk-12, Candida albicans Kl-03, Saccharomyces cerevisiae Ksc-22) were obtained from surgically drained soft tissue abscesses from patients of the hospital of the Transcarpathian Regional Clinical Center of Neurosurgery and Neurology, Uzhhorod, Ukraine. Subsequent confirmation of the cultures was done using conventional microbiological techniques including Gram staining, microscopic examination, mannitol fermentation, β-hemolysis assay using 5% sheep’s blood agar, catalase test using 3% hydrogen peroxide and coagulase test (PlivaLaChema, Chezh Republic). Conventional characterization was further validated by PCR amplification and DNA sequencing of 16srRNA gene (Agilent Technologies, USA). Only those isolates were subjected to molecular characterizations which were confirmed as being Multidrug Resistant (MDR) after screening through Kirby-Bauer Disk Diffusion Assay.
All clinical strains used in this study are preserved in the collection of microorganisms at the Research and Educational Center for Molecular Microbiology and Immunology of Mucous Membranes at Uzhhorod National University. The Ethics Committee of the State Higher Educational Establishment «Uzhhorod National University» approved the study protocol No. 7/3 on 12/22/2021.
Determination of Minimum Inhibitory Concentration (MIC)
Minimum Inhibitory Concentration (MIC) of selected antibiotics and the test compounds against test organisms (0.25–512 μg/mL) was determined by broth microdilution method in accordance with CLSI guidelines (CLSI, 2019). In this assay, antibiotics and test compounds were serially diluted using Mueller Hinton Broth (MHB) (Sigma-Aldrich, USA) in such a way that each well of 96 well microtiter plate (DeltaLab, Spain) contained half the concentration of antibiotic present in preceding well. Finally, 5 × 106 of bacterial cells were inoculated into each well except for wells labeled negative control after 24 h incubation. To enhance the clarity of the growth end points, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT dye) was added to each well at a concentration of 0.5 μg/mL. The sealed plates were then incubated on a shaking incubator at 37°C and 80 rpm for 2–3 hours and subsequently examined for any discernible color changes.
Determination of TP Inhibition Activity
TP/PD-ECGF (E. coli TP (Sigma T6632)) activity was determined by measuring the absorbance at 290 nm spectrophotometrically (DeNovix Inc., USA). The original method is reported in Krenitsky and Bushby.35 In 96 wells, flat bottom, microplate with each well capacity 200 μL, reaction mixture of 200 μL was prepared which contained 145 μL of potassium phosphate buffer (pH 7.4), 20 μL of 1.5 mM Thymidine-5’-monophosphate solution as substrate, 30 μL of enzyme (E. coli TP (Sigma T6632)) at concentration 0.05 and 0.002 U, respectively, were incubated with 5 μL of test materials for 10 min at 25°C in temperature controlled incubator before taking readings by microplate reader ELx800 (BioTek, USA) at 290 nm. The wells containing a reaction mixture without substrate served as blanks, and the mean optical density (OD) of these blank wells was subtracted from wells containing the reaction mixture with substrate. The readings were taken continuously after 10, 20, and 30 min using a microplate reader. All assays were performed in triplicate.
Molecular Docking
Molecular docking studies of molecule 2 that were docked into the crystal structures of thymidine phosphorylase in E. coli with PDB_ID: 4LHM were carried out using Autodock Vina software,36 an open-source molecular docking software. First, we optimized the structure of the enzyme using BIOVIA Discovery Studio 2021 software (https://discover.3ds.com/discovery-studio-visualizer-download). We utilized this software for adding polar hydrogens to the enzyme structure and performing energy minimization. Additionally, we employed HyperChem 7 (http://www.hypercubeusa.com/) to optimize the structure of compound 2. We have generated a grid box with desired parameters around the active site of thymidine phosphorylase (PDB_ID: 4LHM) as center: x = 31.889, y = −12.619, z = 4.741 and grid box size: x = 20, y = 20, z = 20 with 1Å grid spacing. We used the advanced Genetic algorithm method in Vina Protein/TP to generate 10 conformations in each docking output. The molecule input preparations and docking output analysis were carried out using Discovery Studio software.
