Back to Journals » Clinical Epidemiology » Volume 13
Swedish Covid-19 Investigation for Future Insights – A Population Epidemiology Approach Using Register Linkage (SCIFI-PEARL)
Authors Nyberg F , Franzén S, Lindh M, Vanfleteren L , Hammar N, Wettermark B , Sundström J , Santosa A, Björck S, Gisslén M
Received 30 March 2021
Accepted for publication 12 July 2021
Published 30 July 2021 Volume 2021:13 Pages 649—659
DOI https://doi.org/10.2147/CLEP.S312742
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Eyal Cohen
Fredrik Nyberg,1 Stefan Franzén,1,2 Magnus Lindh,3,4 Lowie Vanfleteren,5,6 Niklas Hammar,7 Björn Wettermark,8 Johan Sundström,9,10 Ailiana Santosa,1 Staffan Björck,11 Magnus Gisslén3,12
1School of Public Health and Community Medicine, Institute of Medicine, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden; 2National Diabetes Register, Centre of Registers Västra Götaland, Gothenburg, Sweden; 3Department of Infectious Diseases, Institute of Biomedicine, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden; 4Department of Clinical Microbiology, Sahlgrenska University Hospital, Gothenburg, Sweden; 5COPD Center, Department of Respiratory Medicine and Allergology, Sahlgrenska University Hospital, Gothenburg, Sweden; 6Department of Internal Medicine and Clinical Nutrition, Institute of Medicine, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden; 7Unit of Epidemiology, Institute of Environmental Medicine, Karolinska Institute, Stockholm, Sweden; 8Pharmacoepidemiology & Social Pharmacy, Department of Pharmacy, Uppsala University, Uppsala, Sweden; 9Clinical Epidemiology Unit, Department of Medical Sciences, Uppsala University, Uppsala, Sweden; 10The George Institute for Global Health, University of New South Wales, Sydney, Australia; 11Centre of Registers Västra Götaland, Gothenburg, Sweden; 12Region Västra Götaland, Department of Infectious Diseases, Sahlgrenska University Hospital, Gothenburg, Sweden
Correspondence: Fredrik Nyberg
School of Public Health and Community Medicine, Institute of Medicine, Sahlgrenska Academy, University of Gothenburg, Box 463, Gothenburg, 405 30, Sweden
Tel +46 31 7866289; +46 76 6186289
Email [email protected]
Background: In response to the Covid-19 pandemic, we designed and initiated a nationwide linked multi-register, regularly updated, observational study for timely response to urgent scientific questions.
Aim: To describe the SCIFI-PEARL (Swedish Covid-19 Investigation for Future Insights – a Population Epidemiology Approach using Register Linkage) linked database encompassing essentially all known diagnosed Swedish Covid-19 patients plus a large general population comparison cohort and outline its utility in the current and future phases of the pandemic.
Methods: Individuals with Covid-19 from the entire country are identified on a regularly updated basis, from different sources: all individuals from SmiNet, the national database of notifiable diseases, with positive SARS-CoV-2 polymerase chain reaction (PCR) test results; patients identified in the healthcare system by condition (ICD-10) or procedure codes in the National Patient Register or Cause-of-Death Register; patients identified through several disease-specific national quality registers (NQRs); and in two regions additionally patients identified in primary care. A comparison population was obtained by stratified random sampling from Swedish national population registers. Data from all these registers plus the National Prescribed Drug Register, the Cancer Register, national sociodemographic registers, some additional NQRs, the National Vaccination Register, and further data sources, are then linked to all study subjects (Covid-19 cases and population cohort). New cases in the study population and all data for all subjects are updated every few months, as required.
Conclusion and Utility: The SCIFI-PEARL study cohort captures Swedish residents with Covid-19 on an ongoing basis, includes a representative general population comparison cohort, and links to a broad range of national and regional healthcare data for a comprehensive longitudinal view of the Covid-19 pandemic. By combining high-quality national registers with short time delay and continuous repeated linkage and updating, the project brings timely and internationally relevant data for epidemiological research on SARS-CoV-2. Our efforts provide an example and important learnings for similar efforts internationally in the future.