Results
Synthesis
The starting bis-triazoles were prepared using adipic acid dihydrazide. The synthesis involved refluxing the dihydrazide with the corresponding isothiocyanates in ethanol for one hour, leading to the formation of 5,5’-butane-bis-1,2,4-triazole-3-thiones 1–6. Subsequently, the thiosemicarbazides were subjected to prolonged heating in an aqueous solution of potassium hydroxide, resulting in the formation of triazoles. These triazoles were then treated with a double excess of prenyl bromide, leading to the excellent yield of the alkylated triazoles 1–6 (Scheme 1).
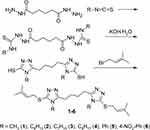 |
Scheme 1 Synthetic route for obtaining target compounds 1–6. |
The NMR spectra of the synthesized compounds exhibited signals that fully matched the assigned structures, confirming the success of the synthesis and the purity of the products. Specifically, the chemical shifts of the aromatic and aliphatic protons were consistent with the predicted structures, and no unexpected peaks were observed. This analysis provided further evidence for the accurate synthesis and high quality of the compounds.
In vitro Antibacterial Activity
To assess the biological activity of the synthesized compounds, we conducted a comprehensive evaluation using a panel of 10 different bacteria, including both Gram-positive and Gram-negative strains. The results obtained from these assays revealed that the alkylated derivatives 1–6 exhibited moderate levels of biological activity. Notably, compounds 1–4, which incorporate alkyl substituents at the 4 position of the 1,2,4-triazole moiety, displayed higher levels of biological activity compared to compounds with aromatic substituents at the same position.
In fact, several of the tested compounds demonstrated comparable or even greater activity when compared to the reference antibiotic. Specifically, compound 2 exhibited notable activity against Escherichia coli, while compound 5 showed significant inhibitory effects against Saccharomyces cerevisiae. Additionally, compounds 1, 2, 5, and 6 displayed promising activity against Pseudomonas aeruginosa. An overview of all the obtained results is presented in Table 1, providing a clear comparison of the biological activity of each compound.
 |
Table 1 Comparison of Minimum Inhibitory Concentration (MIC (μg/mL)) of Compounds 1–6 Against Gram-Positive and Gram-Negative Bacteria |
The observed trends in the biological activity of the alkylated derivatives highlight the importance of the specific substitution pattern at the 4 position of the 1,2,4-triazole moiety. The presence of alkyl substituents in this position appears to enhance the compounds’ antimicrobial potential, underscoring the significance of structural modifications in modulating their biological activity. These findings contribute to a better understanding of the structure–activity relationship and guide future optimization efforts to develop more potent compounds with improved antimicrobial properties.
The evaluation of the synthesized compounds against a diverse panel of bacteria provides valuable insights into their potential as \antimicrobial agents. While the observed biological activity is moderate, it establishes a promising starting point for further optimization and development of more potent derivatives. Subsequent investigations should focus on elucidating the mechanisms of action and conducting additional structure–activity relationship studies to unlock the full therapeutic potential of these compounds as antimicrobial agents.
In order to facilitate a meaningful comparison of the obtained results, we have converted the data for compounds 1–6 into IC50 values in micromolar (μM) concentrations, as presented in Table 2.
 |
Table 2 Comparison of Minimum Inhibitory Concentration (IC50 (μM)) of Compounds 1–6 Against Gram-Positive and Gram-Negative Bacteria |
Based on the antimicrobial activity data of the synthesized compounds, a notable trend can be observed. Compounds 1–4 exhibited greater activity against Gram-positive bacteria, while compounds 5 and 6 displayed enhanced activity against Gram-negative bacteria. This distinction in activity against different bacterial types underscores the potential versatility and broad-spectrum nature of the synthesized compounds.
Overall, the results presented in Table 2 indicate the promising antimicrobial activity of compounds 1–6, with distinct preferences for different types of bacteria. These findings contribute to our understanding of the compounds’ potential as antimicrobial agents and pave the way for future research and development efforts in this field.
Thymidine Phosphorylase Inhibitory Activity
The inhibitory activity of prenyl-alkylated triazoles 1–6 against thymidine phosphorylase was determined using a spectrophotometric protocol,37 and the results are presented as IC50 values. Notably, all the tested compounds demonstrated moderate to excellent inhibitory activity against thymidine phosphorylase.
Among the series of compounds tested, compound 2, which contains a hexyl substituent, exhibited the highest inhibitory activity with an IC50 value of 28.74 ± 0.59 μM. This fact indicates that compound 2 is the most potent inhibitor in the series. Additionally, compound 6, which features a p-nitrophenyl group in the 4th position, also showed significant inhibitory activity with an IC50 value of 34.95 ± 0.73 μM, further highlighting its potential as an effective inhibitor.
The results presented in Table 3 demonstrate the efficacy of the tested compounds in inhibiting thymidine phosphorylase. Further investigations can explore their mechanisms of action, conduct structure–activity relationship studies, and evaluate their efficacy in relevant biological models.
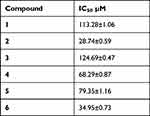 |
Table 3 Inhibition of Thymidine Phosphorylase by Triazoles 1–6 |
Molecular Docking Study
The leading compound 2 was chosen for the molecular docking study to analyze its interaction with the protein target. For this investigation, the crystallographic structure of thymidine phosphorylase (TP) in E. coli, specifically the 4LHM (pdb), was selected as the protein of interest. The selection of this target protein is based on the essential role of TP in E. coli bacteria, making it an intriguing choice to explore the interaction characteristics between the synthesized compounds and the bacterial enzyme.
The affinity value of the interaction between the optimized conformation of compound 2 and the active site of the protein 4LHM was determined to be −6.9 kcal/mol, indicating a favorable binding affinity. This interaction involved nine amino acids in the binding process. Notably, Lys190 participated in the interaction with the triazole ring through hydrogen bonding and electrostatic interactions. Val177 and Phe210 formed hydrophobic bonds with the triazole group, while Met211 engaged in a pi-sulfur bond interaction. His85 was observed to interact with both the hexyl group and the sulfur atom at the 5 position of the triazole ring through pi-alkyl and pi-bonds, respectively. Leu117 contributed to the interaction by forming a hydrophobic bond with the alkyl part of the prenyl group and the hexyl substituent. Additionally, hydrophobic interactions were observed between Tyr168 and the triazole ring, as well as between Ile183 and the butane bridge, and Ile187 and the hexyl group (Figure 2).
 |
Figure 2 Ligand interaction diagram of compound 2 in the active site of 4LHM in 3D (a) and 2D (b) visualization. |
Obtained results provide valuable insights into the specific interactions between compound 2 and the active site of thymidine phosphorylase, shedding light on the potential mechanisms of action and aiding in the understanding of the compound’s inhibitory properties.
Discussion
The variation in the biological activity of the synthesized compounds can be attributed to structural differences in the cell wall of the bacteria. Gram-negative bacteria are characterized by the presence of an outer lipid membrane, which serves as a selective permeation barrier. This outer membrane restricts the entry and access of substances to the periplasmic space, thereby influencing the interaction and activity of the compounds.37 Based on the presence of a p-nitrophenyl substituent at the 4 position of the triazole nucleus in compound 6, it can be inferred that this fragment plays a role in enhancing the compound’s activity against Gram-negative bacteria. The beneficial influence of a p-nitrophenyl moiety on biological properties has been previously described, further supporting its positive impact in this context.38
Our research not only uncovered the antibacterial activity of the synthesized compounds against both Gram-positive and Gram-negative bacteria but also highlighted their potential as antifungal agents. Specifically, our findings indicated that derivatives 1, 2, 3, and 6, which contain methyl, hexyl, heptyl, and p-nitrophenyl substituents at the 4th position of the triazole moiety, exhibited enhanced antifungal activity against Candida albicans. Additionally, compounds 1, 3, 4, 5, and 6, comprising methyl, heptyl, octyl, phenyl, and p-nitrophenyl fragments, respectively, demonstrated activity against the yeast Saccharomyces cerevisiae. These results align with previously published studies that have reported on the antibacterial and antifungal properties of similar compounds.39
Based on the obtained results, it is evident that the synthesized triazole derivatives have the potential to serve as valuable agents for the design and development of novel therapeutic drugs to combat bacterial resistance.