Keywords: SARS-CoV-2, population cohort, data updates, observational research, longitudinal
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) first emerged in Wuhan in December 2019,1 and was soon found to cause the disease now named coronavirus disease 2019 (Covid-19). During 2020, the disease developed into a large-scale pandemic, spreading to most countries in the world, including entering a second or third wave in many places, with cumulative numbers of over 83 million cases and 1.8 million deaths reported globally during the first year until end of 2020.2 By the end of 2020, vaccines had been developed and vaccination campaigns were initiated in multiple countries, including Sweden. The first Covid-19 case in Sweden was reported on 31 January 2020, and in parallel with Europe becoming the epicenter for the epidemic during the Spring of 2020, the disease rapidly expanded in Sweden, causing severe strain on society and the healthcare system.3
It was clear at the time that research both in the short and long term would be required, and the research communities in Sweden and internationally quickly geared up to respond to the challenge. The study described here based on Swedish data was rapidly designed as a large-scale effort to assemble a rich database for the study of Covid-19 epidemiologically in real time, ie, with the requirement that data would be regularly and sufficiently frequently updated. The importance of not only focusing on Covid-19 patients was also recognized, making the inclusion of a large comparison cohort reflecting the general population another key asset.
Even against the background of fast-track processes for ethical approval and data access for Covid-19 research developing at the time,4 this has been quite a complex task without clear precedent in the Swedish registry data ecosystem. This paper describes the database now established under the name of SCIFI-PEARL (Swedish Covid-19 Investigation for Future Insights – a Population Epidemiology Approach using Register Linkage), and outlines its research potential and utility in the ongoing and future phases of the pandemic.
Materials and Methods
Sweden, similar to other Nordic countries, has high quality national health and population registers that can be linked with a high degree of accuracy, providing an extraordinary suitable environment for epidemiologic research. The Swedish Personal Identity Number (PIN; personnummer) uniquely linked to each individual enables compilation and linkage of data from diverse register data to the same individuals, with relative ease and with extremely high accuracy.5 This SCIFI-PEARL study exploits the possibility to assemble a unique large database of individuals with Covid-19 plus a population cohort. After linkage, the database is pseudonymised, according to usual procedures.
Study Population
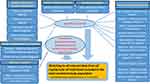
Newly identified individuals with Covid-19 from the entire country are included in the linked database on a regularly updated basis, from different sources (Figure 1). This includes all individuals with a positive SARS-CoV-2 polymerase chain reactions (PCR) test results identified from SmiNet,6 the national register of notifiable communicable diseases managed by the Public Health Agency of Sweden. In addition, patients who may not have a positive test registered but have been identified by the healthcare system are included, eg, by Covid-19 ICD codes (ie, ICD-10 U07.1 = Covid-19, virus demonstrated; U07.2 = Covid-19, virus not shown) or relevant procedure codes on a national basis from the National Patient Register (NPR) for outpatient and hospitalized inpatient specialist care.7 Similarly, individuals recorded in the National Cause-of-Death Register (NCDR)8 as deaths due to Covid-19 are included. In addition, registration of test result and ICD diagnoses made by several disease-specific national quality registers (NQRs) are scrutinized to identify additional potential Covid-19 patients with healthcare contacts related to their particular disease and therefore captured in that register. Further, since there is no national coverage for primary care data, healthcare contacts for Covid-19 in primary care are captured on a regional basis, for Western Sweden (Västra Götaland Region) and the Stockholm Region.
In addition, a general population comparison group was obtained using the national Register of the Total Population (RTB) at Statistics Sweden,9 consisting of approximately 10% of the Swedish population (~1,000,000 individuals), based on individuals of all ages resident in Sweden on 1 January 2020, through stratified random sampling in 20 age (5-year age categories) by 2 sex strata. This population can be used either in analyses as it is, weighted to the Swedish population through sampling weights, or subsampled, as required for different types of statistical analyses and research questions.
Data Acquisition
To this combined study population of individuals with Covid-19 from the whole Swedish population plus the 10% general population comparison group, we linked diverse data from a range of registers with relevant information, to get a comprehensive and detailed view of patient history (Figure 1, Table 1). For practical and ethical reasons the actual linkage is performed by the National Board of Health and Welfare (NBHW), and the PIN for each subject is replaced by an individual study subject identifier, to pseudonymise the data.
 |
 |
 |
Table 1 Source Registers and Types of Data Currently Available in the SCIFI-PEARL Study Database |
The bulk of the data are from different national registers. Date and cause of death are obtained from RTB and the NCDR. In-patient and specialist outpatient healthcare data from 2015 onward, comprising a complete specialist care medical history for all individuals, are obtained from the National Patient Register (NPR). Cancer incidence data from 2015 are extracted from the National Cancer Register (NCR). A complete drug history for prescription drugs from 2018 is obtained from the National Prescribed Drug Register (NPDR).10 Sociodemographic data including education, family situation, income and occupational data from 2018 onward are obtained from the Statistics Sweden Longitudinal Integrated Database for Health Insurance and Labour Market Studies (LISA; Longitudinell Integrationsdatabas för Sjukförsäkrings- och Arbetsmarknadsstudier).11 Finally, data from a range of national quality registers are included.12 These registers are national in scope and provide detailed information on Covid-19 management, as well as relevant comorbidities and risk factors of importance for Covid-19 patients, with high granularity and detail, on all recorded individuals with the respective underlying disease or condition covered by that particular NQR (Table 1). They include registers for acute emergency care and intensive care (particularly important for Covid-19 patient data) and the Swedish Register of Cardiopulmonary Resuscitation (SRCR) with data extracted from 1 Jan 2020, as well as registers for infectious diseases, several cardiovascular disease registers, 2 respiratory disease registers, and the National Diabetes Register (NDR) with data from 2015. The database is also linked to the National Vaccination Register (NVR), which following urgent pandemic legislative changes in Dec 2020 captures information on vaccinations against Covid-19 in Sweden.
In addition, some data not available on a consolidated national basis are included in the database for a more detailed regional view. For Western Sweden (Västra Götaland Region) and the Stockholm Region, with a population of 4,902,162 out of 10,327,589 nationally (39.7%%), healthcare contacts and diagnoses recorded in primary care are captured from 2015. For Western Sweden, a data source with data on SARS-CoV-2 testing from the start of the pandemic (both positive and negative tests), is also included in the database, to provide data on test penetration over time in the population.
All data are updated with the most recent data from all included registers on a regular basis, every few months, as required by the study objectives and the evolution of the pandemic, in an agreed process triggered each time by a request from the study team to all registers. The complex update process from extraction of data at each data source to linkage and delivery of a new complete dataset is designed to take around 1 month.
Data Management
The study population selection, data linkages and pseudonymisation of the data are managed in secure environments at the NBHW and Statistics Sweden. The pseudonymized research database thus produced and delivered is then housed on a dedicated server at the Centre of Registers Västra Götaland, which has extensive experience with large databases, as it houses and manages a number of NQRs. Data transfers use state-of-the-art encryption and the project-specific hardware is maintained with high protection levels relevant for sensitive personal data, with regular backups. Access protection includes disc encryption, network segmentation and two-level firewall protection with differential access rights based on project role.
Ethics
The study, including regular updates until at least end of 2025, has received ethical approval from the Swedish Ethical Review Authority.
Research Objectives
The core initial research objectives outlined in the project plan are relatively broad and cover descriptive, analytical and predictive modeling analyses.
Primary Aims
- To descriptively characterize individuals with Covid-19 (overall, hospitalized, intensive care unit (ICU) treated, deaths, and in important subgroups) in terms of sociodemographics, various risk factors (eg, comorbidity, co-medication), geographic distribution, timing, etc. Further, describe how the epidemic and the characteristics of the affected patient group change over time, ie, “natural history” of the epidemic in relation to the population.
- To quantify the impact and importance of comorbidities (in particular respiratory and cardiovascular, diabetes, obesity), severity of and co-medications for concomitant disease, sociodemographics, and other risk factors, as well as protective measures and vaccinations, on risk of various endpoints. Some anticipated endpoints include developing Covid-19, being hospitalized for Covid-19, emergency/ICU care, treatments, specific care, hospitalization duration, prognosis, death and in survivors study long-term outcomes up to 3 years. Various study designs and methods as appropriate will be employed, including case-cohort, case-control and interrupted time series study designs, and machine learning methods.