Particularly noteworthy is the fact that compounds 2 and 6 exhibited greater TP inhibition activity compared to the standard 7-deazaxanthine. This highlights their promising prospects in targeting and inhibiting the activity of thymidine phosphorylase, an enzyme involved in various physiological processes, and provides evidence that these substances are suitable candidates for further clinical studies as potent anti-tumor drugs.40
The notable correlation between the minimum inhibitory concentration (MIC) values against the E. coli strain and the corresponding TP inhibitory activity values can be attributed to the fact that the crystalline structure of the TP enzyme was obtained from the same bacterial species. This correlation reinforces the potential significance of targeting thymidine phosphorylase as a strategy for combating tumor growth.41–44
Conclusion
We have conducted a comprehensive investigation on a novel series of bis-1,2,4-triazoles, exploring their potential as antibacterial and fungicidal agents, efficient inhibitors of thymidine phosphorylase (TP), and promising anti-tumor compounds. Our results demonstrate that most of the synthesized compounds possess significant biological activity against various bacteria. Notably, the alkylated derivatives displayed excellent TP inhibition, with two compounds exhibiting higher inhibitory activity than the standard reference, 7-deazaxanthine (IC50 = 41.0 ± 1.63 μM). Besides, molecular docking analysis revealed a specific interaction between the lead compound 2 and Lys190 through a hydrogen bond. These results provide valuable insights for the design of advanced compounds with enhanced inhibitory activity against the TP enzyme, thus offering potential prospects for their utilization as effective anti-tumor agents.
Acknowledgments
This study was partially supported by the Ministry of Education and Science of Ukraine (Project SR-0122U000936) and the National Scholarship Programme of the Slovak Republic (Grant ID 39272).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Saini MS, Kumar A, Dwivedi J, Singh R. A review: biological significances of heterocyclic compounds. Int J Pharm Sci Res. 2013;4:66–77.
2. Al-Mulla A. A review: biological importance of heterocyclic compounds. Der Pharma Chem. 2017;13:141–147.
3. Taylor AP, Robinson RP, Fobian YM, Blakemore DC, Jones LH, Fadeyi O. Modern advances in heterocyclic chemistry in drug discovery. Org Biomol Chem. 2016;14:6611–6637. doi:10.1039/C6OB00936K
4. Gomtsyan A. Heterocycles in drugs and drug discovery. Chem Heterocycl Comp. 2012;48(1):7–10. doi:10.1007/s10593-012-0960-z
5. Heravi MM, Zadsirjan V. Prescribed drugs containing nitrogen heterocycles: an overview. RSC Adv. 2020;10(72):44247–44311. doi:10.1039/d0ra09198g
6. Küçükgüzel SG, Çıkla-Süzgün P. Recent advances bioactive 1,2,4-triazole-3-thiones. Eur J Med Chem. 2015;97:830–870. doi:10.1016/j.ejmech.2014.11.033
7. Korol NI, Slivka MV. Recent progress in the synthesis of thiazolo[3,2-b][1,2,4]triazoles (microreview). Chem Heterocycl Comp. 2017;53(8):852–854. doi:10.1007/s10593-017-2136-3
8. Slivka MV, Korol NI, Fizer MM. Fused bicyclic 1,2,4-triazoles with one extra Sulfur atom: synthesis, properties, and biological activity. J Heterocyclic Chem. 2020;57:3236–3254.
9. Korol N, Slivka M, Holovko-Kamoshenkova O. Recent advances in 1,2,4-triazole ring construction via cycloaddition reactions. Heterocycles. 2021;102(11):2099–2117. doi:10.3987/REV-21-958
10. Meixiang G, Qiang D, Feng D, Xiangyang S, Jiaqi X. Bis-triazole-containing compounds with anticancer potential: a short review. Curr Top Med Chem. 2021;21(18):1674–1691. doi:10.2174/1568026621666210728154728