- To develop, and periodically update prediction models for Covid-19 occurrence and prognosis, and evaluate how this impacts healthcare resource use, including the need for ICU treatment. Use of machine learning methods will be important here.
Secondary Aims
Secondary aims will include rapidly upcoming research questions based on primary prevention and healthcare needs, and include:
- Treatment effects of medicines and other healthcare interventions, including novel indications for established treatments and vaccinations, and possible side-effects related to Covid-19.
- Resource use in the healthcare system, at the individual level, and at societal level (hospital resources, measures, medicines, costs, etc.)
- Study the effectiveness of vaccination programs and possible adverse reactions in the population.
- Consequences of the covid-19 pandemic including future vaccination programs on drug use, health care and other societal functions in the population.
- Other upcoming questions and information gaps from society, healthcare and the scientific community (if these are outside the above specified issues, may need additional supplemental ethical application).
Current and Future Activities
A first data delivery of all data was compiled in October 2020. Data merging and data management, as well as analysis variable and analysis database construction, are performed on an ongoing basis. We have received three complete data updates by Spring 2021. The updating process has thus been tested and found to be functional, and regular updates of the study population and data may now be considered routine. A number of research analyses are now in various stages of completion.
Based on the first data delivery, the number of study subjects and their age and sex distribution are described in Table 2 and Figure 2. As the data are updated, the population comparison cohort (as sampled on 31 Dec 2019) will remain initially static, with a view to update at suitable intervals to the current Swedish population, whereas the number of Covid-19 patients will grow successively with each data delivery.
 |
Table 2 Study Sample for SCIFI-PEARL Based on Initial October 2020 Data Delivery, with Age and Sex Distributions |
 |
Figure 2 Age distribution of study subjects in SCIFI-PEARL based on initial October 2020 database for population comparison cohort, test-positive covid-19 patients and hospitalized covid-19 patients. |
Currently, a number of research projects have been started at various stages. The core project team has initiated collaborations with a number of external researchers, and collaboration proposals from other external partners nationally and internationally are encouraged. The study database is a valuable resource that should be used as broadly as feasible for important research to provide epidemiological and clinical evidence to benefit patients, the healthcare system and society.
We are also exploring the potential to link the database to some existing large-scale well-characterized population-based epidemiological studies in Sweden that have more detailed clinical and questionnaire data on specific populations, such as the West Sweden Asthma Study (WSAS) and SCAPIS (the Swedish CArdioPulmonary bioImage Study). These potential spin-off projects will likely require separate setup and ethics review, but may provide interesting insights, also on a more mechanistic level.
An international outlook is also important. We have established collaboration with the Nordic COHERENCE project,13 aiming to facilitate large-scale register-based observational analyses across several Nordic Covid-19 national databases, especially for research questions where larger sample sizes are desirable, or where comparisons across different populations, societies or healthcare systems may be relevant. The analysis strategy will build on a Nordic common data model (CDM) and distributed analyses, retaining data at each country site to protect individual data. Other international collaborations are also encouraged.
Discussion
The Covid-19 pandemic is a significant concern globally as well as in Sweden, given the pace of spread and breadth of health impacts on the population level, posing a massive challenge to global health. A major obstacle to understanding, managing and preventing Covid-19 continues to be inadequate, non-transparent, and out-of-date Covid-19 data as the pandemic progresses. Many scientists continue to search for innovative sources and uses of real-world data for comprehensive and current Covid-19 research. Since knowledge on Covid-19 is constantly evolving, population-based research at a national scale is crucial, using the latest, point-of-care data that can be leveraged to support research and development. Carefully assembled and used, the high-quality national register data available in Sweden will help research immensely, for example, to characterize risk patients, identify and quantify risk factors, develop machine learning tools to predict COVID-19 severity and mortality, and follow vaccination and pandemic developments, critical insights for driving care to highest-risk patients and better managing the outbreak, with broad global applicability. We have therefore assembled a continuously updated database of Covid-19 patients and general population comparator individuals.