11. Richardson K, Brammer KW, Marriott MS, Troke PF. Activity of UK-49,858, a bis-triazole derivative, against experimental infections with Candida albicans and Trichophyton mentagrophytes. Antimicrob Agents Chemother. 1985;27(5):832–835. doi:10.1128/AAC.27.5.832
12. Dawood KM, Abdel‐Wahab BF, Raslan MA. Synthesis and applications of bi‐ and bis‐triazole systems. Arkivoc. 2018;I:
13. Slivka M, Korol N, Rusyn I, Lendel V. Synthesis of [1,3]thiazolo[3,2-b][1,2,4]triazol-7-ium and [1,2,4]triazolo[5,1-b][1,3]thiazin-4-ium salts via regioselective electrophilic cyclization of 3-[(2-alken-1-yl)sulfanyl]-4H-1,2,4-triazoles. Heterocycl Commun. 2015;21(6):397–401. doi:10.1515/hc-2015-0158
14. Slivka M, Korol N, Pantyo V, Baumer V, Lendel V. Regio- and stereoselective synthesis of [1,3]thiazolo[3,2-b][1,2,4]triazol-7-ium salts via electrophilic heterocyclization of 3-S-propargylthio-4Н-1,2,4-triazoles and their antimicrobial activity. Heterocycl Commun. 2017;23(2):109–113. doi:10.1515/hc-2016-0233
15. Slivka M, Korol N, Fizer M, Baumer V, Lendel V. [1,3]Thiazolo[3,2-b][1,2,4]triazol-7-ium salts: synthesis, properties and structural studies. Heterocycl Commun. 2018;24(4):197–203. doi:10.1515/hc-2018-0048
16. Korol N, Slivka M, Fizer M, Baumer V, Lendel V. Halo-heterocyclization of butenyl(prenyl)thioethers of 4,5-diphenyl-1,2,4-triazol-3-thiole into triazolo[5,1-b][1,3]thiazinium systems: experimental and theoretical evolution. Monatsh Chem. 2018;151(2):191–198. doi:10.1007/s00706-019-02545-w
17. Slivka M, Onysko M. The use of electrophilic cyclization for the preparation of condensed heterocycles. Synthesis. 2021;53(19):3497–3512. doi:10.1055/s-0040-1706036
18. Kuca K, Kassa J. A Comparison of the ability of a new bispyridinium oxime—1-(4-hydroxyiminomethylpyridinium)-4-(4-carbamoylpyridinium)butane dibromide and currently used oximes to reactivate nerve agent-inhibited rat brain acetylcholinesterase by in vitro methods. J Enzyme Inhib Med Chem. 2003;18(6):529–535. doi:10.1080/14756360310001605552
19. Friedkin M, Roberts D. The enzymatic synthesis of nucleosides: I. Thymidine phosphorylase in mammalian tissue. J Biol Chem. 1954;207(1):245–256. doi:10.1016/S0021-9258(18)71264-7
20. Matsumae G, Shimizu T, Tian Y, et al. Targeting thymidine phosphorylase as a potential therapy for bone loss associated with periprosthetic osteolysis. Bioeng Transl Med. 2021;6(3):e10232. doi:10.1002/btm2.10232
21. Zeng Y, Huang G, Li J, et al. Thymidine phosphorylase is increased in patients with infection and could be a prognostic marker for sepsis and disseminated intravascular coagulation. Res Square. 2021. doi:10.21203/rs.3.rs-610842/v1
22. Li W, Yue H. Thymidine phosphorylase is increased in COVID-19 patients in an acuity-dependent manner. Front Med. 2021;22:653773. doi:10.3389/fmed.2021.653773
23. Iftikhar F, Yaqoob F, Tabassum N, et al. Design, synthesis, in-vitro thymidine phosphorylase inhibition, in-vivo antiangiogenic and in-silico studies of C-6 substituted dihydropyrimidines. Bioorg Chem. 2018;80:99–111. doi:10.1016/j.bioorg.2018.05.026
24. Ullah H, Rahim F, Taha M, et al. Synthesis, molecular docking study and in vitro thymidine phosphorylase inhibitory potential of oxadiazole derivatives. Bioorg Chem. 2018;78:58–67. doi:10.1016/j.bioorg.2018.02.020
25. Shahzad SA, Yar M, Khan ZA, et al. Identification of 1,2,4-triazoles as new thymidine phosphorylase inhibitors: future anti-tumor drugs. Bioorg Chem. 1029;85:209–220. doi:10.1016/j.bioorg.2019.01.005
26. Zaman K, Rahim F, Taha M, et al. Synthesis, thymidine phosphorylase, angiogenic inhibition and molecular docking study of isoquinoline derivatives. Bioorg Chem. 2019;89:102999. doi:10.1016/j.bioorg.2019.102999
27. Taha M, Aldhamin EAJ, Almandil NB, et al. Synthesis of indole based acetohydrazide analogs: their in vitro and in silico thymidine phosphorylase studies. Bioorg Chem. 2020;98:103745. doi:10.1016/j.bioorg.2020.103745