Some learnings from this initiative are broadly relevant in an international setting. Assembling and accessing data in real time from many data sources can be challenging and very time consuming due to process, legal and other aspects – even in a country such as Sweden - and therefore good planning is essential. Plans for such efforts should be made in any country that aims for better future pandemic preparedness, tailored to the particular situation in that area. It is particularly important to pay close attention to local legal frameworks when establishing processes for data sharing. This is unfortunately not the last time there will be urgent need of scientifically robust data on a new health concern, and it will be essential to plan in advance to have rich data available for research use when the need arises.
This initiative has several strengths. The unique PIN in Sweden allows for high-quality linkage of many registers, assembling rich data for each person. Swedish national healthcare registers and national quality registers are generally of very high quality, with reliable data. The complete and accurate national population register (RTB) that is frequently updated and available for sampling enables us to sample essentially all identifiable individuals with Covid-19 in Sweden as well as a very large stratified random sample of general population comparison subjects that can provide context and comparison for planned analyses. This cannot be easily done outside the Nordic countries. The study has a large sample size and we have also built in regular updating of the database with new Covid-19 subjects and current data over time during the pandemic, to ensure optimal power and data availability.
The study builds on real life clinical data from a health care system with essentially equal access to all residents in Sweden. Thus, unequal or biased testing/diagnosis/hospital care for eg, cost reasons is a limited problem. However, other factors such as resource limitations and prioritization, especially during the most intensive pandemic phases, may still affect access to care, and should be adequately considered to avoid bias in any research performed.
Potential weaknesses include those common to most observational data. Data quality is dependent on the completeness and accuracy of the information registered. There are inherent weaknesses related to eg, ICD10 coding and the way different conditions are classified, recorded and grouped. Similar considerations apply to procedure and drug coding. Some data are just not available, eg, despite a high-quality prescription drug register being available, in-hospital drug treatment is not readily available in Sweden. Register research is by its very nature retrospective in the sense that it relies on data collected primarily for another purpose, which may affect content, accuracy, and completeness of data. This can also be a strength, eg, for inference and potential relevance, and avoidance of some types of bias. Finally, register data most often is used for observational research, which, irrespective of study designs – descriptive, case-cohort, case control etc. – may suffer from residual bias even after use of standardization, weighting or adjustment to eliminate confounding, and therefore as high quality as possible of the underlying register data is important.
A potential weakness that needs to be considered in studies of COVID-19 is the uncertainty regarding who is tested for Covid-19 and how this affects estimates of disease occurrence or severity in different groups.14 Rich data to help address this is present in our study. It is also clear that the probability of being infected varies across groups depending on eg, housing conditions and social and behavioral factors, although to the extent that these can be captured in the database this will be taken into account or specifically studied. However, these weaknesses are present everywhere in epidemiologic research on Covid-19 and may in many cases be less prominent in Sweden than in some other countries with greater socioeconomic differences and more restricted access to health care. This is also not unique for Covid-19. For almost any disease there is uneven diagnosis with both over- and underdiagnosis, which varies by other factors. Similarly, most diseases lack stable gold standard criteria that can be applied in clinical practice over time, and observational research must rely on the actual clinical diagnostic efforts and clinical reality.
Conclusions
The SCIFI-PEARL (Swedish Covid-19 Investigation for Future Insights – a Population Epidemiology Approach using Register Linkage) study cohort captures essentially all Swedish residents with Covid-19 on an ongoing basis, includes a representative general population comparison cohort, and links to a broad range of national and regional healthcare data for a comprehensive longitudinal view of the Covid-19 pandemic. By combining high quality national registers with short time delay and continuous repeated linkage updating, the project will bring timely and internationally unique data for epidemiological research on SARS-CoV-2. Broad research efforts are ongoing and further collaborative efforts are invited. Finally, our experience in establishing the project and database carries important lessons for building data repositories elsewhere to address urgent health problems.
Funding
The core study is currently financed by grants from the Swedish state under the agreement between the Swedish government and the county councils, the ALF-agreement (Avtal om Läkarutbildning och Forskning/Medical Training and Research Agreement) grant ALFGBG-938453 and from FORMAS (Forskningsrådet för miljö, areella näringar och samhällsbyggande/Research Council for Environment, Agricultural Sciences and Spatial Planning), a government research council for sustainable development, grant 2020-02828. Additional grants supporting different aspects of ongoing research within the study include: the Swedish Heart Lung Foundation (20210030), Knut och Alice Wallenbergs Stiftelse/SciLifeLab (KAW 2021.0010), the Swedish Research Council (2021-05045, 2021-05450), and the Swedish Social Insurance Agency (FK 2021/011186).