28. Tong JB, Feng Y, Wang TH, Luo D. Investigation of quantitative structure activity relationship of isatin-based oxadiazole derivatives as thymidine phosphorylase inhibitors. Chin J Anal Chem. 2021;49(4):e21046–e21054. doi:10.1016/S1872-2040(21)60095-6
29. Akiyama S-I, Furukawa T, Sumizawa T, Takebayashi Y, Nakajima Y, Shimaoka S. The role of thymidine phosphorylase, an angiogenic enzyme, in tumor progression. Cancer Sci. 2004;95(11):851–857. doi:10.1111/j.1349-7006.2004.tb02193.x
30. Shahzad SA, Yar M, Bajda M, et al. Synthesis and biological evaluation of novel oxadiazole derivatives: a new class of thymidine phosphorylase inhibitors as potential anti-tumor agents. Bioorg Med Chem. 2014;22(3):1008–1015. doi:10.1016/j.bmc.2013.12.043
31. Kanyama H, Tomita N, Yamano T, et al. Enhancement of the anti-tumor effect of 5′-deoxy-5-fluorouridine by transfection of thymidine phosphorylase gene into human colon cancer cells. Jpn J Cancer Res. 2005;90:454–459. doi:10.1111/j.1349-7006.1999.tb00769.x
32. Emura T, Suzuki N, Fujioka A, Ohshimo H, Fukushima M. Potentiation of the antitumor activity of α,α,α-trifluorothymidine by the co-administration of an inhibitor of thymidine phosphorylase at a suitable molar ratio in vivo. Int J Oncol. 2005;27:449–455.
33. Takiguchi N, Ishii R, Koda K, Oda K, Miyazaki M. Thymidine phosphorylase expression correlates with malignant potential and anti-tumor effect of doxifluridine on gastric cancer: multivariate analysis for adjuvant chemotherapy doxifluridine vs. 5-fluorouracil. Oncol Rep. 2003;10:1105–1111.
34. Tamotsu Y, Susumu S, Kenji A, Kiyoshi H. One-pot synthesis of isothiocyanates from primary amines synthesis using cyanamide. Org Prep Proced Int. 1991;24:346–349.
35. Krenitsky TA, Bushby SRM. US Patent 4, 178, 212, 1–8. Research Triangle Park (NC): BurroughsWelcome CO.; 1979.
36. Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–461.
37. Champlin FR, Ellison ML, Bullard JW, Conrad RS. Effect of outer membrane permeabilisation on intrinsic resistance to low triclosan levels in Pseudomonas aeruginosa. Int J Antimicrob Agents. 2005;26(2):159–164. doi:10.1016/j.ijantimicag.2005.04.020
38. Hasan S, Danishuddin M, Adil M, Singh K, Verma PK. Efficacy of E. officinalis on the cariogenic properties of Streptococcus mutans: a novel and alternative approach to suppress quorum-sensing mechanism. PLoS One. 2012;7(7):e40319. doi:10.1371/journal.pone.0040319
39. Dhanavath R, Dharavath R, Kothula D, et al. Synthesis and biological evaluation of novel 2‐arylquinoline‐3‐fused thiazolo[2,3‐c]1,2,4‐triazole heterocycles as potential antiproliferative and antimicrobial agents. J Heterocycl Chem. 2022;59(7):1198–1212. doi:10.1002/jhet.4460
40. Desgranges C, Razaka G, Rabaud M, Picard P, Dupuch F, Bricaud H. The human blood platelet: a cellular model to study the degradation of thymidine and its inhibition. Biochem Pharmacol. 1982;31(17):2755–2759. doi:10.1016/0006-2952(82)90129-0
41. Schwartz M. Thymidine phosphorylase from Escherichia coli. Eur J Biochem. 1971;21(2):191–198. doi:10.1111/j.1432-1033.1971.tb01455.x
42. Schwartz M. [59] Thymidine phosphorylase from Escherichia coli. Methods Enzymol. 1978;51:441–445.
43. Walter R, Cook WJ, Cole LB, et al. Three-dimensional structure of thymidine phosphorylase from Escherichia coli at 2.8 A resolution. J Bio Chem. 1990;265(23):14016–14022. doi:10.1016/S0021-9258(18)77450-4
44. Esteban-Gamboa A, Balzarini J, Esnouf R, De Clercq E, Camarasa M-J, Pérez-Pérez M-J. Design, synthesis, and enzymatic evaluation of multisubstrate analogue inhibitors of Escherichia coli thymidine phosphorylase. J Med Chem. 2000;43(5):971–983. doi:10.1021/jm9911377
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