Disclosure
Dr. Nyberg reports prior employment at AstraZeneca until 2019, and ownership of some AstraZeneca shares. Dr. Vanfleteren reports grants and personal fees from AstraZeneca, personal fees from Novartis, GSK, Chiesi, Menarini, Pulmonx, Resmed, Boehringer, Verona Pharma, AGA Linde outside the submitted work. Dr. Sundström reports ownership in companies providing services to Itrim, Amgen, Janssen, Novo Nordisk, Eli Lilly, Boehringer, Bayer, Pfizer and AstraZeneca, outside the submitted work. Dr. Gisslén reports personal fees from Gilead, personal fees from GSK/ViiV, personal fees from MSD, other from Gilead, other from GSK/ViiV, personal fees from Biogen, personal fees from Novocure, personal fees from Amgen, personal fees from Novo Nordisk, outside the submitted work. Dr. Lindh, Dr. Santosa, Dr. Franzén, Dr. Wettermark, Dr Björck and Dr. Hammar have nothing to disclose.
References
1. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi:10.1016/S0140-6736(20)30183-5
2. World Health Organization. Coronavirus disease (Covid-19) weekly epidemiological Update January 5, 2020. Available from: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports/.
3. Ludvigsson JF. The first eight months of Sweden’s COVID-19 strategy and the key actions and actors that were involved. Acta Paediatr. 2020;109(12):2459–2471. doi:10.1111/apa.15582
4. Glasziou PP, Sanders S, Hoffmann T. Waste in covid-19 research. BMJ. 2020;369:m1847. doi:10.1136/bmj.m1847
5. Ludvigsson JF, Otterblad-Olausson P, Pettersson BU, Ekbom A. The Swedish personal identity number: possibilities and pitfalls in healthcare and medical research. Eur J Epidemiol. 2009;24(11):659–667. doi:10.1007/s10654-009-9350-y
6. Rolfhamre P, Janson A, Arneborn M, Ekdahl K. SmiNet-2: description of an internet-based surveillance system for communicable diseases in Sweden. Euro Surveill. 2006;11(5):626. doi:10.2807/esm.11.05.00626-en
7. Ludvigsson JF, Andersson E, Ekbom A, et al. External review and validation of the Swedish national inpatient register. BMC Public Health. 2011;11(1):450. doi:10.1186/1471-2458-11-450
8. Brooke HL, Talbäck M, Hörnblad J, et al. The Swedish cause of death register. Eur J Epidemiol. 2017;32(9):765–773. doi:10.1007/s10654-017-0316-1
9. Ludvigsson JF, Almqvist C, Bonamy A-KE, et al. Registers of the Swedish total population and their use in medical research. Eur J Epidemiol. 2016;31(2):125–136. doi:10.1007/s10654-016-0117-y
10. Wettermark B, Hammar N, Fored CM, et al. The new Swedish prescribed drug register–opportunities for pharmacoepidemiological research and experience from the first six months. Pharmacoepidemiol Drug Saf. 2007;16(7):726–735. doi:10.1002/pds.1294
11. Ludvigsson JF, Svedberg P, Olén O, Bruze G, Neovius M. The longitudinal integrated database for health insurance and labour market studies (LISA) and its use in medical research. Eur J Epidemiol. 2019;34(4):423–437. doi:10.1007/s10654-019-00511-8
12. Emilsson L, Lindahl B, Köster M, Lambe M, Ludvigsson JF. Review of 103 Swedish healthcare quality registries. J Intern Med. 2015;277(1):94–136. doi:10.1111/joim.12303
13. NordForsk. 2020. Available from: https://www.nordforsk.org/sv/projects/nordic-collaborative-health-register-network-covid-19-epidemiology-nordic-coherence.
14. Pottegård A, Kurz X, Moore N, Christiansen CF, Klungel O. Considerations for pharmacoepidemiological analyses in the SARS-CoV-2 pandemic. Pharmacoepidemiol Drug Saf. 2020;29(8):825–831. doi:10.1002/pds.5029
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.